Introduction
Neoplasms, or abnormal growths, are complex
diseases. They result from an accumulation of genetic and
epigenetic abnormalities, leading to uncontrolled cell
proliferation, infiltration and metastasis (1–3). Among
these diseases, head and neck squamous cell carcinoma (HNSCC)
stands out due to its prevalence. It is the sixth most common
neoplasm worldwide, making up ~4% of all malignancies (4–6).
Despite significant progress in cancer research, the outlook for
patients with advanced HNSCC remains poor. The 5-year survival rate
is ~50% (7–10). This highlights the urgent need for
new therapeutic targets for HNSCC. One promising area of focus is
cancer stem cells (CSCs). They are considered to drive tumor
initiation, progression and recurrence (6,11–15).
CSCs are a small subset of cells within a tumor that possess the
capacity for self-renewal and the ability to give rise to the
heterogeneous lineages of cancer cells that comprise the tumor.
They are considered to be responsible for the initiation,
propagation and recurrence of cancer. CSCs are also often resistant
to conventional chemotherapeutic drugs and radiation therapy, which
can lead to the recurrence of tumors after treatment.
CD44 is the most frequently reported CSC marker. Its
expression is associated with poor prognosis and resistance to
therapy (13–16). The CD44 antigen is a multifunctional
cell surface glycoprotein involved in cell-cell interactions, cell
adhesion, and migration. It is encoded by the CD44 gene
located on chromosome 11 in humans. CD44 is a receptor for
hyaluronic acid (HA) and can also interact with other ligands, such
as osteopontin, collagens and matrix metalloproteinases (17,18).
CD44 interaction with HA plays a significant role in numerous
biological activities including lymphocyte activation,
recirculation and homing, hematopoiesis and tumor metastasis
(19). In the context of HNSCC,
CD44 is the most frequently reported CSC marker. CD44-positive (+)
cells in HNSCC have been revealed to exhibit stem cell properties,
such as self-renewal and tumorigenicity. The expression of CD44 in
HNSCC is associated with poor prognosis and resistance to therapy.
Nevertheless, the pursuit of supplementary indicators to delineate
CSCs and elucidate their characteristics persists. This endeavor
transcends mere scholarly interest, as it holds the potential to
engender groundbreaking and efficacious therapies for cancer,
capable of eradicating neoplastic cells and impeding their
reappearance.
One such potential marker is the stage-specific
embryonic antigen 3 (SSEA3). This glycosphingolipid molecule is an
important marker for mammalian cells that display pluripotent and
stem cell-like characteristics (20–22).
However, the expression and function of SSEA3 in HNSCCs remain
unclear. This gap forms the basis of the present study. The
objective of the present study was to examine the expression of
SSEA3 and its potential role in modulating CSC properties. This
includes cellular proliferation stemness, and drug resistance in
CD44(+) cells within an oral cancer cell line. The present study is
crucial as it could contribute to enhance the understanding of the
role of SSEA3 in HNSCC; thus, patients suffering from this
life-threatening disease will be able to improve their outcomes and
survival rates.
Materials and methods
Cell lines
The human oral neoplastic cell lines, OSC-19 (cat.
no. 0198), OSC-20 cells (cat. no. 0197), SCC4 (cat. no. 9118), SAS
(cat. no. 0260), HSC2 (cat. no. 0622), HSC3 (cat. no. 0623), HSC4
(cat. no. 0624), KOSC-2 (cat. no. 0126.1), KON (cat. no. 0194),
Ca9-22 (cat. no. 0625), HO-1-u-1 (cat. no. 0828), HO-1-N-1 (cat.
no. 0831), SAT (cat. no. 1027) and SKN-3 (cat. no. 1039) were
obtained from the Japanese Collection of Research Bioresources
(JCRB) Cell Bank. The aforementioned cell lines were authenticated
by JCRB Cell Bank using STR methods. The other human oral
neoplastic cell lines, OLC-01 (23)
and OSC-70 (24) were gifted from
Dr S. Kawashiri (Kanazawa University, Kanazawa, Japan). All cell
lines were cultured in high glucose DMEM (Sigma-Aldrich;
MilliporeSigma) supplemented with 10% heat-inactivated fetal bovine
serum (FBS) at 37°C in a humidified atmosphere containing 5%
CO2.
Flow cytometric assessment of CD44 and
SSEA3
The assessment of CD44 and SSEA3 was conducted
through a process of flow cytometry. This process was facilitated
by the use of BD FACSVerse™ Flow Cytometer (cat. no. 651155,
version 1.1), which was sourced from BD Biosciences. Before the
assessment, the cells were dissociated with Accutase (cat. no.
AT104; Invitrogen; Thermo Fisher Scientific, Inc.) and washed with
ice cold PBS. A number of the cells was adjusted to a concentration
of 1.0×106 cells/ml in ice cold FACS Buffer (PBS, 1%
BSA; cat. no. BR-220701651; Biomedical Science, Co., Ltd.). A total
of 500 µl cell suspension was stained with a primary antibody that
is a rat anti-human SSEA3 monoclonal antibody (MC-631; 1:100; cat.
no. MA1-020; Thermo Fisher Scientific, Inc.) and incubated for 30
min at 4°C. After incubation, the cells were washed with ice cold
PBS by centrifugation at 200 × g for 5 min and resuspended in 500
µl ice cold FACS Buffer. The cells were stained with the following
secondary antibodies: allophycocyanin (APC)-labeled mouse anti-CD44
monoclonal antibody (MEM-85; 1:500; cat. no. ab239294; Abcam) and
fluorescein isothiocyanate (FITC)-labeled goat anti-rat IgM
polyclonal antibody (1:500; cat. no. 112-095-075; Jackson
ImmunoResearch Laboratories, Inc.) and kept at 4°C in a fridge for
30 min. After incubation and washing with PBS, the cells were
stained with SYTOX™ Blue Dead Cell Stain (1:500: cat. no. S34857;
Invitrogen; Thermo Fisher Scientific, Inc.) to differentiate
between viable and nonviable cells. In the flow cytometric
analysis, purified rat IgM k isotype control monoclonal antibody
(1:100; cat. no. 400801; BioLegend, Inc.) and purified mouse IgG2b
k isotype control monoclonal antibody (1:100; cat. no. 202201;
BioLegend, Inc.) were used as control primary antibody. These
controls were obtained from BD Pharmingen; BD Biosciences. This
comprehensive process of flow cytometric assessment allowed for a
detailed and accurate appraisal of CD44 and SSEA3 in the cells.
Each positive cell population in the entire oral cancer cell
population has been analyzed. Values were expressed as
percentages.
Isolation of SSEA3 (+) and SSEA3
negative (−) cell populations via cell sorting
The isolation of SSEA3 positive (+) and SSEA3
negative (−) cell populations was achieved through a process of
cell sorting. This process was facilitated by the use of a Sony
SH800S Cell Sorter (Sony Biotechnology Inc.). The sorter was
equipped with a 130-µm sorting chip, which was crucial for the
segregation of cells. The SCC4 cell lines were prepared for sorting
by staining them with a rat anti-human SSEA3 monoclonal antibody
(Thermo Fisher Scientific, Inc.) and a goat anti-rat IgM (FITC)
antibody (Jackson ImmunoResearch Laboratories, Inc.). This staining
process was carried out strictly in accordance with the
manufacturer's guidelines provided by Sony Biotechnology. Following
the staining, the purity of the partitioned SSEA3(+) and SSEA3(−)
cells was assessed. This assessment was carried out through flow
cytometry, employing a FACS Verse instrument. The cells were
labeled again with the rat anti-human SSEA3 monoclonal antibody and
the goat anti-rat IgM FITC antibody for this assessment. Before the
sorting process, the stained cells were filtered into a conical
tube. A 35-µm nylon filter was used for this filtration process.
The cell proliferation, cell cycle, reverse
transcription-polymerase chain reaction and chemoresistance assay
data reflecting SSEA3 expression were differentiated via cell
sorting from SCC4. This comprehensive process ensured the isolation
of the specific cell populations for further study.
Cell proliferation assessment
The study of cellular proliferation was conducted to
compare the growth rates between various cellular samples. The
procedure began with seeding 1×105 cells in each well of
a six-well plate. The cell culture conditions were maintained at
37°C and atmosphere standard for cell growth, specifically within a
5% CO2 incubator. The growth medium utilized was DMEM,
supplemented with 10% FBS to provide necessary nutrients for the
cells. These conditions were maintained for a period ranging
between 1 to 3 days, allowing for cells to undergo several rounds
of proliferation. Post-incubation, the counting of cells was
carried out using a Countess Automated Cell Counter (Invitrogen;
Thermo Fisher Scientific, Inc.). This device facilitated the
accurate and reliable quantification of cells in the sample. To
enhance the reliability of the count and to reduce sample bias, a
minimum of four different cell images from each well were randomly
selected and captured with a phase-contrast microscope for
independent verification. This ensured that the cell count was
representative of the entire well, providing a more accurate
measurement of cellular proliferation. These methods provided a
comprehensive and precise examination of the cellular proliferation
rates, eliminating any potential for misunderstanding and ensuring
the reproducibility of the experiments.
Cell cycle analysis
The cell cycle analysis was conducted on SSEA3(+)
and negative (−) cells, which were sorted from SCC4 cells. The
sorted cells were first resuspended in a solution. This solution
was prepared by adding 5 µl of Cell Cycle Assay Solution Blue kit
(Dojindo Molecular Technologies, Inc.) to 500 µl of
Phosphate-Buffered Saline. This resuspension process was carried
out at a controlled temperature of 37°C for a duration of 15 min,
strictly adhering to the guidelines provided by the manufacturer of
the Cell Cycle Assay Solution Blue. After the resuspension, the
cells were stained using the same solution. The stained cells were
then assessed using a specific instrument known as BD FACSVerse™
Flow Cytometer. The assessment was followed by a filtration
process. In this process, the cells were filtered into a conical
tube. A 35-µm nylon filter was used for this purpose to ensure the
collection of only the cells of interest. The entire process was
carried out under sterile conditions to prevent any contamination.
Percentage of cells in the G0/G1, S, and G2/M phases on SSEA3(+) or
(−) cells from SCC4 cells has been analyzed.
RNA extraction, cDNA synthesis, and
reverse transcription-quantitative PCR
The mRNA expression levels of brother of the
imprinted site regulator (BORIS), sex-determining region Y-box 2
(SOX2), homeobox protein NANOG (Nanog), and octamer-binding
transcription factor 4, also known as POU Class 5 Homeobox 1
(POU5F1) were analyzed using a Rotor-Gene Q 2plex System (Qiagen
GmbH) with FAM/ZEN/IBFQ probes (Integrated DNA Technologies, Inc.).
DNA sequences of pre-designed BORIS primer/probe (ref. 106829301,
Hs.PT.58.50443006, NM_001269040, Integrated DNA Technology),
pre-designed SOX2 primer/probe (ref. 105825433, Hs.PT.58.237897.g,
NM_003106, Integrated DNA Technologies, Inc.), pre-designed Nanong
primer/probe (ref. 105825437, Hs.PT.58.21480849, NM_024865,
Integrated DNA Technology) and pre-designed POU5F1 primer/probe
(ref. 105825441, Hs.PT.58.14648152.g, NM_203289, Integrated DNA
Technology) were not available. Assay IDs and RefSeq accession
numbers of the tested genes were provided. Total RNA was extracted
from SCC4 cells using TRI Reagent (Merck KGaA) and a PureLink RNA
Mini kit (Thermo Fisher Scientific, Inc.). cDNA was obtained using
a PrimeScript 1st strand cDNA Synthesis kit (Takara Bio, Inc.). All
reactions were performed in accordance with the manufacturer's
instructions. All PCR reactions were performed on a Qiagen
Rotor-Gene Q real-time PCR system with the following thermocycling
conditions: Pre-incubation at 94°C for 2 min, followed by 40 cycles
of 15 sec at 95°C and 1 min at 60°C. A total of 10 ng cDNA sample
were used for the RT-qPCR reaction following the manufacturer's
instructions. The fluorescent signals were analyzed by Rotor-Gene Q
software version 2.3.1 (Qiagen). For the 2−ΔΔCq method
for qPCR to be valid, the efficiency of the target amplification
and the efficiency of the reference amplification must be ~equal.
Thus, Hypoxanthine Phosphoribosyltransferase 1 (Hs.PT.58v.45621572,
NM_000194), beta 2-microglobulin (Hs.PT.58v.18759587, NM_004048),
TATA-Box Binding Protein (Hs.PT.58v.39858774, NM_003194),
Glucuronidase Beta (Hs.PT.58v.27737538, NM_000181), Actin Beta
(ACTB) (Hs.PT.39a.22214847, NM_001101), RNA Polymerase II Subunit A
(Hs.PT.39a.19639531, NM_000937), Ribosomal Protein Lateral Stalk
Subunit P0 (Hs.PT.39a.22214824, NM_001002) and Peptidylprolyl
Isomerase A (Hs.PT.58v.38887593.g, NM_021130) were analyzed as
potential internal standards. Based on these validation
experiments, ACTB was selected as an internal standard using
HEX/ZEN/IBFQ probes (Integrated DNA Technologies, Inc.). Relative
expression levels were calculated using the 2−ΔΔCq
method for qPCR (25), which
presents the data as fold differences in expression relative to a
calibrator sample, in this case represented by the mean expression
of three experimental measurements.
Chemoresistance assay
The chemoresistance assay was conducted using
SSEA3(+) or (−) cells that were isolated from SCC4 cells. These
cells were seeded at 2.5×104 cells/well and inoculated
in 96-well plates and then cultivated with varying concentrations
of cisplatin (0, 0.01, 0.1, 1, 5, 10 µM), paclitaxel (0, 0.01, 0.1,
1, 5, 10 nM) and docetaxel (0, 0.01, 0.1, 1, 5, 10 nM) for 72 h.
Following this cultivation period, the number of cells and their
viability were measured. The percentage of viability was
determined. This was achieved by utilizing the Cell Counting Kit-8
(Dojindo Molecular Technologies, Inc.), following the
manufacturer's protocol. A total of 10 µl Cell Counting Kit-8
solution was added to each well, then incubated for 30 min. The
optical density at 490 nm was measured using a microplate reader
(iMark™ Microplate Absorbance Reader; Bio-Rad Laboratories, Inc.).
The mean IC50 values, which represent the concentration
of a drug that gives half-maximal response, were derived from three
independent experiments. These values were calculated based on
discrete sigmoidal curves that represent the growth inhibition
activity in response to cisplatin, paclitaxel, and docetaxel. The
cisplatin used in the aforementioned assay was procured from
Nichi-Iko Pharmaceutical Co., Ltd., while the paclitaxel and
docetaxel were sourced from Nipro Corporation. The data are
presented as the mean ± standard deviation.
Statistical analysis
Data were analyzed by one-way analysis of variance
and bilateral Student's t-test using EZR (Saitama Medical Center,
Jichi Medical University, version 2.13.0), which is a graphical
user interface for R (The R Foundation for statistical computing)
(26). P≤0.05 was considered to
indicate a statistically significant difference.
Results
SSEA3 is expressed in CD44(+) cells
within an oral cancer cell line
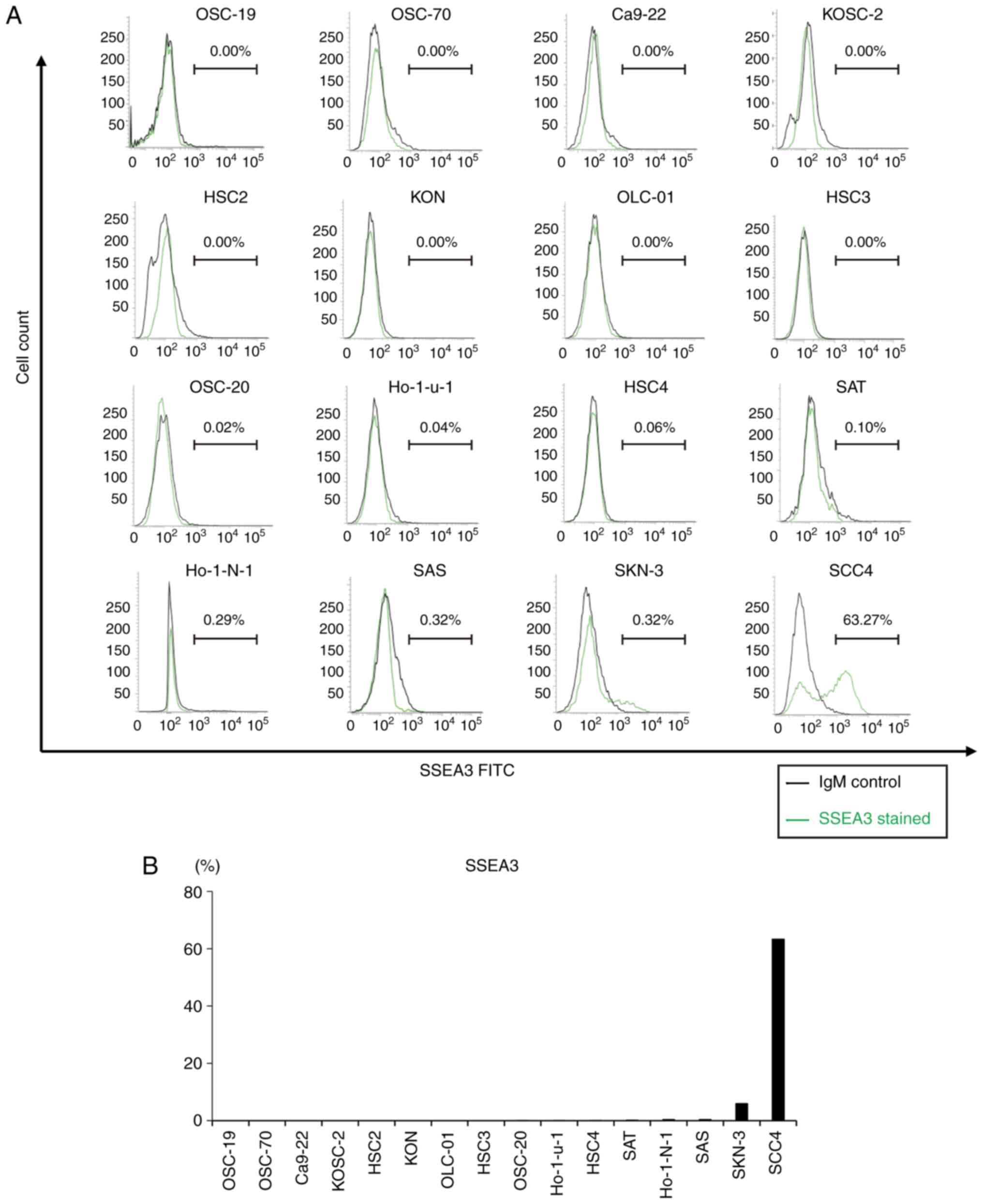
To evaluate SSEA3 expression in oral cancer cell
lines, flow cytometric analysis was conducted on 16 oral cancer
cell lines. This revealed the presence of a small number of
SSEA3(+) cells in the several oral cancer cell lines, ranging from
0.02 to 5.88% in OSC-20, Ho-1-u-1, HSC4, SAT, Ho-1-N-1, SAS and
SKN-3. SCC4 exhibited the highest expression levels among the
tested oral cancer cell lines (Fig. 1A
and B).
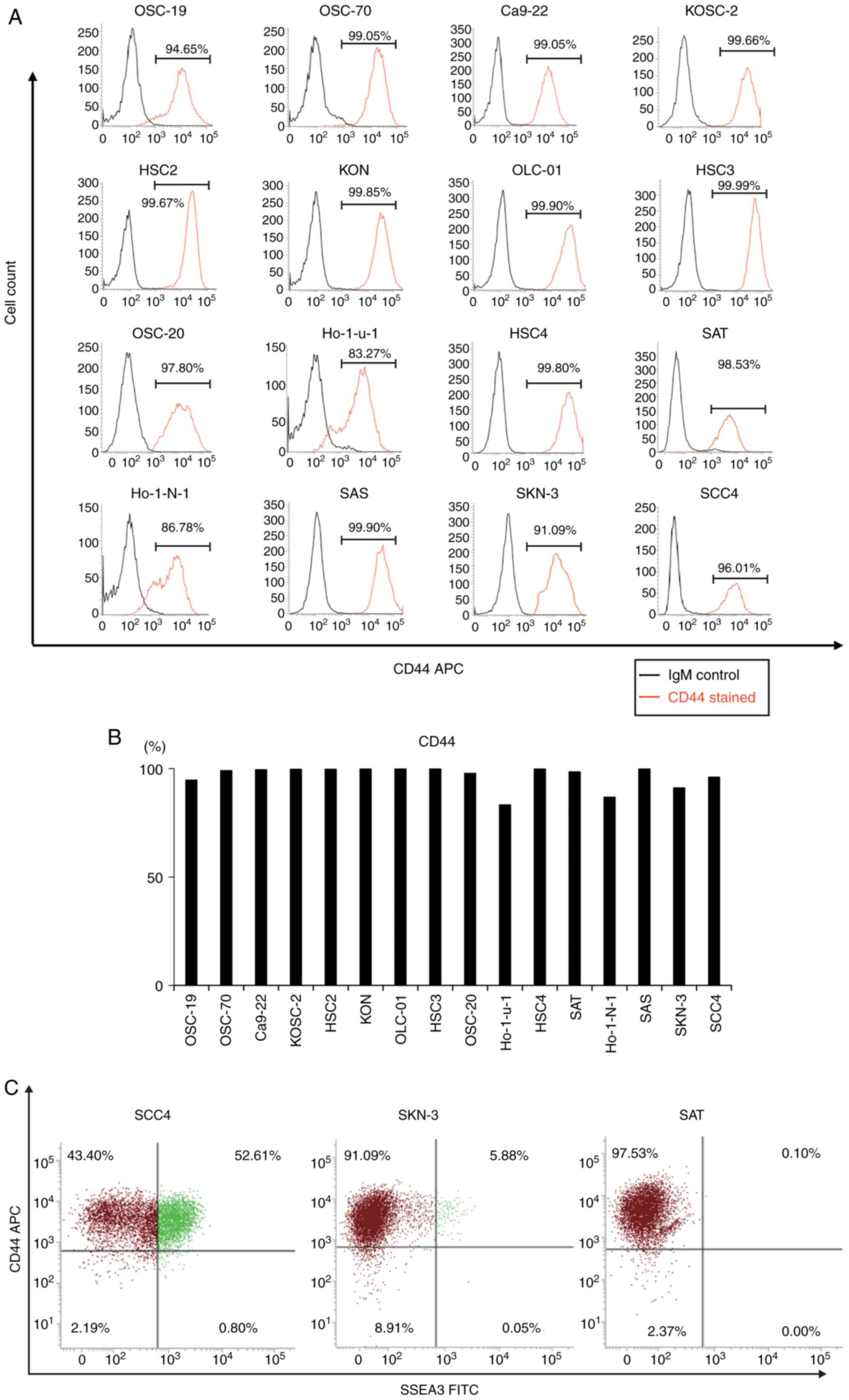
Flow cytometric analysis revealed a profusion of
CD44(+) cells in 16 oral cancer cell lines, ranging from 83.27 to
99.99% (Fig. 2A and B). To
elucidate the association between SSEA3 and CD44, SCC4, SKN-3 and
SAT were double-stained with CD44 and SSEA3. In the CD44(+)
fraction, 52.61% of SCC4 cells, 5.88% of SKN-3 cells, and 0.10% of
SAT cells were SSEA3(+) (Fig. 2C).
Consequently, further experiments with SCC4 were conducted in order
to demonstrate the characteristics of SSEA3 in an oral cancer cell
line.
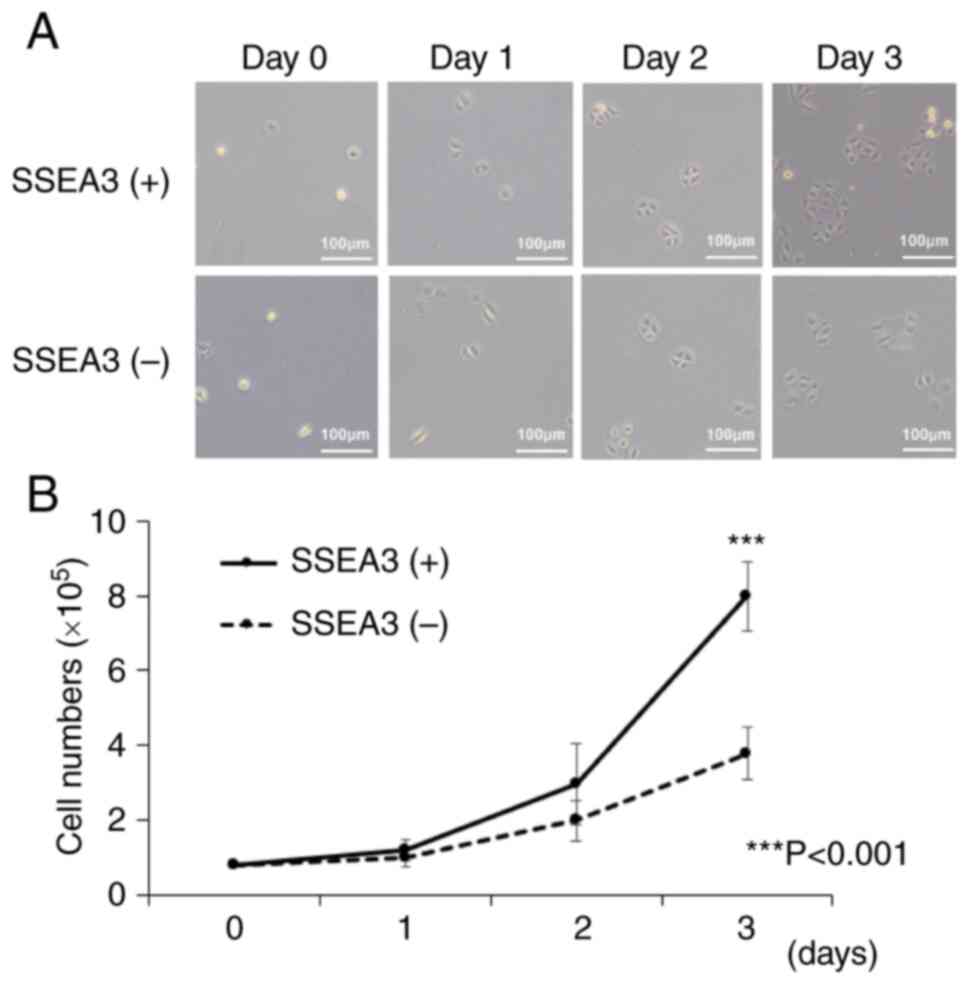
SSEA3 positive (+) SCC4 cells exhibit
elevated proliferation activity and cyclical cell cycle status
To elucidate the basis for the difference in
tumorigenic activity, the cell proliferation capacity of SSEA3(+)
or (−) SCC4 cells was investigated. The cell proliferation assay
indicated that SSEA3(+) SCC4 cells displayed significantly higher
cell proliferation activity compared with SSEA3(−) cells after a
72-h incubation (Fig. 3A and B).
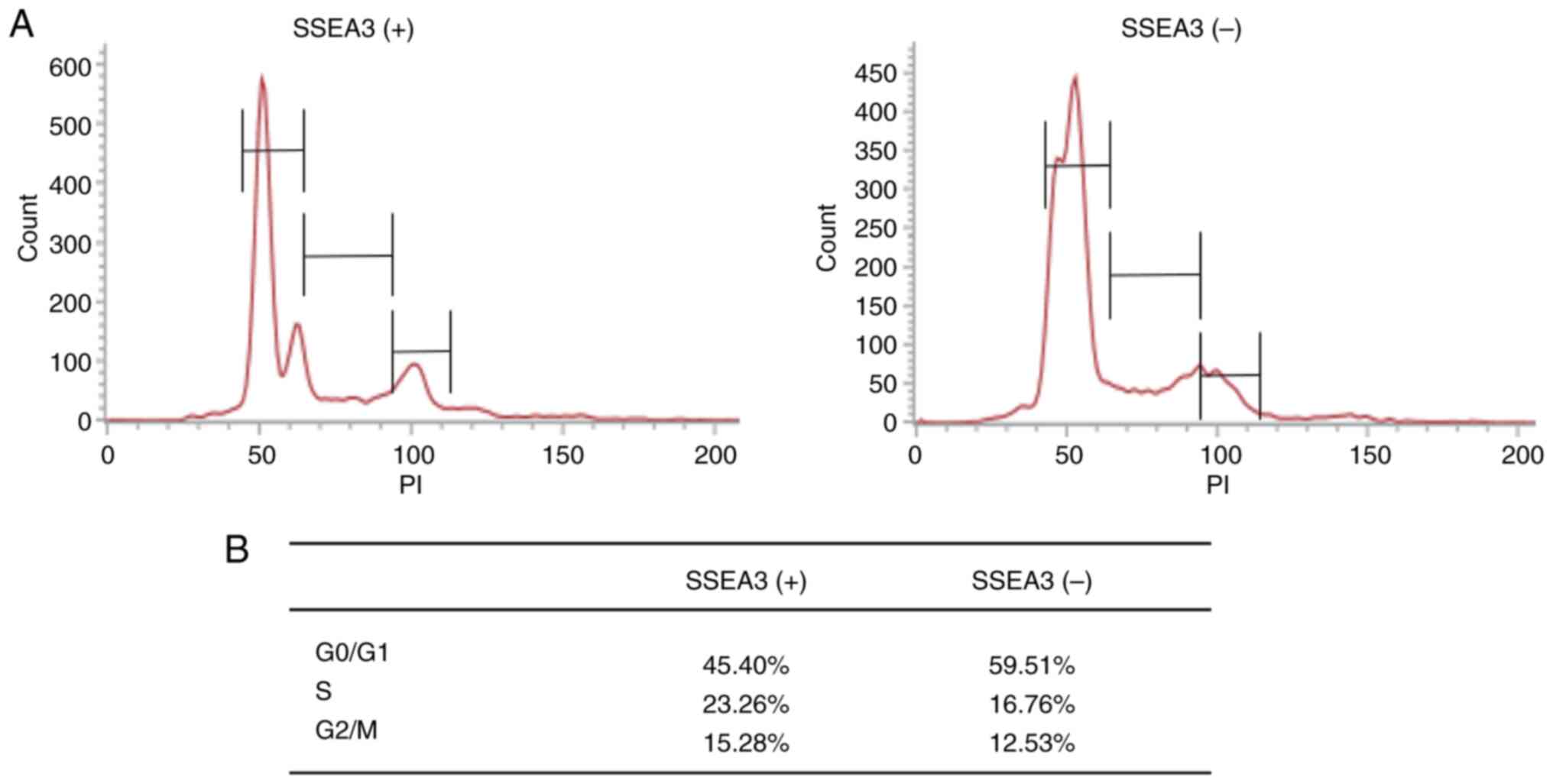
SSEA3(+) cells in ES cells and HCT116 cells demonstrated rapid cell
cycling (27,28). To determine the cause of the high
proliferative capacity of SSEA(+) SCC4 cells, the cell cycle status
was examined. Cell cycle analysis revealed that the proportion of
SSEA3(+) SCC4 cells in the S and G2/M phases was higher than that
of SSEA3 negative (−) cells (Fig. 4A
and B).
SSEA3(+) SCC4 cells express elevated
levels of stem cell-related markers
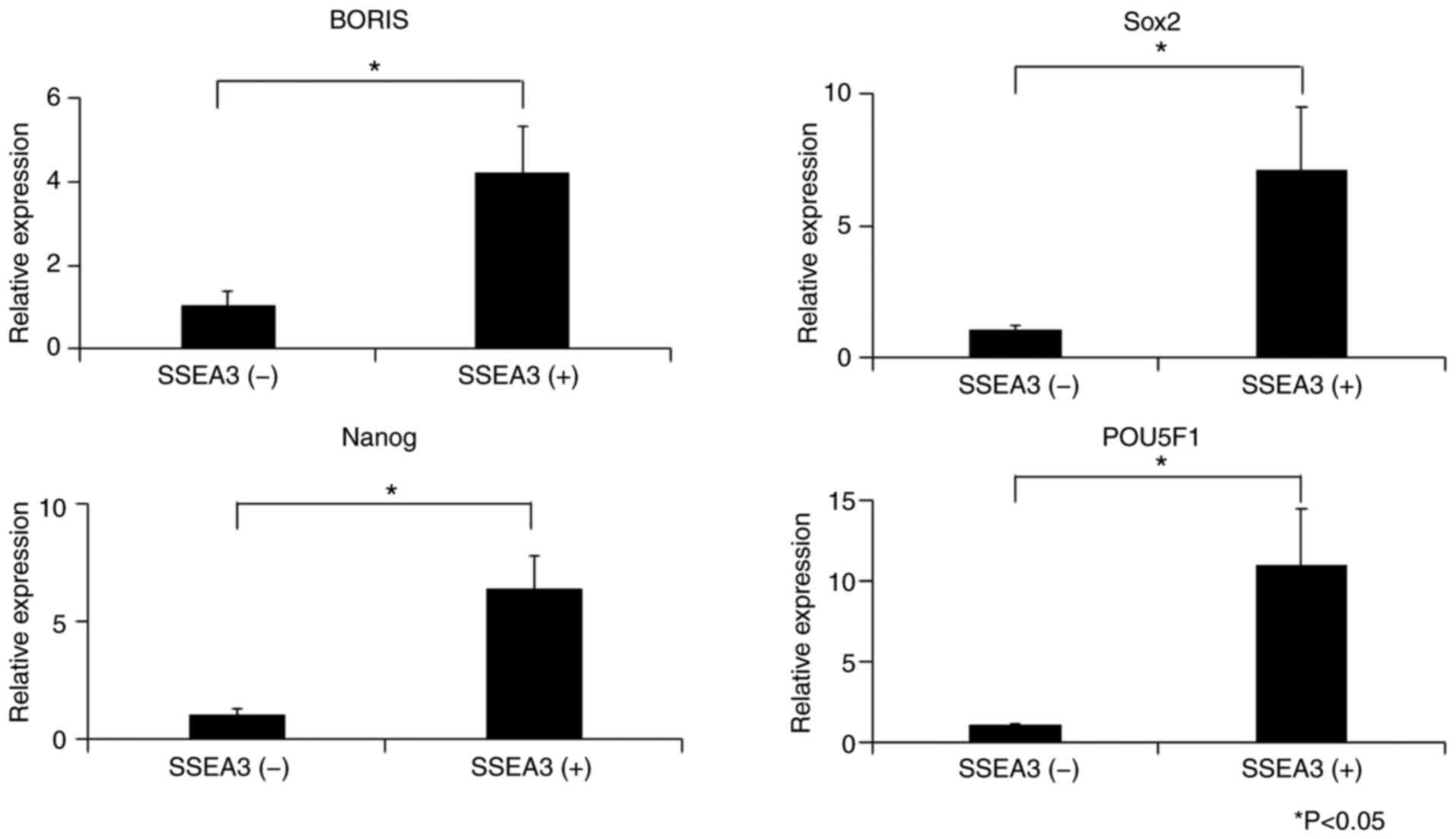
qPCR was performed to validate the stem cell-related
markers in SSEA3(+) or (−) SCC4 cells. The expression levels of
BORIS, SOX2, Nanog and POU5F1 in the SSEA3(+) SCC4 cells were
4.18-, 7.09-, 6.33- and 10.92-fold higher than those in the
SSEA3(−) SCC4 cells, respectively (Fig.
5).
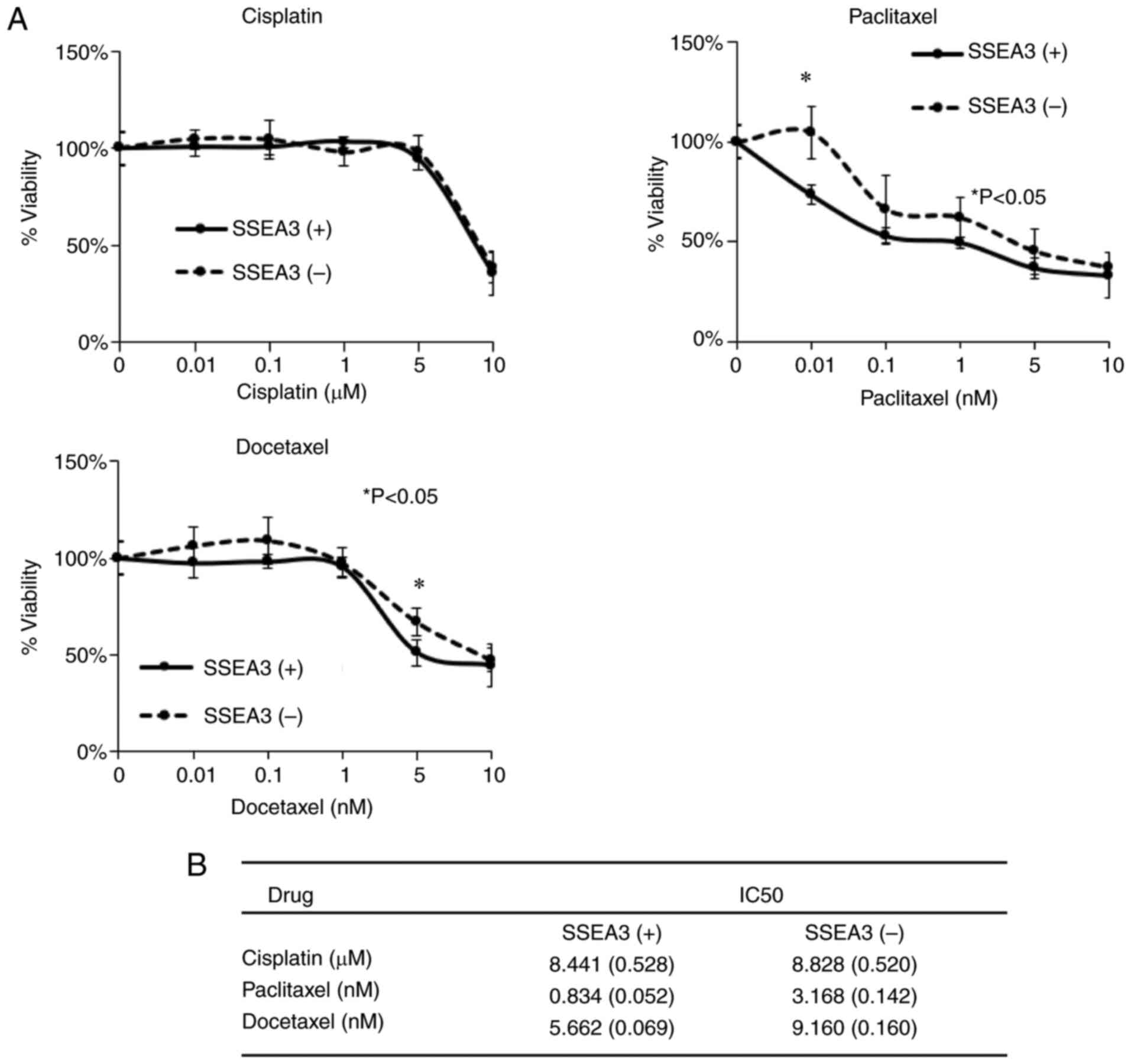
Taxane-based anticancer agents inhibit
the growth of SSEA3(+) SCC4 cells in vitro
The chemo-resistant properties of SSEA3(+) or (−)
SCC4 cells were assessed by determining their sensitivity to
anticancer drugs, including cisplatin, paclitaxel and docetaxel,
utilizing the Cell Counting Kit-8 (Fig.
6A). The IC50 of cisplatin in SSEA3(+) SCC4 cells
was 8.441±0.528, compared with 8.828±0.520 in SSEA3(−) SCC4 cells
(Fig. 6B). SSEA3(+) SCC4 cells
exhibited significantly inhibited cell growth with paclitaxel
(0.834±0.052) and docetaxel (5.662±0.069) compared with SSEA3(−)
SCC4 cells with paclitaxel (3.168±0.142) and docetaxel
(9.160±0.160) (Fig. 6B).
Discussion
The present study investigated the expression and
characteristics of SSEA3 in oral cavity cancer cell lines. SSEA3
has also been explored for its potential role in promoting
tumorigenicity and chemoresistance. The present study addressed
several knowledge gaps in the field of CSC research. The specific
objective of the present study was to determine whether there is an
association between SSEA3 expression and the expression of other
stem cell-associated markers in oral cancer cells. The effect of
these factors on the properties of CSCs was also examined.
Furthermore, the present study examined the response of SSEA3(+)
and SSEA3(−) cells to common anticancer agents, such as cisplatin,
paclitaxel and docetaxel, which are frequently used in oral cancer
treatment. In addition, the investigation of oral cavity cancer
cell lines in the present study revealed the presence and
characteristics of SSEA3. A particular association was observed
between SSEA3 and CD44, as well as other markers associated with
stem cells. Compared with SSEA3(−) cells, SSEA3(+) cells revealed
increased proliferative activity and stem cell-related gene
expression. Additionally, the present study demonstrated a higher
susceptibility to taxane-based medications, suggesting that SSEA3
may be involved in the enhancement of tumorigenicity and
chemoresistance in oral cancer. The hypothesis of the present study
was further supported by the detection of SSEA3 expression in a
small subset of CD44(+) cells in most of the oral cancer cell lines
examined. Moreover, SSEA3(+) cells exhibited increased
proliferative activity and stem cell-related gene expression
compared with SSEA3(−) cells. This is consistent with the role of
SSEA3 as a stem cell marker (28–30).
Previous studies have identified the presence of
SSEA3 in various cancers, including breast and colorectal cancers
(20,21,27,31).
However, to the best of the authors' knowledge, the present study
is the first to investigate the expression of SSEA3 in oral cancer.
The results of the present study revealed that SSEA3(+) cells
exhibit increased proliferation activity and a high level of stem
cell-related gene expression, which is consistent with previous
studies (28–30). Interestingly, SSEA3(+) cells are
more sensitive to taxanes. This novel discovery suggests that
targeting SSEA3 could potentially enhance the effectiveness of
taxane-based chemotherapy in patients with oral cavity cancer
(32–35). The results of the present study
demonstrated several important implications for oral cancer
research. First, SSEA3 is expressed in oral cancer cell lines and
may contribute to promoting tumorigenicity and chemoresistance.
This adds to the existing body of knowledge about the role of SSEA3
in cancer (20,21,27,31).
Second, the findings of the present study underscore the need for
further investigation into the expression of stem cell-related
markers such as SSEA3 and CD44 in oral cancer. These markers could
potentially serve as targets for the development of new anticancer
therapies. The observation that taxanes effectively inhibit the
growth of SSEA3(+) cells could be valuable in devising chemotherapy
regimens for oral cancer.
On the other hand, it is possible that SSEA3
expression may not directly contribute to tumorigenicity and
chemoresistance. Instead, it could be a byproduct of increased cell
proliferation, serving merely as an indicator of a cell subtype
responsible for these characteristics. CSCs are often characterized
by their ability to self-renew and differentiate into various tumor
cell populations. While they typically exhibit cell cycle arrest
and resistance to chemotherapy (36), certain conditions may lead to cancer
cells expressing stem cell markers revealing increased
proliferation and heightened susceptibility to chemotherapy. The
reasons for this increased proliferation and sensitivity to
chemotherapy in cancer cells expressing stem cell markers are
complex and multifactorial. They depend on a variety of biological
and environmental factors. These factors can also influence the
heterogeneity observed within the CSC population. Therefore, it is
crucial to understand the specific context of each cancer type and
patient when developing targeted therapies. In this case, future
research could shed light on several possibilities by examining the
functional role of SSEA3 in oral cancer. Further investigations may
also explore the relationship between SSEA3 and other stem cell and
tumorigenicity markers. In addition, the potential therapeutic
implications of targeting SSEA3 in oral cancer could be
examined.
Nonetheless, the present study demonstrated some
limitations. Firstly, it was conducted in vitro,
necessitating further validation through animal models or clinical
trials to establish the applicability of the findings to humans.
Secondly, it concentrated solely on SSEA3, while other stem
cell-related markers or mechanisms may also participate in
promoting tumorigenicity and chemoresistance in oral cancer.
Thirdly, the present study was conducted on cell lines that may not
entirely capture the intricacy of in vivo tumors and possess
limited representativeness of the actual tumor. Finally, the
mechanisms underpinning the association between SSEA3 and CD44
expression or the specific signaling pathways implicated in
SSEA3-mediated proliferation and chemoresistance have not been
explored and remain enigmatic.
In summary, the present study provided several key
insights into the role of SSEA3 in oral cancer. SSEA3 expression
appeared to be associated with increased cell proliferation
activity, heightened expression of stem cell markers, and altered
drug responsiveness in oral cancer cells. Moreover, the present
study sheds light on the potential role of SSEA3 in modulating CSC
properties in oral cancer. This suggests that SSEA3 could
potentially serve as a therapeutic target in the development of new
cancer treatments.
Acknowledgements
The authors are grateful to the members of the
Department of Oral and Maxillofacial Surgery of Ryukyu University
for their helpful suggestions and assistance.
Funding
The present study was supported by a Grants-in-Aid for
Scientific Research from the Ministry of Education, Culture,
Sports, Science and Technology (grant no. 23K09358).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HN was responsible for the conception and design of
the analyses. KI, TK and JS conducted the experiments. EN and HS
confirm the authenticity of all the raw data. KI, EN and HN wrote
the manuscript. TI, HS, SM, MS and YS provided conceptual advice
for the study. EN and HN edited the manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CSC
|
cancer stem cells
|
|
SSEA3
|
stage-specific embryonic antigen 3
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
References
|
1
|
Jones PA and Baylin SB: The Epigenomics of
Cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Macaluso M, Paggi MG and Giordano A:
Genetic and epigenetic alterations as hallmarks of the intricate
road to cancer. Oncogene. 22:6472–6478. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Madhukar G and Subbarao N: Current and
future therapeutic targets: A review on treating head and neck
squamous cell carcinoma. Curr Cancer Drug Targets. 21:386–400.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gingerich MA, Smith JD, Michmerhuizen NL,
Ludwig M, Devenport S, Matovina C, Brenner C and Chinn SB:
Comprehensive review of genetic factors contributing to head and
neck squamous cell carcinoma development in low-risk,
nontraditional patients. Head Neck. 40:943–954. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hunter KD, Parkinson EK and Harrison PR:
Profiling early head and neck cancer. Nat Rev Cancer. 5:127–135.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Conley BA: Treatment of advanced head and
neck cancer: What lessons have we learned? J Clin Oncol.
24:1023–1025. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dubey P, Gupta R, Mishra A, Kumar V,
Bhadauria S and Bhatt MLB: Evaluation of correlation between CD44,
radiotherapy response, and survival rate in patients with advanced
stage of head and neck squamous cell carcinoma (HNSCC). Cancer Med.
11:1937–1947. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Demurtas S, Ingargiola R, Orlandi E and
Locati LD: How can we address the challenge of distant metastases
in HNSCC prognosis? Expert Rev Anticancer Ther. 22:781–783. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takebe N, Harris PJ, Warren RQ and Ivy SP:
Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog
pathways. Nat Rev Clin Oncol. 8:97–106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sayed SI, Dwivedi RC, Katna R, Garg A,
Pathak KA, Nutting CM, Rhys-Evans P, Harrington KJ and Kazi R:
Implications of understanding cancer stem cell (CSC) biology in
head and neck squamous cell cancer. Oral Oncol. 47:237–243. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Trapasso S and Allegra E: Role of CD44 as
a marker of cancer stem cells in head and neck cancer. Biologics.
6:379–383. 2012.PubMed/NCBI
|
|
14
|
Joshua B, Kaplan MJ, Doweck I, Pai R,
Weissman IL, Prince ME and Ailles LE: Frequency of cells expressing
CD44, a head and neck cancer stem cell marker: Correlation with
tumor aggressiveness. Head Neck. 34:42–49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chikamatsu K, Takahashi G, Sakakura K,
Ferrone S and Masuyama K: Immunoregulatory properties of CD44+
cancer stem-like cells in squamous cell carcinoma of the head and
neck. Head Neck. 33:208–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kokko LL, Hurme S, Maula SM, Alanen K,
Grénman R, Kinnunen I and Ventelä S: Significance of site-specific
prognosis of cancer stem cell marker CD44 in head and neck
squamous-cell carcinoma. Oral Oncol. 47:510–516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Spring FA, Dalchau R, Daniels GL,
Mallinson G, Judson PA, Parsons SF, Fabre JW and Anstee DJ: The Ina
and Inb blood group antigens are located on a glycoprotein of
80,000 MW (the CDw44 glycoprotein) whose expression is influenced
by the In (Lu) gene. Immunology. 64:37–43. 1988.PubMed/NCBI
|
|
18
|
Nakamura H, Suenaga N, Taniwaki K, Matsuki
H, Yonezawa K, Fujii M, Okada Y and Seiki M: Constitutive and
induced CD44 shedding by ADAM-like proteases and membrane-type 1
matrix metalloproteinase. Cancer Res. 64:876–882. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Misra S, Heldin P, Hascall VC, Karamanos
NK, Skandalis SS, Markwald RR and Ghatak S: Hyaluronan-CD44
interactions as potential targets for cancer therapy. FEBS J.
278:1429–1443. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kannagi R, Cochran NA, Ishigami F,
Hakomori S, Andrews PW, Knowles BB and Solter D: Stage-specific
embryonic antigens (SSEA-3 and −4) are epitopes of a unique
globo-series ganglioside isolated from human teratocarcinoma cells.
EMBO J. 2:2355–2361. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pornsuriyasak P and Demchenko AV:
Synthesis of cancer-associated glycoantigens: Stage-specific
embryonic antigen 3 (SSEA-3). Carbohydr Res. 341:1458–1466. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuroda Y, Kitada M, Wakao S, Nishikawa K,
Tanimura Y, Makinoshima H, Goda M, Akashi H, Inutsuka A, Niwa A, et
al: Unique multipotent cells in adult human mesenchymal cell
populations. Proc Natl Acad Sci USA. 107:8639–8643. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kobayashi Y, Kitahara H, Hirai M, Tanaka
A, Jokaji R, Kobayashi K, Bou-Gharios G, Nakamura H and Kawashiri
S: Selectively high efficacy of eribulin against high-grade
invasive recurrent and/or metastatic squamous cell carcinoma of the
head and neck. Oncol Lett. 17:5064–5072. 2019.PubMed/NCBI
|
|
24
|
Kobayashi JI, Hirohashi Y, Torigoe T,
Michifuri Y, Yamamoto T, Tamura Y, Kamiguchi K, Miyazaki A,
Yamaguchi A, Hariu H, et al: Clonal diversity of cytotoxic T
lymphocytes that recognize autologous oral squamous cell carcinoma.
Hum Immunol. 70:89–95. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suzuki Y, Haraguchi N, Takahashi H, Uemura
M, Nishimura J, Hata T, Takemasa I, Mizushima T, Ishii H, Doki Y,
et al: SSEA-3 as a novel amplifying cancer cell surface marker in
colorectal cancers. Int J Oncol. 42:161–167. 2013.PubMed/NCBI
|
|
28
|
Yang Z, Liu J, Liu H, Qiu M, Liu Q, Zheng
L, Pang M, Quan F and Zhang Y: Isolation and characterization of
SSEA3(+) stem cells derived from goat skin fibroblasts. Cell
Reprogram. 15:195–205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suila H, Pitkänen V, Hirvonen T, Heiskanen
A, Anderson H, Laitinen A, Natunen S, Miller-Podraza H, Satomaa T,
Natunen J, et al: Are globoseries glycosphingolipids SSEA-3 and −4
markers for stem cells derived from human umbilical cord blood? J
Mol Cell Biol. 3:99–107. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leng Z, Sun D, Huang Z, Tadmori I, Chiang
N, Kethidi N, Sabra A, Kushida Y, Fu YS, Dezawa M, et al:
Quantitative analysis of SSEA3+ cells from human umbilical cord
after magnetic sorting. Cell Transplant. 28:907–923. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang WW, Lee CH, Lee P, Lin J, Hsu CW,
Hung JT, Lin JJ, Yu JC, Shao LE, Yu J, et al: Expression of Globo H
and SSEA3 in breast cancer stem cells and the involvement of
fucosyl transferases 1 and 2 in Globo H synthesis. Proc Natl Acad
Sci USA. 105:11667–11672. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abal M, Andreu J and Barasoain I: Taxanes:
Microtubule and centrosome targets, and cell cycle dependent
mechanisms of action. Curr Cancer Drug Targets. 3:193–203. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hennenfent KL and Govindan R: Novel
formulations of taxanes: A review. Old wine in a new bottle? Ann
Oncol. 17:735–749. 2006.PubMed/NCBI
|
|
34
|
Schrijvers D and Vermorken JB: Taxanes in
the treatment of head and neck cancer. Curr Opin Oncol. 17:218–224.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rowinsky EK: The development and clinical
utility of the taxane class of antimicrotubule chemotherapy agents.
Annu Rev Med. 48:353–374. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ishii H, Iwatsuki M, Ieta K, Ohta D,
Haraguchi N, Mimori K and Mori M: Cancer stem cells and
chemoradiation resistance. Cancer Sci. 99:1871–1877. 2008.
View Article : Google Scholar : PubMed/NCBI
|