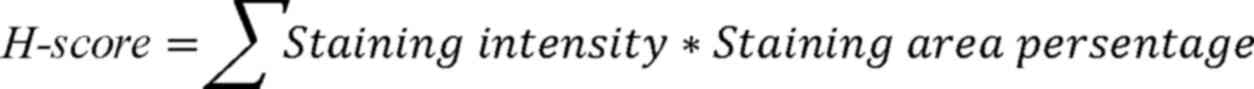Introduction
Bladder cancer (BC) is the 12th most common type of
cancer worldwide, which imposes substantial financial burden to
societies. According to the statistics, there were 573,278 new BC
cases in 2020 worldwide (1). Of the
BC cases, ~75% are non-muscle-invasive BC (NMIBC) and the remainder
are muscle-invasive BC (MIBC). Currently, cystoscopy is the gold
standard diagnostic procedure, and the American Joint Committee on
Cancer (AJCC) TNM system is commonly used to predict the clinical
outcomes of patients with BC (2,3).
However, the individual genetic heterogeneity often results in
divergent clinical outcomes, which stresses the vital necessity to
identify biomarkers for BC.
With advancements being made in transcriptomics, a
number of biomarkers for BC prognosis have emerged, such as urinary
extracellular vesicles (4),
telomerase reverse transcriptase promoter mutations (5) and nuclear matrix protein 22 (6). The forkhead box (FOX) superfamily
consists of 43 evolutionarily conserved transcriptional regulators
that participate in DNA repair, cell lineage, embryogenesis and
longevity (7,8). The forkhead domain enables the
combination of members with target DNA, and is responsible for
their promotive or suppressive effects on gene transcription. A
number of FOX members have been proven to be differentially
expressed in various BC subtypes, and the dysregulation of FOX
genes may be involved in bladder tumor development and progression
(9,10).
As a transcription factor in the hedgehog signaling
pathway (11), FOXF1 plays critical
roles in gastrointestinal tract development, cancer-associated
fibroblasts (12), endothelia
progenitor activation (13) and
VEGF signaling regulation (14).
Recently, the effects of FOXF1 in the antitumor process and its
association with the prognosis of patients have been demonstrated.
In papillary thyroid cancer, hepatocellular carcinoma and lung
adenocarcinoma, FOXF1 is downregulated in tumor tissue compared
with pericarcinomatous tissues; the decreased expression of FOXF1
has been found to be associated with more malignant phenotypes and
a poorer survival, indicating its predictive ability in the
prognosis of patients (15–17). However, the effects of FOXF1 in BC
have not yet been fully elucidated.
The present study aimed to investigate the
expression patterns of FOXF1 in BC, evaluate the association
between FOXF1 expression levels and the clinicopathological
features of patients with BC, and explore the antitumor mechanisms
of FOXF1 in BC. The results described herein demonstrate that FOXF1
expression is downregulated in BC tissues, its activation inhibits
cell proliferation and induces cell apoptosis via the caspase
signaling pathway. The decreased expression level of FOXF1 is
associated with a more severe clinical stage, muscle invasion,
lymphatic infiltration and a poorer prognosis of patients with BC.
The expression level of FOXF1 may thus have robust predictive
ability in the clinical outcome of patients with BC.
Materials and methods
Patients and clinical information
The present study recruited 64 patients with BC
undergoing cystectomy at Ruijin Hospital (Shanghai, China) between
January, 2007 and February, 2022 (Ruijin Cohort). The present study
was performed in accordance with the Declaration of Helsinki. The
studies involving human participants were reviewed and approved by
the Ethics Committee of Ruijin Hospital, School of Medicine,
Shanghai Jiao Tong University (PA23030202). Informed consent was
obtained from each patient. The patient specimens were
pathologically diagnosed using three independent experts. The BC
samples and matched normal bladder mucosae were embedded into a
tissue microarray (Shanghai Outdo Biotech Co., Ltd.). Clinical and
pathological data were recorded, including sample ID, age, sex,
operation data, follow-up period, status, tumor grade, tumor size,
metastatic lesion and lymphatic infiltration, followed by
determining their stages according to the 8th AJCC staging system
(18).
Data collection and preprocessing
Gene chips were first selected from the Gene
Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The inclusion
criteria for validating the FOXF1 expression patterns are as
follows: i) The biospecimens were obtained from human primary
bladder cells or tissues; ii) transcriptomic data; iii) samples
contained para-cancerous samples; iv) contained at least six
samples in each group. In total, eight independent GEO datasets,
GSE121711, GSE13507, GSE188715, GSE3167, GSE37815, GSE38264,
GSE40355 and GSE42089 that met the requirements were collected
using the ‘GEOquery’ package (19).
The FOXF1 expression levels of pan-cancer tissues were acquired
from the GEPIA1 website (http://gepia2.cancer-pku.cn). Moreover, to validate
the FOXF1 expression levels in different types of BC, the RNA
sequencing (RNA-seq) data were downloaded and the clinical
information of 411 patients with BC was obtained from The Cancer
Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/).
The inclusion criteria for validating the prognostic
predictive performance of FOXF1 are as described below: i) The
biospecimens were obtained from human primary bladder cells or
tissues; ii) transcriptomic data; iii) the survival information was
available; iv) a sufficient sample size was included for survival
analysis. Four independent GEO datasets (GSE31684, GSE48075,
GSE169455 and GSE13507) and an immunotherapy cohort (IMvigor210)
with their corresponding survival data were obtained from the GEO
website, ‘IMvigor210CoreBiologies’ and ‘IOBR’ packages (20). The cut-off values for the FOXF1
expression level in these datasets were determined using ‘X-tile’
software (version 3.6.1; http://medicine.yale.edu/lab/rimm/research/software/).
Immunohistochemistry staining and
scoring of FOXF1
Tissues were fixed with 4% paraformaldehyde for 48 h
at room temperature, then paraffin-embedded and prepared into a
tissue chip by Shanghai Outdo Biotech Co., Ltd.
Immunohistochemistry (IHC) of the tissue microarray was performed
using streptavidin-peroxidase methods. Specifically, the slides
were deparaffinized with xylene for 30 min at room temperature
followed by rehydration using a series of graded alcohols (100, 95,
80 and 70%, 5 min for each step, at room temperature). Heat-induced
epitope retrieval (with Tris/EDTA pH 9.0 buffer) was then performed
using a induction heater for 20 min at 60°C. Non-specific binding
sites were blocked using Immunol Staining Blocking Buffer (Beyotime
Institute of Biotechnology) for 15 min at room temperature.
Subsequently, the microarray was incubated with primary antibody
against human FOXF1 (1:100 dilution; cat. no. PAB30083, Abnova)
overnight at 4°C. The following day, the microarray was washed with
TBST (Wuhan Servicebio Technology Co., Ltd.) for 30 min at room
temperature and incubated with the HRP-conjugated goat anti-rabbit
lgG antibody (ready to use; cat. no. D110073, Sangon Biotech Co.,
Ltd.) for 1 h at room temperature. The slides were then stained
using diaminobenzidine (Beyotime Institute of Biotechnology) for 5
min at room temperature and re-stained with hematoxylin (Beyotime
Institute of Biotechnology) for 10 min at room temperature. Finaly,
a series of graded alcohols (70, 80, 90 and 100%, 10 sec for each
step, at room temperature) followed by xylene (10 sec, at room
temperature) were used for dehydration of the tissues. The slides
were mounted by neutral balsam (Wuhan Servicebio Technology Co.,
Ltd.) and scanned using Pannoramic MIDI automatic digital slide
scanner (3DHISTECN Ltd.). The negative controls were treated using
the same experimental process, but the primary antibody was rabbit
IgG (1:200; cat. no. 30000-0-AP, Proteintech Group, Ltd.). Positive
staining was considered when staining was predominantly located in
the nucleus and cytoplasmic staining was regarded as non-specific
staining. The expression level of FOXF1 was quantified by the
proportion of positive cell nucleus and their staining intensity as
follows:
The staining intensity was graded into four levels
as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong);
the staining area percentage was defined as the percentage of cell
nucleus with a corresponding staining intensity (0–100). Three
pathologists, who were blinded to the clinical information of the
patients, scored the immunoreactivity of FOXF1 independently on an
Aperio ImageScope (magnification, ×400; Leica Microsystems GmbH);
the final H-score was the average of their scores.
Evaluation of the prognostic
predictive performance of FOXF1 expression
For the patients in TCGA dataset, their survival
outcomes were compared with the aid of the KMplotter online
database (https://kmplot.com/analysis/). For the patients in the
GEO datasets and the IMvigor210 cohort, their survival information
was acquired from the corresponding website. All patients in each
dataset were divided into the FOXF1-high and FOXF1-low groups based
on a cut-off value produced using X-tile' software. Survival risk
differences between the two groups were determined using
Kaplan-Meier survival analysis and the log-rank test. Univariable
and multivariable Cox regression analyses were performed to examine
the independency of FOXF1 in predicting the overall survival (OS)
of patients with BC. Boxplots were used for revealing the
association between FOXF1 expression and the clinicopathological
features of the patients.
Cells, cell culture and chemical
reagents
Human bladder cancer cell lines 5637 (HTB-9), J82
(HTB-1), T24 (HTB-4) and the normal uroepithelium cell line
SV-HUC-1 (CRL-9520), were acquired from ATCC. The EJ cell line
(YS1803C) was obtained and STR-authenticated by Shanghai Yaji
Biological Technology Co., Ltd.. The SV-HUC-1 cells were cultured
in Ham's F-12K medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher
Scientific, Inc.); the 5637, J82, EJ and T24 cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS. All cells were cultured in humidified
incubator with 5% CO2 at 37°C. Ac-DEVD-CHO (Beyotime
Institute of Biotechnology) at a final concentration of 50 µM was
used to inhibit caspase-3 activity 12 h after dsRNA
transfection.
self-amplifying RNA (saRNA) design,
transfection with double-stranded RNA (dsRNA) and infection with
recombinant lentivirus
The promoter sequence (1 kb) of FOXF1 was downloaded
from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gene/). An Excel
macro template was used to read the promoter sequence and the
putative saRNA target sites were scanned as previously described
(21). Following manual screening,
four saRNAs and a non-specific negative control (dsControl) were
synthesized by GenePharma Co., Ltd.; the sequences of the five
dsRNAs are listed in Table SI.
Transfection with the dsRNAs (final concentration, 50 nM) was
performed using Lipofectamine RNAiMax (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. RNA
was isolated from the BC cells at 48 h following transfection, and
total proteins were extracted from the BC cells at 72 h following
transfection; the time durations between transfection and other
phenotypic experiments are specified in the relevant subsections
below.
The efficiencies of the four saRNAs were examined
using reverse transcription-quantitative PCR (RT-qPCR) and western
blot analysis. The most efficient saRNA and dsControl were selected
to create short hairpin RNA (shDNA) and packaged into lentivirus
(Lenti-dsFOXF1-367 or Lenti-dsControl, GenePharma Co., Ltd.). The
cells were infected by the lentivirus at a confluency of 60–70%,
and the medium was replaced 24 h later. Cells with the stable
activation of FOXF1 were selected with 3 µg/ml puromycin (Beyotime
Institute of Biotechnology).
RT-qPCR
Total RNA was isolated from the BC cells, carcinoma
and adjacent tissue samples using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The reverse
transcription of 1,000 ng RNA was carried out using the First
Strand cDNA Synthesis kit (cat. no. D7178; Beyotime Institute of
Biotechnology) according to the manufacturer's instructions, the
reaction system was incubated for 30 min at 42°C, then heated for
10 min at 80°C. The gene expression level was detected using
SYBR-Green qPCR Mix (cat. no. D7262; Beyotime Institute of
Biotechnology) and quantified using the 2−ΔΔCq method
(22). GAPDH was used as the
reference gene. Primers were produced by Biosune Biotechnology
(Shanghai) Co.; their sequences are presented in Table SI.
RNA-seq and protein-protein
interaction (PPI) network analysis
The RNA-seq library construction was completed by
Shanghai Biotree Biotech Co., Ltd.. The negative control (nc) and
FOXF1-activated (sa) group of BC cells were established, and the
quantity and purity of the RNA were analyzed using a Bioanalyzer
2100 and RNA 6000 Nano LabChip kit (Agilent Technologies, Inc.).
The mRNAs were purified using oligo (dT)25 magnetic
beads (Invitrogen; Thermo Fisher Scientific, Inc.), followed by
fragmenting to ~200 bp (magnesium RNA fragmentation module, New
England Biolabs) and reverse transcription (random hexamer priming
method, Invitrogen; Thermo Fisher Scientific, Inc.). After adding
A-base, ligating adaptor and PCR amplifying, the products libraries
were sequenced on an Illumina platform (Novaseq™ 6000) in
accordance with the vendor's recommended protocol.
Differentially expressed genes (DEGs)
and PPI analysis
DEGs were identified based on a threshold set as
fold change (FC) >2 or <0.5, and adjusted P-value <0.05.
The PPI network was analyzed on the STRING website (https://cn.string-db.org/) and using Cytoscape
software (version 3.9.1; http://cytoscape.org/); the degree of protein
interaction was calculated using the maximum neighborhood component
(MNC) method.
Western blot analysis
Total proteins were extracted from the BC cells
using RIPA buffer (NCM Biotech) supplemented with protease
inhibitor cocktail (NCM Biotech). The concentrations of the
proteins were determined using the BCA method (Epizyme). The
supernatants containing 30 µg protein were separated by 7.5%
SDS-PAGE and then blotted onto PVDF membranes (MilliporeSigma). The
membranes were incubated with 5% skim milk for 2 h at room
temperature in order to block non-specific binding sites. After
washing with TBST (Wuhan Servicebio Technology Co., Ltd.) for 10
min at room temperature, primary antibodies included FOXF1 (1:3,000
dilution; cat. no. ab168383, Abcam), Bax (1:1,000 dilution; cat.
no. R22708), caspase-9 (1:1,000 dilution; cat. no. 381336), cleaved
caspase-3 (1:1,000 dilution; cat. no. R23727), poly (ADP-ribose)
polymerase (PARP; 1:1,000 dilution; cat. no. R25279) (all from
ZenBio, Inc.) and GAPDH (1:2,000 dilution; cat. no. GB15004, Wuhan
Servicebio Technology Co., Ltd.) were incubated with the membranes
overnight at 4°C. The second day, the membranes were washed with
TBST (Wuhan Servicebio Technology Co., Ltd.) for 30 min at room
temperature, and incubated with HRP-conjugated secondary antibodies
(1:10,000 dilution; cat. no. C31460100, Invitrogen; Thermo Fisher
Scientific, Inc.) for 1 h at room temperature. The protein
expression level of each gene was detected using the ECL Pico Light
Chemiluminescence kit (cat. no. SQ202, Epizyme). The gray level of
each blot was measured by ImageJ software (version 1.53t,
https://imagej.nih.gov/ij/, National
Institutes of Health).
Immunofluorescence (IF) staining
The cells were fixed with 4% paraformaldehyde
(Beyotime Institute of Biotechnology) for 30 min at room
temperature and permeabilized with 0.5% Triton X-100 (Wuhan
Servicebio Technology Co., Ltd.) for 30 min at room temperature,
followed by blocking with 5% BSA (Wuhan Servicebio Technology Co.,
Ltd.) for 1 h at room temperature. FOXF1 antibody (1:500 dilution;
cat. no. ab168383, Abcam, in 5% BSA) was used to incubated the
cells overnight at 4°C, followed by incubation with CoraLite594
conjugated goat anti-rabbit IgG (H+L) (1:500 dilution; cat. no.
SA00013-4, Proteintech Group, Inc., in 5% BSA) for 1 h at room
temperature. After mounting with Antifade Mounting Medium with DAPI
(Beyotime Institute of Biotechnology), cell images were captured
using an inverted confocal microscope equipped with a 400X lens
objective (Zeiss AG, Germany).
Flow cytometry
Flow cytometry was used to assess the proportion of
cells undergoing apoptosis. At 72 h after transfection, the cells
were co-stained with Annexin V-FITC and PI (cat. no. A211; Nanjing
Vazyme Biotech Co. Ltd.) according to the manufacturer's
instructions. Cells with Annexin V-FITC-positive and PI-negative
staining were considered to be in early stage of apoptosis, while
cells with double-positive staining were considered to be in the
late stage of apoptosis.
Clonogenic capacity, and Cell Counting
Kit-8 (CCK-8) and 5-ethynyl-2′-deoxyuridine (EdU) assays
The cells were harvested 24 h following dsRNA
transfection. For colony formation assay, 1,000 cells/well were
seeded in a six-well plate with 2 ml complete medium and cultured
for 10 days. The proliferating colonies were fixed with 4%
paraformaldehyde (Beyotime Institute of Biotechnology) for 30 min
at room temperature and stained by 0.5% crystal violet (Beyotime
Institute of Biotechnology) for 2 h at room temperature. The
numbers of colonies were countered using ImageJ software (version
1.53t, https://imagej.nih.gov/ij/, National
Institutes of Health), and the colony formation rate was calculated
as the number of colonies/1,000 cells.
A total of 5,000 cells/well were suspended in
96-well plate with 100 µl complete medium. The CCK-8 kit (1:10
dilution, 10 µl CCK-8 reagent and 90 µl complete medium per well,
cat. no. C6030, NCM Biotech) was used to analyze cell growth by
measuring the absorbance value at 24, 48, 72 and 96 h at 450 nm.
For EdU assay, the cells were cultured with 50 µM EdU reagent (cat.
no. C10310; Guangzhou Ribobio Co., Ltd.) for 2 h at 37°C, then
fixed with 4% paraformaldehyde (Beyotime Institute of
Biotechnology) for 30 min at room temperature and permeabilized
with 0.5% Triton X-100 (Wuhan Servicebio Technology Co., Ltd.) for
30 min at room temperature. Subsequently, the EdU-stained cells
were labeled using Apollo567 reagent (Guangzhou Ribobio Co., Ltd.)
for 30 min at room temperature while the cell nuclei were stained
using DAPI (Wuhan Servicebio Technology Co., Ltd.) for 10 min at
room temperature. Fluorescence images were captured using an
inverted fluorescent microscope equipped with a 200X lens objective
(Zeiss AG, Germany) and the cell proliferative ability was measured
by calculating the proportion of EdU-labeled cells.
Cell migration and invasion
assays
72 h following transfection, 4×104 cells
were suspended in 200 µl serum-free RPMI-1640 medium and seeded
into the upper chambers of a Transwell insert (24-well, 8 µm pore
size, Corning, Inc.) to measure cell migration. For the invasion
assay, the upper chambers were pre-coated with Matrigel (Corning,
Inc., 1:5 dilution in serum-free RPMI-1640 medium). As a
chemoattractant, 600 µl complete medium supplemented with 10% FBS
was added to the lower chamber. The plates were cultured in
incubator for 24 h at 37°C, non-motile cells on the upper surface
were removed. Following fixation with 4% paraformaldehyde (Beyotime
Institute of Biotechnology; 30 min, room temperature) and staining
with 0.5% crystal violet (Beyotime Institute of Biotechnology; 2 h,
room temperature), the membranes were photographed under an upright
fluorescent microscope microscope (×100 magnification; Zeiss AG,
Germany), random fields were selected and the cell numbers were
counted.
Subcutaneous xenograft tumor
model
The studies involving human participants or animals
were reviewed and approved by the Ethics Committee of Ruijin
Hospital, School of Medicine, Shanghai Jiao Tong University
(PA23030202). The animal experiments followed the ARRIVE checklist,
and the animals were housed under pathogen-free, 20–26°C, 40–70%
humidity, 12-h light/dark circle conditions, with free access to
water and food. The health and behavior of the animals were
monitored every day; an animal would be euthanized if the length of
the tumor was >17 mm. Four-week-old male BALB/c nude mice
(Beijing Vital River Laboratory Animal Technology Co., Ltd.) were
randomly divided into two groups (5 mice per group). EJ cells
(~6×106) infected with Lenti-dsFOXF1-367 or
Lenti-dsControl were dissociated into 200 µl suspension and
injected subcutaneously into the right flank of each mouse. As the
procedure produced only mild pain, no anesthetic was used. The
length and width of the tumors were measured using calipers every 5
days, and tumor volumes were calculated as 0.5 × width2
× length. All mice were sacrificed by cervical dislocation 30 days
later, and after confirming that the animals had no breathing or
heartbeat, the tumors were dissected for further analysis. The
maximum observed tumor diameter in the animals in the present study
was 7 mm.
Statistical analysis
Statistical analysis was carried out using R
software (version 4.1.3; http://www.R-project.org/) and Origin 2023 software
(https://www.originlab.com/). The
experimental data were derived from three independent experiments
and are presented as the mean ± standard deviation (SD). For
continuous variables, the Wilcoxon-Mann-Whitney (WMW) test or
Student's t-test were used to examine the significance of the
differences between two groups. The Kruskal-Wallis test (with
Dunn's post hoc test) or one-way ANOVA (with Tukey's post hoc test)
were used to determine whether or not there was a statistically
significant difference among multiple groups containing
non-parametric ranked data or parametric data. For categorical
variables, assumptions were analyzed using the Chi-squared test or
Fisher's exact test. P-value <0.05 was considered to indicate a
statistically significant difference for all analyses.
Results
Patient characteristics
In the present study, 64 patients with BC that had
undergone cystectomy at Ruijin hospital were recruited, 59 of them
had corresponding survival information (Ruijin Cohort) and 46 of
them had matched para-cancerous bladder tissues. There were 50
males and 9 females involved in this cohort, with a mean age of
68.46 years (range, 44–85 years). Their clinicopathological
information was recorded and the median follow-up period was 38
months (range, 3–82 months; Tables
SII and SIII). For the
validation group, FOXF1 expression levels were obtained in eight
GEO datasets containing tumor and normal bladder tissues, as well
as in five independent BC datasets with prognostic information
(IMvigor210 cohort and four GEO datasets) (23). The detailed information of these
datasets is presented in Table
SIV.
FOXF1 is downregulated in BC
tissues
The present study aimed to explore whether FOXF1 is
associated with the development of BC. FOXF1 expression was first
analyzed in pan-cancer scope using TCGA database. As shown in
Fig. 1A, FOXF1 expression was
downregulated in numerous types of tumors. Furthermore, in BC
(BLCA), FOXF1 expression exhibited a distinct difference between
normal bladder and carcinoma tissues. The FOXF1 expression patterns
in BC and adjacent normal tissues in the Ruijin cohort were then
investigated using IHC. The staining intensity in the nuclei of the
bladder tumor cells was lighter than that in the matched adjacent
normal urothelial cells from T stage I to IV (Fig. S1A-H). An overview of the tissue
microarray is illustrated in Fig.
S1I, and the assignment of the tumor and para-cancerous tissues
is presented in Fig. S1J. The
H-scores of BC tissues were significantly lower than those of their
matched normal bladder mucosae (P<0.001, Fig. 1B). In order to verify the
universality of this downregulated FOXF1 expression in urothelial
carcinoma, the FOXF1 expression levels were compared between BC
tissues and normal bladder mucosae in all required GEO datasets. As
shown in Fig. S2, FOXF1 expression
was significantly higher in normal tissues than in BC samples
(P<0.001 in GSE42089, GSE121711, GSE13507, GSE188715, GSE32864
and GSE40355; P=0.024 in GSE3167; P=0.036 in GSE37815).
 | Figure 1.FOXF1 expression is downregulated in
tumor tissues and is associated with a poor prognosis of patients
with BC. (A) Expression of FOXF1 in pan-cancer and (B) BC tissues
in the Ruijin Cohort. The association of the FOXF1 expression
levels with (C) tumor type, (D) pathological grade, (E) muscle
invasion, (F) T stage and (G) American Joint Committee on Cancer
stage in the Ruijin Cohort. (H) The H-score curve and (I) survival
status of FOXF1-low group and FOXF1-high group in the Ruijin
Cohort; patients were listed in an order of increased FOXF1
expression level; the dotted line represents the cut-off value
between the two groups. (J) Kaplan-Meier analysis of overall
survival in the Ruijin Cohort and (K) in the GSE48075 dataset.
FOXF1, forkhead box F1; BC, bladder cancer; ACC, adrenocortical
carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast
invasive carcinoma; CESC, cervical squamous cell carcinoma and
endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme;
HNSC, head and neck squamous cell carcinoma; KICH, kidney
chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG,
brain lower grade glioma; LIHC, liver hepatocellular carcinoma;
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV,
ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma;
PCPG, pheochromocytoma and paraganglioma; PRAD, prostate
adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM,
skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT,
testicular germ cell tumors; THCA, thyroid carcinoma; THYM,
thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine
carcinosarcoma. |
Association between FOXF1 expression
and the clinicopathological characteristics of patients with
BC
Based on the cut-off value of H-score, the patients
were divided into the FOXF1-high and FOXF1-low groups. The detailed
clinicopathological information is presented in Table I. The Chi-squared test was used to
examine the association between FOXF1 expression and the
clinicopathological characteristics of the patients in the two
groups. Compared to FOXF1-high group, FOXF1-low group had a higher
proportion of patients with muscle invasion (P=0.019), worse
clinical stage (P<0.001 for T stage, P<0.001 for AJCC stage)
and lymphatic metastasis (P=0.011). In addition, an unfavorable
survival status (P=0.003) and a shorter survival time (P=0.001)
were observed in patients with a lower FOXF1 expression. There were
no significant differences in sex (P=0.995), pathological grade
(P=0.824) and subtype (P=0.356) between two groups (Table I). The same approach was then
applied to TCGA dataset (Table
SV), which contained 406 patients with BC. These results
revealed that a low expression of FOXF1 was significantly
associated with clinical stage (P=0.029 for AJCC stage and
P<0.001 for T stage) and lymphatic metastasis (P<0.001).
 | Table I.Association between FOXF1 expression
and the clinicopathological characteristics of patients with
BC. |
Table I.
Association between FOXF1 expression
and the clinicopathological characteristics of patients with
BC.
| Clinical
features | Patients with
BC | FOXF1-high | FOXF1-low | P-value |
|---|
| No. of
patients | 59 | 23 | 36 |
|
| Mean age (years),
mean (SD) | 68.46 (10.04) | 67.83±9.81 | 67.58±11.01 | 0.703 |
|
|
| 68.86±10.30 |
|
|
| Sex, n (%) |
|
|
| 0.995 |
|
Male | 50 (84.7) | 20 (87.0) | 30 (83.3) |
|
|
Female | 9 (15.3) | 3 (13.0) | 6 (16.7) |
|
| Stage, n (%) |
|
|
| <0.001 |
|
0is | 3 (5.1) | 2 (8.7) | 1 (2.8) |
|
| Stage
i | 8 (13.6) | 7 (30.4) | 1 (2.8) |
|
| Stage
ii | 11 (18.6) | 8 (34.8) | 3 (8.3) |
|
| Stage
iii | 15 (25.4) | 2 (8.7) | 13 (36.1) |
|
| Stage
iv | 8 (13.6) | 0 (0) | 8 (22.2) |
|
|
Unknown | 14 (23.7) | 4 (17.4) | 10 (27.8) |
|
| Grade, n (%) |
|
|
| 1.000 |
| Low
grade | 3 (5.1) | 1 (4.3) | 2 (5.6) |
|
| High
grade | 56 (94.9) | 22 (95.7) | 34 (94.4) |
|
| Pathological grade,
n (%) |
|
|
|
|
|
I–II | 3 (5.1) | 1 (4.3) | 2 (5.6) | 0.824 |
| II | 17 (28.8) | 8 (34.8) | 9 (25.0) |
|
|
II–III | 27 (45.8) | 9 (39.1) | 18 (50.0) |
|
|
III | 12 (20.3) | 5 (21.7) | 7 (19.4) |
|
| Subtype, n (%) |
|
|
| 0.356 |
|
Papillary | 13 (22.0) | 7 (30.4) | 6 (16.7) |
|
|
Non-papillary | 46 (78.0) | 16 (69.6) | 30 (83.3) |
|
| Muscle invasion, n
(%) |
|
|
|
|
|
MIBC | 40 (67.8) | 11 (50.0) | 29 (82.9) | 0.019 |
|
NMIBC | 17 (28.8) | 11 (50.0) | 6 (17.1) |
|
|
Unknown | 2 (3.4) | 1 (4.3) | 1 (2.8) |
|
| Pathological T
stage, n (%) |
|
|
| <0.001 |
|
Tis | 5 (8.5) | 2 (9.1) | 3 (8.6) |
|
| T1 | 11 (18.6) | 9 (40.9) | 2 (5.7) |
|
| T2 | 16 (27.1) | 9 (40.9) | 7 (20.0) |
|
| T3 | 22 (37.3) | 2 (9.1) | 20 (57.1) |
|
| T4 | 3 (5.1) | 0 (0) | 3 (8.6) |
|
|
Unknown | 2 (3.4) | 1 (4.3) | 1 (2.8) |
|
| Pathological M
stage, n (%) |
|
|
| / |
| M0 | 59 (100.0) | 23 (100.0) | 36 (100.0) |
|
| Pathological N
stage, n (%) |
|
|
| 0.011 |
| N0 | 37 (62.7) | 19 (82.6) | 18 (50.0) |
|
| N1 | 7 (11.9) | 0 (0) | 7 (19.4) |
|
|
Unknown | 15 (25.4) | 4 (17.4) | 11 (30.6) |
|
| Survival status, n
(%) |
|
|
| 0.003 |
|
Alive | 28 (47.5) | 17 (73.9) | 11 (30.6) |
|
|
Deceased | 31 (52.5) | 6 (26.1) | 25 (69.4) |
|
| Mean
survival (months), mean (SD) | 36.20 (22.64) | 48.26 (20.13) | 28.50 (20.93) | 0.001 |
Furthermore, the expression levels of FOXF1 were
compared among different BC types. The FOXF1 expression level was
not significantly associated with the pathological type (P=0.71,
Fig. 1C) and grade (P=0.94,
Fig. 1D) of BC. However, it was
highly expressed in tumors without muscle invasion (P=0.026,
Fig. 1E) and with early
pathological stages (P=0.001 for T stage and P<0.001 for AJCC
stage; Fig. 1F and G). All these
results suggested that FOXF1 may exert an antitumor effect on
BC.
Prognostic value of FOXF1 for the
clinical outcome of patients with BC
To estimate the prognostic value of the FOXF1
expression level in the Ruijin cohort, the distribution of the
H-score and survival outcomes were first evaluated. Of note, it was
found that the FOXF1-high group had an improved survival rate and a
longer OS (Fig. 1H and I).
Furthermore, patients with high FOXF1 expression levels exhibited
significantly enhanced OS than those with low FOXF1 expression
levels, as determined by Kaplan-Meier analysis (P=0.00047, Fig. 1J). The mean survival time for the
FOXF1-high group was 48.26 months, while for the FOXF1-low group it
is 28.50 months (P=0.001, Table
I).
Additionally, we attempted to verify the
universality of the obtained conclusion. After screening the public
databases, the following datasets were selected and analyzed to
validate the predictive ability of FOXF1: TCGA-BLCA dataset,
IMvigor210 cohort and four GEO datasets that contained survival
data of patients with BC. In GSE48075 (P=0.011, Fig. 1K), GSE13507 (P=0.023, Fig. S3A), GSE31684 (P=0.047, Fig. S3B), IMvigor210 (P=0.043, Fig. S3C) and GSE169455 (OS: P=0.04,
Fig. S3D; recurrence-free
survival: P=0.037, Fig. S3E),
compared with the FOXF1-low groups, the FOXF1-high groups all
exhibited more favorable clinical outcomes. In TCGA dataset, to be
consistent with Ruijin cohort, the OS of the patients with T2 or
higher disease was compared. The patients with lower FOXF1
expression levels had poorer survival than those with higher FOXF1
expression levels, but the P-value was not significant, which might
be the result of insufficient number of events and the effect of
other disturbances (for instance, comorbidity or different
treatment) on OS (Fig. S3F).
Subsequently, the authors examined whether FOXF1 was
an independent prognostic indicator of BC. Univariate Cox
regression analysis confirmed that the FOXF1 expression level
[hazard ratio (HR), 0.23; 95% confidence interval (CI), 0.085–0.64;
P=0.0049], AJCC stage (HR, 1.9; 95% CI, 1.2–3.1; P=0.0045) and N
stage (HR, 3.7, 95% CI, 1.5–9.3; P=0.0048) were significantly
associated with the prognosis of patients with BC (Fig. S3G). Multivariate Cox regression
analysis revealed that FOXF1 could almost predict the OS of
patients with BC independently (P=0.065, Fig. S3H). The aforementioned results
demonstrated that the downregulated expression of FOXF1 was an
unfavorable indicator of the prognosis of patients with BC; thus,
it has a robust predictive ability.
Expression patterns of FOXF1 in
bladder tumor specimens and cells
Following the tissue array analysis and the
verification of public datasets, it was hypothesized that FOXF1
exerted a suppressive effect on BC. The analysis of the expression
level of FOXF1 in 10 human bladder tissues (5 tumor specimens and 5
normal mucosae) and different cell lines revealed that FOXF1 was
downregulated in bladder tumor samples and BC cells compared with
para-cancerous bladder tissues and the normal uroepithelium cell
line, SV-HUC-1, respectively (Fig. 2A
and B).
To further investigate the antitumor role of FOXF1
in BC cells, candidate saRNAs that could activate FOXF1 expression
were screened; the T24 and EJ cells were selected for use in
subsequent experiments due to their lower FOXF1 expression levels,
and dsControl was applied to avoid the off-target effect. Among the
four designed saRNAs, dsFOXF1-367 (complementary to sequence
position −367 relative to the TSS of FOXF1) led to a 5.39- and
6.85-fold induction of FOXF1 mRNA expression in the T24 and EJ
cells at 72 h following transfection, respectively (Fig. S4A and B); the induction effects on
FOXF1 protein expression were further compared using western blot
analysis (Fig. S4C and D).
According to the gray value, dsFOXF1-367 led to a 2.15- and
2.49-fold increase in FOXF1 protein levels in the T24 and EJ cells,
respectively (Fig. 2C and D). IF
staining also confirmed the elevated protein expression of FOXF1,
and FOXF1 protein was mainly located in the cell nuclei (Fig. 2E and F).
Activation of FOXF1 expression in BC
cell lines suppresses cell proliferation and induces apoptosis
First, CCK-8 assays were carried out to examine the
suppressive effect of FOXF1 on BC cell proliferation (Fig. 3A and B). Compared with the
dsControl, cell growth was significantly inhibited by dsFOXF1-367
from 2 days after reseeded (P<0.01, 72 h following
transfection). The results of EdU assays also revealed the
suppressive effects of FOXF1 on the DNA synthesis of BC cells
(Fig. 3C-E). Subsequently, the
growth mode of the BC cells following FOXF1 activation was examined
using colony formation assay. As shown in Fig. 3F and G, FOXF1 attenuated the colony
formation ability in both colony areas and colony formation rates.
To determine whether FOXF1 activation affects on BC cell
metastasis, Transwell assays with or without Matrigel were
conducted 72 h following transfection. FOXF1 markedly suppressed
cell migration (Fig. 3H) and
invasion (Fig. 3I); the number of
migrated and invaded cells were significantly reduced by
dsFOXF1-367 (P<0.01, Fig. 3J and
K).
 | Figure 3.dsFOXF1-367 suppresses cell
proliferation, migration, invasion and induces apoptosis in BC.
CCK-8 assays were used to examine the viability of (A) T24 and (B)
EJ cells. EdU assays revealed that dsFOXF1-367 inhibited (C) T24
and (D) EJ cell proliferation. The replicating DNA were marked with
Apollo567 (orange), while nuclei were stained with DAPI (blue;
magnification, ×200). (E) Proportion of replication DNA in T24 and
EJ cells. (F) dsFOXF1-367 impaired the clonogenic capacity of T24
and EJ cells; (G) the colony formation rate in each group was
compared in the histogram. (H and I) Representative images
(magnification, ×200) of Transwell assays (H) with or (I) without
Matrigel in T24 and EJ cells; the numbers of (J) migrated and (K)
invaded cells were compared. Flow cytometry of (L and M) T24 cells
following transfection with (L) dsControl or (M) dsFOXF1-367, and
(N and O) EJ cells following transfection with (N) dsControl or (O)
dsFOXF1-367. Percentages of (P) T24 and (Q) EJ cells in early and
late apoptosis. (R) The heatmap revealed 1,178 protein coding genes
among 34,715 genes which were possibly regulated by FOXF1
activation. (S) Protein-protein interaction network of potential
hub genes and (T) the interaction degree of top 10 genes. (U) The
protein expression levels of apoptosis-related genes. *P<0.05,
**P<0.01 and ***P<0.001, vs. dsControl. FOXF1, forkhead box
F1; BC, bladder cancer. |
Subsequently, flow cytometry was employed to assess
the effects of FOXF1 on cell apoptosis. The induction of FOXF1
promoted both the early and late stages of apoptosis of BC cells
compared with the negative control (T24 cells: Fig. 3L, M and P; EJ cells: Fig. 3N, O and Q).
FOXF1 induces the apoptosis of BC
cells via the caspase signaling pathway
With the aim of elucidating the mechanisms of FOXF1
in BC cell proliferation, RNA-Seq was performed to evaluate the
impact of induction on the transcriptome. T24 dsControl (nc) and
T24 dsFOXF1-367 (sa) were set with three replicates of each group;
34,715 genes were differentially expressed in these two groups and
1,178 of these were protein coding genes. Through the heatmap of
DEGs (Fig. 3R) and PPI (min score
65, Fig. 3S), it was noted that
caspase-3 was significantly affected in the sa-vs-nc group, with
log2 FC=1.27 and an adjusted P-value of
1.85×10−74; in the PPI network, caspase-3 was also found
as the hub gene with an interaction degree of 145 (Fig. 3T). Based on these findings, we
hypothesized that FOXF1 promotes the apoptosis of BC cells via the
caspase signaling pathway; this was then confirmed using western
blot analysis (Fig. 3U).
Furthermore, the promoting effect of FOXF1 on cell apoptosis was
reversed by the inhibitor of caspase-3, and the reduced cell
viability in the dsFOXF1-367-transfected groups was also recovered
by this inhibitor (Fig. 4).
 | Figure 4.Promoting effect of FOXF1 on the
apoptosis of T24 and EJ cells is reversed by caspase-3 inhibitor.
Apoptosis assays of T24 cells treated with (A) dsControl, (B)
dsControl plus caspase inhibitor, (C) dsFOXF1-367 and (D)
dsFOXF1-367 plus caspase inhibitor, respectively. (E) Percentage of
T24 cells undergoing apoptosis and (F) cell viability in each
group. The apoptosis assays of EJ cells treated with (G) dsControl,
(H) dsControl plus caspase inhibitor, (I) dsFOXF1-367 and (J)
dsFOXF1-367 plus caspase inhibitor, respectively. (K) Percentage of
EJ cells undergoing apoptosis and (L) cell viability in each group.
**P<0.01 and ***P<0.001, vs. dsControl or as indicated. The
cell apoptotic rates in the dsFOXF1-367(+) Ac-DEVD-CHO(−) groups
were compared with those in the dsControl(+) Ac-DEVD-CHO(−) groups,
and the cell apoptotic rates in the dsFOXF1-367(+) Ac-DEVD-CHO(+)
groups were compared with those in the dsFOXF1-367(+)
Ac-DEVD-CHO(−) groups, respectively. FOXF1, forkhead box F1. |
FOXF1 suppresses bladder tumor growth
in vivo
To investigate the antitumor effects of FOXF1 in
vivo, dsFOXF1-367 was packaged into lentivirus to construct a
cell line with the stable activation of FOXF1. As dsFOXF1-367 had a
more prominent activation effect in the EJ cells (Fig. S4E and F), subcutaneous xenograft
tumor models were developed with Lenti-dsFOXF1-367- or
Lenti-dsControl-infected EJ cells. The procedure of injection,
weighting and measurements are illustrated in Fig. 5A. Lenti-FOXF1-367 impaired tumor
formation; the tumor volume, weight and growth rates in the
Lenti-FOXF1-367 group were significantly decreased compared with
the Lenti-dsControl group (P<0.05, Fig. 5B-E). Furthermore, the volume of the
xenograft models developed from Lenti-FOXF1-367 began to decline on
day 25 (Fig. 5D). The mechanisms of
the antitumor signaling of dsFOXF1-367 in bladder tumors are shown
in Fig. 6. The antitumor effect of
FOXF1 in vitro and in vivo demonstrated its promising
potential in bladder cancer target therapy.
Discussion
In the present study, the downregulated expression
of FOXF1 in BC tissue was revealed using IHC, and the low
expression levels were associated with more aggressive tumor
phenotypes. The decreased expression of FOXF1 is an unfavorable
predictor in patients with BC. The expression level of FOXF1 may be
used to stratify patients with BC into groups with distinct tumor
invasiveness and clinical outcomes. Moreover, the downregulated
expression of FOXF1 was detected in bladder tumor samples of both
Ruijin Cohort and GEO datasets. The clinical stages, invasiveness,
lymphoid infiltrates and OS were significantly more severe in
patients with BC with a low FOXF1 expression compared with patients
with a high FOXF1 expression, confirming the stratifying ability of
FOXF1. In the T24 and EJ cells, dsFOXF1-367 activated the
expression level of FOXF1, this activation was dependent on the
binding of the dsRNA and FOXF1 promoter sequence. FOXF1 exerted a
suppressive effect on BC cells, and caspase-3 was found to be one
of the downstream genes of FOXF1 through RNA seq and PPI network
analyses; FOXF1 may induce BC cell apoptosis via the caspase
signaling pathway.
The subcellular localization and role of FOXF1 in
various types of cancers is controversial. In the present study, it
was found that FOXF1-positive staining was mainly localized in the
nucleus using IHC and IF staining. The nuclear expression of FOXF1
has also been found in hilar cholangiocarcinoma or metastatic
pancreatic ductal adenocarcinoma (24). However, inconsistent results were
observed in other types of cancer. Lo et al (25) revealed that FOXF1 protein was
predominately expressed in the cytoplasm of epithelial cells in
colorectal adenocarcinoma tissue, while positive staining was only
identified in the stroma of adjacent normal tissue. Their findings
illustrated that FOXF1 expression was increased and mis-localized
in epithelial cells of colorectal adenocarcinoma (25).
Similar to the findings presented herein, other
studies have proven that a low FOXF1 expression level is also
associated with the malignant types of other tumors. In breast
cancer, the increased expression of FOXF1 was shown to induce G1
phase arrest through the inactivation of the CDK2-RB-E2F cascade,
thus suppressing tumor cell growth (26). In lung cancer, FOXF1 was found to be
underexpressed not only in tumor samples, but also in cancer cell
lines. In the manufactured FOXF1-overexpressing lung cancer cell
line, the cell proliferative and migratory abilities were
inhibited, accompanied by G1 phase arrest (27). In hepatocellular carcinoma, the
overexpression of FOXF1 was found to impair the stemness of cancer
cells, and the prognosis of patients was positively associated with
the expression level of FOXF1 (16). However, Wang et al (28) indicated that the upregulated
expression of FOXF1 was related to angiogenesis, as well as a
number of aggressive clinical features in colorectal cancer (CRC).
They inferred that FOXF1 could function as a promoter for the
transcription of vascular endothelial growth factor A1 (VEGFA),
thus altering tumor progression. In their another study, they
demonstrated that the upregulation of FOXF1 expression in CRC
transcriptionally elevated SNAI1 expression, promoting
epithelial-mesenchymal transition by downregulating the expression
level of epithelial markers (29).
These studies suggest that FOXF1 might have tumor suppressor and
tumor enhancer dual functions, and that it plays differential roles
in various types of cancer.
FOXF1 may be a prognostic indicator in some types of
cancer. In papillary thyroid cancer, the mRNA expression level of
FOXF1 has been found to be significantly lower in tumor tissue than
in the normal thyroid gland (15).
Patients with a downregulated FOXF1 mRNA expression have been shown
to have more malignant cancer phenotypes and a shorter
recurrence-free survival than patients with a higher FOXF1 mRNA
expression (15). Furthermore, Zhao
et al (16) found that FOXF1
suppressed hepatocellular carcinoma in vivo; the
FOXF1-overexpressing xenografts had lower tumor weights and PCNA
expression levels than the control xenografts, and patients whose
tumors had more positive FOXF1 IHC staining in the nuclei had
significantly better survival outcomes than patients with a lower
FOXF1 expression level (16).
However, the prognostic value of FOXF1 in cancer warrants further
investigation.
The caspase family consists of a number of
proteolytic enzymes, their levels will culminate during apoptosis
(30). According to their
functions, they can be divided into two groups as follows:
Initiators (includes caspase-2, −8, −9 and −10) and effectors
(includes caspase-3, −6 and −7) (31,32).
Caspases are usually inactive, and their activation plays a central
role in the signaling pathway of apoptosis. The effector caspases
(such as caspase-3) can be activated by initiator caspases (such as
caspase-9) or Bax/Bak, while initiator caspases are self-activated
(32,33). In the present study, the activation
of FOXF1 expression induced cell apoptosis and elevated the
expression level of Bax, caspase-9, caspase-3 and PARP. Moreover,
the cell apoptotic rate was suppressed by caspase inhibitor. These
results suggest that FOXF1 may contribute to a mitochondrial
caspase-dependent apoptotic pathway in BC.
The resistance of apoptosis is a hallmark of cancer,
as tumor cells are able to limit apoptosis through a number of
strategies (34). However, in
certain types of cancer, by regulating the tumor microenvironment,
apoptosis also functions as a pro-oncogenic factor, potentiating
angiogenesis, metastasis and the invasion of cancer (35). Apoptosis-driven growth and repair
can stimulate the generation of the tumor microenvironment,
repopulating tumors with surviving cells (36). Caspase-3 is crucial in linking the
regeneration and repopulation processes. It has been reported that
caspase-3 inhibitor enhances the efficacy of radiotherapy, and
caspase-3 activation can predict the sensitivity of tumors to
adjuvant treatment (37). This
paradox of cell death may explain the ambivalent effects of FOXF1
in cancer; the ‘yin and yang’ of apoptosis in tumorigenesis needs
further investigations.
To the best of our knowledge, the present study is
the first to analyze the expression patterns, prognostic value and
antitumor mechanisms of FOXF1 in BC. FOXF1 can not only strength
the current staging systems, but can also assist clinical
judgement. The apoptotic promoting effects of FOXF1 on BC cells
suggest its potential application in tumor targeted therapy.
However, there are several limitations to the present study. First,
limited by the surgical quantity of cystectomy, only 59 patients
were recruited in the present study. The predictive ability of
FOXF1 in patients with BC remains to be validated in a larger
cohort. Therefore, public datasets were utilized to validate the
predictive ability of FOXF1 and its association with the clinical
data of patients with BC. In TCGA dataset, 406 patients with BC
were divided into the FOXF1-high and FOXF1-low groups; the patients
with a higher FOXF1 expression exhibited more favorable
clinicopathological features than the patients with a lower FOXF1
expression (Table SV).
Furthermore, in the GSE13507, GSE31684, GSE48075, GSE169455 and
IMvigor210 cohort, patients with BC in the FOXF1-low groups all had
significantly poorer survival outcomes than patients in the
FOXF1-high groups (Figs. 1K and
S3A-E), confirming the
universality of the conclusions reached herein. Second, the present
study recruited patients with BC who had undergone cystectomy, and
the majority of these had high-grade urothelium carcinoma (56 out
of 59 patients); thus, the expression pattern of FOXF1 in low-grade
BC remains to be evaluated. Therefore, further multicenter studies
with a greater number of patients and higher tumor stages are
required. Third, the exact mechanisms of RNA activation, as well as
the interaction between FOXF1 and caspase-3 remain unclear; thus,
further studies are warranted to investigate this pathway in more
detail.
In conclusion, the present study confirmed that the
downregulated expression of FOXF1 was associated with unfavorable
clinical stages and types in BC. FOXF1 can be used to stratify
patients with BC with significantly different survival outcomes,
and its expression level has a robust predictive ability as regards
the prognosis of patients with BC. FOXF1 exerts antitumor effects
on BC cells, as it can induce cell apoptosis in a caspase-dependent
manner. This finding may be of value to staging systems, and it may
assist clinical decisions for adjuvant therapies and follow-up
after surgery. The activation of FOXF1 expression may be used as a
novel strategy in tumor therapeutics.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National
Natural Science Foundation of China (nos. 81602215 and 82173045)
and Guangci Distinguished Young Scholars Training Program (nos.
GCQN-2018-A10).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The RNA-Seq data have been submitted to the GEO repository
(accession no. GSE229810).
Authors' contributions
YH, CW and DX conceived and designed the study. YH
and CW contributed to the execution of the experiments, statistical
analysis of the data and in the drafting of the manuscript. WH, HW,
WR, FS and YZ performed the surgeries. YH and CW confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki. The studies involving human
participants or animals were reviewed and approved by the Ethics
Committee of Ruijin Hospital, School of Medicine, Shanghai Jiao
Tong University (PA23030202). All patients/participants provided
their written informed consent to participate in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BC
|
bladder cancer
|
|
MIBC
|
muscular invasive bladder cancer
|
|
NMIBC
|
non-muscular invasive bladder
cancer
|
|
AJCC
|
American Joint Committee on Cancer
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GEO
|
Gene Expression Omnibus
|
|
DEGs
|
differentially expressed genes
|
|
FC
|
fold change
|
|
OS
|
overall survival
|
|
IHC
|
immunohistochemistry
|
|
TNN
|
tumor node metastasis stage of
tumors
|
|
FOXF1
|
forkhead box F1
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
RNA-seq
|
RNA sequencing
|
|
PPI
|
protein-protein interaction
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Witjes JA, Bruins HM, Cathomas R, Compérat
EM, Cowan NC, Gakis G, Hernández V, Espinós EL, Lorch A, Neuzillet
Y, et al: European association of urology guidelines on
muscle-invasive and metastatic bladder cancer: Summary of the 2020
guidelines. Eur Urol. 79:82–104. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Babjuk M, Burger M, Capoun O, Cohen D,
Compérat EM, Escrig JL, Gontero P, Liedberg F, Masson-Lecomte A,
Mostafid AH, et al: European association of urology guidelines on
non-muscle-invasive bladder cancer (Ta, T1, and Carcinoma in Situ).
Eur Urol. 81:75–94. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Oliveira MC, Caires HR, Oliveira MJ,
Fraga A, Vasconcelos MH and Ribeiro R: Urinary biomarkers in
bladder cancer: Where do we stand and potential role of
extracellular vesicles. Cancers (Basel). 12:14002020. View Article : Google Scholar
|
|
5
|
Allory Y, Beukers W, Sagrera A, Flández M,
Marqués M, Márquez M, van der Keur KA, Dyrskjot L, Lurkin I,
Vermeij M, et al: Telomerase reverse transcriptase promoter
mutations in bladder cancer: High frequency across stages,
detection in urine, and lack of association with outcome. Eur Urol.
65:360–366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moonen PM, Kiemeney LA and Witjes JA:
Urinary NMP22 bladderchek test in the diagnosis of superficial
bladder cancer. Eur Urol. 48:951–956. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lam EWF, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Katoh M, Igarashi M, Fukuda H, Nakagama H
and Katoh M: Cancer genetics and genomics of human FOX family
genes. Cancer Lett. 328:198–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamashita H, Amponsa VO, Warrick JI, Zheng
Z, Clark PE, Raman JD, Wu XR, Mendelsohn C and DeGraff DJ: On a FOX
hunt: Functions of FOX transcriptional regulators in bladder
cancer. Nat Rev Urol. 14:98–106. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
11
|
Mahlapuu M, Ormestad M, Enerback S and
Carlsson P: The forkhead transcription factor Foxf1 is required for
differentiation of extra-embryonic and lateral plate mesoderm.
Development. 128:155–166. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Paulsson J and Micke P: Prognostic
relevance of cancer-associated fibroblasts in human cancer. Semin
Cancer Biol. 25:61–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sturtzel C, Lipnik K, Hofer-Warbinek R,
Testori J, Ebner B, Seigner J, Qiu P, Bilban M, Jandrositz A,
Preisegger KH, et al: FOXF1 mediates endothelial progenitor
functions and regulates vascular sprouting. Front Bioeng
Biotechnol. 6:762018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren X, Ustiyan V, Pradhan A, Cai Y,
Havrilak JA, Bolte CS, Shannon JM, Kalin TV and Kalinichenko VV:
FOXF1 transcription factor is required for formation of embryonic
vasculature by regulating VEGF signaling in endothelial cells. Circ
Res. 115:709–720. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gu Y and Hu C: Bioinformatic analysis of
the prognostic value and potential regulatory network of FOXF1 in
papillary thyroid cancer. Biofactors. 45:902–911. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao ZG, Wang DQ, Hu DF, Li YS and Liu SH:
Decreased FOXF1 promotes hepatocellular carcinoma tumorigenesis,
invasion, and stemness and is associated with poor clinical
outcome. Onco Targets Ther. 9:1743–1752. 2016.PubMed/NCBI
|
|
17
|
Herrera-Merchan A, Cuadros M, Rodriguez
MI, Rodriguez S, Torres R, Estecio M, Coira IF, Loidi C, Saiz M,
Carmona-Saez P and Medina PP: The value of lncRNA FENDRR and FOXF1
as a prognostic factor for survival of lung adenocarcinoma.
Oncotarget. 11:1172–1185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Magers MJ, Lopez-Beltran A, Montironi R,
Williamson SR, Kaimakliotis HZ and Cheng L: Staging of bladder
cancer. Histopathology. 74:112–134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Davis S and Meltzer PS: GEOquery: A bridge
between the gene expression omnibus (GEO) and bioconductor.
Bioinformatics. 23:1846–1847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zeng D, Ye Z, Shen R, Yu G, Wu J, Xiong Y,
Zhou R, Qiu W, Huang N, Sun L, et al: IOBR: Multi-omics
immuno-oncology biological research to decode tumor
microenvironment and signatures. Front Immunol. 12:6879752021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang V, Qin Y, Wang J, Wang X, Place RF,
Lin G, Lue TF and Li LC: RNAa is conserved in mammalian cells. PLoS
One. 5:e88482010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hoffman-Censits JH, Grivas P, Van Der
Heijden MS, Dreicer R, Loriot Y, Retz M, Vogelzang NJ, Perez-Gracia
JL, Rezazadeh A, Bracarda S, et al: IMvigor 210, a phase II trial
of atezolizumab (MPDL3280A) in platinum-treated locally advanced or
metastatic urothelial carcinoma (mUC). J Clin Oncol. 34 (Suppl
2):S3552016. View Article : Google Scholar
|
|
24
|
Hrudka J, Prouzová Z, Mydlíková K,
Jedličková K, Holešta M, Whitley A and Havlůj L: FOXF1 as an
immunohistochemical marker of hilar cholangiocarcinoma or
metastatic pancreatic ductal adenocarcinoma. Single institution
experience. Pathol Oncol Res. 27:16097562021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lo PK, Lee JS, Chen H, Reisman D, Berger
FG and Sukumar S: Cytoplasmic mislocalization of overexpressed
FOXF1 is associated with the malignancy and metastasis of
colorectal adenocarcinomas. Exp Mol Pathol. 94:262–269. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lo PK, Lee JS, Liang X, Han L, Mori T,
Fackler MJ, Sadik H, Argani P, Pandita TK and Sukumar S: Epigenetic
inactivation of the potential tumor suppressor gene FOXF1 in breast
cancer. Cancer Res. 70:6047–6058. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu CY, Chan CH, Dubey NK, Wei HJ, Lu JH,
Chang CC, Cheng HC, Ou KL and Deng WP: Highly expressed FOXF1
inhibit non-small-cell lung cancer growth via inducing tumor
suppressor and G1-phase cell-cycle arrest. Int J Mol Sci.
21:32272020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Xiao Z, Hong Z, Jiao H, Zhu S,
Zhao Y, Bi J, Qiu J, Zhang D, Yan J, et al: FOXF1 promotes
angiogenesis and accelerates bevacizumab resistance in colorectal
cancer by transcriptionally activating VEGFA. Cancer Lett.
439:78–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang S, Yan S, Zhu S, Zhao Y, Yan J, Xiao
Z, Bi J, Qiu J, Zhang D, Hong Z, et al: FOXF1 induces
epithelial-mesenchymal transition in colorectal cancer metastasis
by transcriptionally activating SNAI1. Neoplasia. 20:996–1007.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Creagh EM, Conroy H and Martin SJ:
Caspase-activation pathways in apoptosis and immunity. Immunol Rev.
193:10–21. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Van Opdenbosch N and Lamkanfi M: Caspases
in cell death, inflammation, and disease. Immunity. 50:1352–1364.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi Y: Mechanisms of caspase activation
and inhibition during apoptosis. Mol Cell. 9:459–470. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vince JE, De Nardo D, Gao W, Vince AJ,
Hall C, McArthur K, Simpson D, Vijayaraj S, Lindqvist LM, Bouillet
P, et al: The mitochondrial apoptotic effectors BAX/BAK activate
caspase-3 and −7 to trigger NLRP3 inflammasome and caspase-8 driven
IL-1β activation. Cell Rep. 25:2339–2353.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ford CA, Petrova S, Pound JD, Voss JJ,
Melville L, Paterson M, Farnworth SL, Gallimore AM, Cuff S, Wheadon
H, et al: Oncogenic properties of apoptotic tumor cells in
aggressive B cell lymphoma. Curr Biol. 25:577–588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Morana O, Wood W and Gregory CD: The
apoptosis paradox in cancer. Int J Mol Sci. 23:113282022.
View Article : Google Scholar
|
|
37
|
Huang Q, Li F, Liu X, Li W, Shi W, Liu FF,
O'Sullivan B, He Z, Peng Y, Tan AC, et al: Caspase 3-mediated
stimulation of tumor cell repopulation during cancer radiotherapy.
Nat Med. 17:860–866. 2011. View Article : Google Scholar : PubMed/NCBI
|