Introduction
Cervical cancer is one of the most common
malignancies of the female reproductive system and seriously
threatens the life and health of women. Approximately 570,000 new
cases of cervical cancer are diagnosed annually, and ~311,000
patients succumb to the disease worldwide (1). Several investigations have indicated
that the incidence of cervical cancer is closely related to human
papillomavirus (HPV) infection (2,3). Owing
to the continuous improvements in cervical cancer detection and
diagnosis technologies and standardized HPV screening, the
incidence of this disease has decreased in countries that have HPV
screening and vaccination programs. The major issue with cervical
cancer is the lack of such programs in low-income countries. Women
in these countries require new therapies, which again are likely to
be too expensive, but hopefully, a target can be identified.
Therefore, developing novel diagnostic and therapeutic biomarkers
to enhance the overall survival of patients with cervical cancer is
the need of the hour.
Follistatin-like protein 1 (FSTL1), a type of
secretory glycoprotein, is expressed in most mammalian cells, with
the exception of peripheral lymphocytes. FSTL1 is localized on
chromosome 3q13.33 and comprises 308 amino acids. The protein
belongs to the secreted protein acidic and rich in cysteine (SPARC)
family (4). However, the functions
and biological characteristics of FSTL1 are different from those of
other members of the SPARC family (5). In recent years, several studies have
confirmed that FSTL1 functions as a crucial regulator in various
biological processes and that it has a key effect on the occurrence
and development of cancer. Epithelial ovarian cancer (6), non-small cell lung cancer (7), clear cell renal cell carcinoma
(8), nasopharyngeal carcinoma
(9), esophageal squamous cell
carcinoma (10), gastric cancer
(11), prostate cancer (12), and hepatocellular carcinoma are a
few examples (13). FSTL1 plays a
role in several cancer-related signaling pathways. For example, the
protein can retard the progression of clear cell renal cell
carcinoma by inhibiting the NF-kB and HIF-2α signaling pathways
(8). In hepatocellular carcinoma
(13), FSTL1 promotes tumor growth
by activating AKT via regulation of TLR4/CD14. With respect to
cervical cancer, only one study has reported that FSTL1 could
significantly inhibit the proliferation and invasion of cervical
cancer cells through negative regulation of the BMP4/Smad1/5/9
signaling pathway (14). A previous
study by the authors confirmed that FSTL1 is a direct target of
miR-9, which is involved in the migration of cervical cancer cells
(15). Nonetheless, clarity on the
clinical significance and mechanism of FSTL1 is lacking in cervical
cancer.
Hence, in the present study, the expression,
function, and molecular mechanisms of FSTL1 in cervical cancer were
investigated. The expression of FSTL1 in cervical tissues was
examined. Subsequently, the effects of abnormal expression of FSTL1
on cervical cancer cells were assessed using in vivo animal
experiments and functional in vitro experiments. Finally,
the molecular mechanisms associated with FSTL1 were examined in
HeLa and SiHa cells. The results indicated the downregulation of
FSTL1 in cervical cancer tissues. FSTL1 inhibited the
proliferation, migration, and invasion of cervical cancer cells and
promoted their apoptosis. This protein is likely to be involved in
the regulation of the IGF-1R/PI3K/AKT/BCL-2 signaling pathway in
cervical cancer.
Materials and methods
Clinical samples and cell lines
A total of 117 cervical cancer tissues were
collected from the People's Hospital of Guangxi Zhuang Autonomous
Region (Nanning, China) between September 2012 and June 2017. The
non-tumor tissues were composed of 29 tumor-adjacent tissues and 43
normal cervical tissues, including uterine prolapse (n=4),
endometriosis (n=19), and hysteromyoma (n=20), which were collected
during the same period. None of the patients received any treatment
before the operation, as confirmed by pathologists after the
operation. The clinical data of all patients with cervical cancer
are shown in Table I. HeLa and SiHa
cells were obtained from the Shanghai Cell Bank, Chinese Academy of
Sciences. HeLa and SiHa cells were maintained in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
cow serum (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
1% penicillin and streptomycin at 37°C in a humidified atmosphere
containing 5% CO2. The present study complied with the
guidelines outlined in the Declaration of Helsinki and was approved
(approval no. KY-LW-2017-5) by the Ethics Committee of the People's
Hospital of Guangxi Zhuang Autonomous Region (Nanning, China).
Written informed consent was obtained from all patients.
 | Table I.Associations between FSTL1 expression
and clinicopathological characteristics in cervical cancer
patients. |
Table I.
Associations between FSTL1 expression
and clinicopathological characteristics in cervical cancer
patients.
|
| Immunoreactivity
score of FSTL1 protein expression | FSTL1 mRNA
expression |
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | Total cases
(n=84) | Number of low cases
(n=45) | Number of high
cases (n=39) | P-value | Total cases
(n=77) | Number of low cases
(n=38) | Number of high
cases (n=39) | P-value |
|---|
| Age |
|
|
| 0.899 |
|
|
| 0.424 |
|
0.<50 | 36 | 19 | 17 |
| 39 | 21 | 18 |
|
|
0.≥50 | 48 | 26 | 22 |
| 38 | 17 | 21 |
|
| Pathologic
types |
|
|
| 0.142 |
|
|
| 0.05 |
|
0.Squamous carcinomas | 70 | 35 | 35 |
| 60 | 33 | 27 |
|
|
0.Adenocarcinoma | 14 | 10 | 4 |
| 15 | 4 | 11 |
|
|
0.Other | - | - | - |
| 2 | 1 | 1 |
|
| Degree of
differentiation |
|
|
|
|
|
|
|
|
|
0.High | 17 | 4 | 13 |
| 19 | 9 | 10 |
|
|
0.Middle | 49 | 26 | 23 |
| 32 | 17 | 15 |
|
|
0.Low | 18 | 15 | 3 |
| 26 | 12 | 14 |
|
| Maximum diameter
(cm) |
|
|
| 1 |
|
|
| 0.104 |
|
0.>4 | 10 | 5 | 5 |
| 20 | 13 | 7 |
|
|
0.≤4 | 74 | 40 | 34 |
| 57 | 25 | 32 |
|
| FIGO stage |
|
|
| 0.517 |
|
|
| 0.645 |
|
0.IA | - | - | - |
| 4 | 3 | 1 |
|
|
0.IB | 61 | 34 | 27 |
| 44 | 20 | 24 |
|
|
0.II | 23 | 11 | 12 |
| 20 | 11 | 9 |
|
|
0.≥III | - | - | - |
| 9 | 4 | 5 |
|
| Infiltration
deptha |
|
|
| 0.021 |
|
|
| 0.037 |
|
0.>50 | 52 | 33 | 19 |
| 44 | 26 | 18 |
|
|
0.≤50 | 32 | 12 | 20 |
| 33 | 12 | 22 |
|
| lymphatic
metastasis |
|
|
| 0.262 |
|
|
| 0.842 |
|
0.Negative | 69 | 35 | 34 |
| 58 | 29 | 29 |
|
|
0.Positive | 15 | 10 | 5 |
| 19 | 9 | 10 |
|
| Vascular
invasion |
|
|
| 0.851 |
|
|
| 0.707 |
|
0.Negative | 40 | 21 | 19 |
| 47 | 24 | 23 |
|
|
0.Positive | 44 | 24 | 20 |
| 30 | 14 | 16 |
|
| Nerve
infiltration |
|
|
| 1 |
|
|
| 0.665 |
|
0.Negative | 77 | 41 | 36 |
| 70 | 34 | 36 |
|
|
0.Positive | 7 | 4 | 3 |
| 7 | 4 | 3 |
|
| Uterus and its
attachments |
|
|
| 0.491 |
|
|
| 0.564 |
|
0.Negative | 44 | 22 | 22 |
| 55 | 26 | 29 |
|
|
0.Positive | 40 | 23 | 17 |
| 22 | 12 | 10 |
|
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted by using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and 500 ng of
purified RNA was reverse transcribed into cDNA by using the Takara
Reverse Transcription kit, following the manufacturer's
instructions. RT-qPCR was performed by using the SYBR Green PCR
Master Mix Kit on the ABI 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Briefly, after an
initial denaturation step at 95°C for 15 min, the amplifications
were conducted with 40 cycles at a melting temperature of 95°C for
10 sec, followed by an annealing temperature of 60°C for 23 sec.
GAPDH was selected as the housekeeping gene. The relative
expression of FSTL1 was calculated by using the standard
2−ΔΔCq method (16). The
special primer sequences used were as follows: FSTL1 forward,
5′-GAGGGCAAGAGTACACCA-3′ and reverse, 5′-TACGGCATAGACGACAGC-3′;
GAPDH forward, 5′-CATGAGAAGTATGACAACAGCCT-3′ and reverse,
5′-AGTCCTTCCACGATACCAAAG-3′. The FSTL1 mRNA expression was divided
into high-expression and low-expression groups based on the median
expression of FSTL1 mRNA.
Immunohistochemistry (IHC)
The cervical tissues were fixed in 4%
polyoxymethylene for 24 h at room temperature, embedded in paraffin
and made into 3.5-µm thick sections. All cervical tissue sections
were dewaxed and hydrated by an automatic dewaxing machine (ST5020;
Leica Microsystems GmbH). The slides were treated under high
pressure for antigen retrieval and 3% hydrogen peroxide to remove
endogenous peroxidase. Subsequently, the sections were incubated
with specific primary antibodies against FSTL1 (1:250; cat. no.
ab71548; Abcam) at 4°C overnight. Known positive tissues (prostate
cancer) were assigned to the positive-control group and
phosphate-buffered saline (PBS) was used instead of the primary
antibody in the negative-control group. The slides were then
incubated with goat-anti-mouse/rabbit secondary antibodies (cat.
no. PV-9000; OriGene Technologies, Inc.) at 37°C for 20 min,
followed by staining by using the DAB detection kit (cat. no.
ZLI9018; OriGene Technologies, Inc.). The slides were sequentially
stained with hematoxylin, followed by differentiation in 0.5%
hydrochloric acid alcohol, 1% ammonia water anti-blue, gradient
alcohol dehydration, and neutral gum sealing. Finally, the slides
were examined using a light microscope (BX51; Olympus Corporation).
The proportion of stained cells was scored as follows: 0–25%
(1), 25–50% (2), 50–75% (3), and 75–100% (4). The intensity of the stained cells was
scored as follows: negative (0), weak (1), moderate (2) and strong (3). The final immunoreactivity score (IRS)
was counted by summing the proportion and intensity scores
(17). The IRS was finally
confirmed by two independent pathologists from the People's
Hospital of Guangxi Zhuang Autonomous Region. The FSTL1 expression
at the protein level was classified as high expression or low
expression group based on the median of IRS.
Hematoxylin-eosin (H&E)
staining
All cervical tissue slides were dewaxed and hydrated
in an automatic dewaxing machine (ST5020; Leica Microsystems GmbH),
followed by rinsing in distilled water for 5 min and staining with
hematoxylin. Next, the slides were immersed for differentiation in
0.5% hydrochloric acid alcohol and 1% ammonia water anti-blue for a
few seconds. Subsequently, the slides were washed with distilled
water for 5 min and then stained with 0.5% eosin for 2 min at room
temperature. Finally, the slides were subjected to dehydration in a
gradient series of alcohol and then sealed with neutral gum,
followed by observation using a light microscope (BX51; Olympus
Corporation).
Construction, infection, and screening
of lentiviral overexpression (OE) of FSTL1
Lentiviral vector harboring FSTL1 was constructed
and packaged by Shanghai GeneChem Co., Ltd. Briefly, cervical
cancer cells (3–6×104 cells/well) were seeded into
six-well plates and cultured at 37°C for 24 h under a humidified
atmosphere of 5% CO2. On the next day, the cells were
infected with lentivirus-FSTL1-puro or lentivirus-negative control
(NC)-puro [multiplicity of infection (MOI) was as follows: HeLa,
2×107 TU/ml; SiHa: 1×107 TU/ml] using
polybrene or enhanced infection solution (Shanghai GeneChem Co.,
Ltd.). The culture medium was replaced after 12 h. Subsequently,
puromycin (1.0 µg/ml) was added to the culture medium at 72 h after
infection. The stable expression cell lines of FSTL1 were selected
with puromycin for 2 weeks. The FSTL1 expression at the mRNA and
protein levels in cervical cancer cells was detected by RT-qPCR and
western blotting, respectively.
Knockout (KO) of FSTL1 by using
CRISPR/Cas9 technology
A total of three small-guide RNAs (sgRNAs) targeting
FSTL1 were designed and used to construct CRISPR/CRISPR-associated
(Cas)9-FSTL1 lentiviral particles. The three sgRNA-FSTL1 sequences
were as follows: sgRNA-1(KO1): AGTGTCCATCGTAATCAACC; sgRNA-2(KO2):
ACTTACCTCAATGCAGAGAC; and sgRNA3(KO3): GTAAGTCCATCTGCCAGCCC.
Screening of biologically active sgRNA was performed by the
Cruiser™ method. Cas9 and sgRNA lentiviral particles
were constructed and packaged by Shanghai GeneChem Co., Ltd.
Briefly, the cells (3–6×104 cells/well) were seeded into
six-well plates and cultured at 37°C for 24 h in a humidified
atmosphere of 5% CO2. On the next day, the cells were
infected with lenti-cas9-puro lentiviral particles using polybrene
or enhanced infection solution (Shanghai GeneChem Co., Ltd.). The
culture medium was replaced after 12 h. Then, puromycin (1.2 µg/ml)
was added to the culture medium at 72 h after infection. The stable
expression cell lines of FSTL1 were selected with puromycin for 2
weeks. Subsequently, cervical cancer cells were infected with
lenti-cas9-puro lentiviral particles, the latter being infected
with lentivirus-sgRNA-FSTL1 or lentivirus-sgRNA-NC, respectively.
The expression of FSTL1 at the protein level was detected by
western blotting.
Screening for active sgRNA
Genomic DNA was extracted by using a nucleic acid
extraction kit (cat. no. D3121; Magen Biotechnology Co., Ltd.).
Active sgRNA was screened using the KO and mutation detection kit.
The PCR reaction was conducted in accordance with the following
system to obtain a hybrid DNA product. Briefly, after an initial
denaturation step at 95°C for 2 min, amplifications were conducted
with 35 cycles at a melting temperature of 95°C for 20 sec and 55°C
for 20 sec, followed by an annealing temperature of 72°C for 5 min.
Then, 3 µl of hybrid DNA product, 2 µl of detecase buffer, 1 µl of
detecase and 4 µl of water were mixed and allowed to react for 20
min at 45°C, to which 2 µl of stop buffer was finally added. The
final PCR product was detected by 2% agarose gel electrophoresis.
The gel was stained with 0.5 µg/ml ethidium bromide solution for 20
min at room temperature and decolorized with deionized water for 10
min. The gel imaging analysis system (Tanon-2500; Shanghai Tianneng
Technology Co., Ltd.) was used to observe and analyze the results.
The special primer sequences used for this reaction were as
follows: sgRNA-1(KO1) forward, 5′-TATCCATAACTGCACAAACATTCTC′ and
reverse, 5′-GGCAAACCCAGCAGGCTCATAAG-3′; sgRNA-2(KO2) forward,
5′-GTATGTTTTAGGAAGAGCTAAGGAG-3′ and reverse,
5′-CCATGCTTTAAAAATCAGGAATCTG-3′; sgRNA-3(KO3) forward,
5′-ACATGTAGAGCAGCTGTAATCCTAG-3′ and reverse,
5′-TCTGACCCAACCAGCTGCTTCAATT-3′.
Colony formation assay and
3-(4,5-dimethylthiazol-2-y1)-2,5-diphenyltetrazolium bromide (MTT)
assay
For the cell colony formation assay, HeLa or SiHa
cells (1×103 cells/well) were seeded into a six-well
plate and cultured for 6–10 days. The cells were then washed twice
with PBS and fixed with methanol for 30 min. Subsequently, the
cells were stained with 1% crystal violet for 20 min at room
temperature (Red Rock Reagent Factory). Finally, the fixed cells
were washed twice with PBS for 10 min and enumerated manually under
an inverted microscope (Olympus Corporation) when the colony
contained ≥50 cells. For the MTT assay, HeLa or SiHa cells
(3×103 cells/well) were seeded and cultured in a 96-well
plate for 24, 48, 72, 96 and 120 h. Next, 20 µl of PBS containing
0.5% MTT (5 mg/ml, MilliporeSigma) was added to each well and
incubated at 37°C for 4 h. Then, the culture medium was discarded
and 150 µl of dimethyl sulfoxide was added to dissolve the formazan
crystals. Next, 96-well plates containing cells were vibrated at
37°C for 10 min. Finally, the absorbance of each well was
determined at 490 nm wavelength by using a microplate reader
(Biotek Synergy H1; Agilent Technologies, Inc.).
Wound-healing assay and Transwell
assay
For the wound-healing assay, the experimental cells
(3×105 cells/well) were seeded into a culture insert
(cat. no. 80209; Ibidi GmbH) and cultured at 37°C for 24 h in a
humidified atmosphere of 5% CO2. After 24 h, a wound was
created by removing the culture insert. The cells were carefully
washed twice in cold PBS and incubated with RPMI-1640 medium
without fetal bovine serum at 37°C for 24 h. The Transwell assay
was conducted in a 24-well Boyden chamber (cat. no. 3422; Corning,
Inc.). Briefly, 3×104 cells were suspended in a
serum-free medium and seeded into the upper chamber. Subsequently,
600 µl of 15% fetal bovine serum was added to the lower chamber.
After the cells were cultured at 37°C for 12 h, the invasive cells
were fixed with methanol for 30 min and then stained with 1%
crystal violet for 20 min. Finally, the invasive cells were counted
under a light microscope. For the invasion assay, Matrigel®
Basement Membrane Matrix (BD Biosciences) and serum-free medium in
the ratio of 1 to 3 were pre-coated to the membrane of upper
chambers at 4°C and then placed at 37°C for 4–5 h to solidify.
After the cells were cultured at 37°C for 24 h, the invasive cells
were fixed with methanol for 30 min and stained with 1% crystal
violet for 20 min. Other measurements were the same as for the
migration assay.
Flow cytometry
Briefly, 1×105 cells were collected and
the apoptotic rate was detected by using the APC Annexin V/PI
Apoptosis Detection kit (cat. no. KGA1022; Nanjing KeyGen Biotech
Co., Ltd.) following the manufacturer's instructions. The cells
were sequentially incubated with Annexin V-APC for 15 min and then
with PI for 10 min in the dark. Subsequently, the cells were
detected by FACS CantoII (BD Biosciences) and analyzed by Flowjo
V10 (FlowJo LLC).
Mouse xenograft models
A total of 72 female BALB/C nude mice (age 4–6 weeks
old) were purchased from the Animal Experiment Center of Guangxi
Medical University and housed in the specific-pathogen-free (SPF)
animal facility under controlled temperature and humidity with
alternating 12/12-h light and dark cycles. SPF food and sterile
water were provided ad libitum. The body weight of the mice
ranged from 13.52–18.64 g. The nude mice were randomly assigned to
the experimental, negative-control group, and mock groups of 6 nude
mice each. Briefly, 2×106 [1×107/l (0.2 ml)]
cells were subcutaneously injected into the corresponding axillary
region of nude mice. The size of the tumor was measured using
vernier calipers every 3 days and then calculated using the
following formula: (volume=0.5× long diameter × short
diameter2). All nude mice were sacrificed by cervical
dislocation at 27 days. The animal experiments were approved
(approval no. 2201805166) by The Animal Care & Welfare
Committee of Guangxi Medical University (Nanning, China).
Gene chip analysis
Gene chip analysis was performed in SiHa cell lines
overexpressing FSTL1 when compared with negative controls. The
assay and data analyses were performed at the Shanghai GeneChem
Co., Ltd. in accordance with the protocols of the Affymetrix Gene
Chip system. Total RNA was extracted from the cells using TRIzol
reagent (Pufei Biotechnology Co., Ltd.). First, a mixture of poly-a
RNA and total RNA was synthesized into first-strand cDNA. Second,
first-strand cDNA was further synthesized to second-strand cDNA, as
the procedure of gene chip as aforementioned by Shanghai GeneChem
Co., Ltd. Then, the second-strand cDNA and IVT were applied to
synthesize biotin-labeled aRNA. Finally, the data were obtained
through purification, hybridization, dyeing and scanning. The
results of the gene chip were analyzed and integrated with the
Ingenuity Pathway Analysis (IPA) software 23.0 (Qiagen).
Protein extraction and western
blotting
The proteins were extracted using the RIPA buffer
(cat. no. P0013C; Beyotime Institute of Biotechnology) containing
protease inhibitors (P0100; Beijing Solarbio Science &
Technology Co., Ltd.). The protein concentration was determined by
using the BCA protein assay kit (cat. no. P001S; Beijing Solarbio
Science & Technology Co., Ltd.). Equal amounts of proteins (25
µg) were subjected to 12% sodium dodecyl-sulfate polyacrylamide gel
electrophoresis (SDS-PAGE) and then transferred onto
PVDF membranes (Beijing Solarbio Science & Technology Co.,
Ltd.). The membranes were incubated with 5% non-fat milk at room
temperature for 1 h. Subsequently, the membranes were incubated
with specific primary antibodies against FSTL1 (1:500; cat. no.
ab71548; Abcam), IGF-1R (1:500; cat. no. 3027; Cell Signaling
Technology, Inc.), PI3K (1:250; cat. no. 4255; Cell Signaling
Technology, Inc.), AKT (1:250; cat. no. 9272; Cell Signaling
Technology, Inc.), phosphorylated (P-)AKT (1:250; cat. no. 4060;
Cell Signaling Technology, Inc.), BCL-2 (1:500; cat. no. ab32503;
Abcam), BAX (1:500; cat. no. ab32124; Abcam) and GAPDH (1:1,000;
cat. no. 5174; Cell Signaling Technology, Inc.) at 4°C overnight,
followed by incubation with horseradish peroxidase (HRP)-conjugated
secondary antibody (1:15,000; cat. no. 7074; Cell Signaling
Technology, Inc.) at 37°C for 2 h. The immunoreactive bands were
visualized using the ECL kit (cat. no. P0018; Beijing Solarbio
Science & Technology Co., Ltd.) and analyzed using ImageJ
software (version 2.1.4.7; National Institutes of Health). The
relative expression of the target proteins was normalized to
GAPDH.
Statistical analysis
All the data were analyzed using SPSS Statistics
V22.0 software (IBM Corp.). The measurement data was tested for
normal distribution using the Shapiro-Wilk method. If the data
showed a normal distribution, it was expressed as the mean ±
standard deviation (SD). The differences between the two groups and
among the three groups were analyzed using two-independent sample
Student's t-test and one-way analysis of variance (ANOVA),
respectively. The least significant difference (LSD) method was
applied by following ANOVA. If the data did not conform to a normal
distribution, it was expressed as a median (interquartile range).
The non-parametric test method was applied for the difference
between the groups. Count data was described in relative numbers.
The expression of FSTL1 at the mRNA and protein levels was analyzed
using the Chi-square test. The mock group was equivalent to a blank
control, and nothing was added to the blank group. P≤0.05 was
considered to indicate a statistically significant difference
(two-tailed).
Results
FSTL1 expression is downregulated at
the mRNA and protein levels in cervical cancer tissues
FSTL1 expression at the mRNA and protein levels was
detected in cervical cancer tissues using RT-qPCR and IHC,
respectively. The findings indicated that the expression of FSTL1
at the mRNA and protein levels was downregulated in cervical cancer
tissues compared with tumor-adjacent or normal cervical tissues
(P<0.001, Fig. 1A and D).
According to the observations, the expression of FSTL1 mRNA was
gradually increased in cervical cancer tissues, tumor-adjacent
tissues, and normal cervical tissues (P<0.001, Fig. 1A). In addition, the expression of
FSTL1 mRNA in the matched cancer tissues was downregulated compared
with the tumor-adjacent tissues (P<0.001, Fig. 1B), and that in the non-matching
cancer tissues was downregulated compared with the normal cervical
tissues (P<0.001, Fig. 1C).
FSTL1 protein expression was lower in the cervical cancer tissues
compared with that in the normal cervical tissues (P=0.006).
Moreover, the expression was predominantly localized in the
nucleus, but some expression was also detected in the cytoplasm
(Fig. 1D).
FSTL1 expression is associated with
the clinical features of patients with cervical cancer
To determine the relationship between FSTL1
expression and clinical features in patients with cervical cancer,
the cervical cancer tissues were categorized into high- and
low-expression groups based on the median expression of FSTL1 mRNA.
The low expression of FSTL1 mRNA was observed to be associated with
the infiltration depth (P=0.037) and pathologic type (P=0.05)
(Table I). Subsequently, the
cervical cancer tissues were classified into high- and
low-expression groups based on the median IRS of FSTL1 protein
expression. The findings demonstrated that the low expression of
FSTL1 at the protein level was linked to the infiltration depth
(P=0.021) and degree of differentiation (P=0.002). On the contrary,
the expression of FSTL1 at the mRNA and protein levels was not
related to age, FIGO stage, or lymph node metastasis (P>0.05)
(Table I).
Screening and verification of stable
strains of cervical cancer cells with FSTL1 OE or KO
HeLa and SiHa cells were infected with
lentivirus-FSTL1 or lentivirus-NC. Stable strains of cervical
cancer cells with FSTL1 OE or KO were selected with puromycin for 2
weeks. The fluorescence efficiency of stable strains of cervical
cancer cells was observed under a fluorescence microscope, which
revealed that the lentiviral infection efficiency of cells with OE
or KO of FSTL1 was >80% (Fig. 2A and
E). RT-qPCR assay suggested that FSTL1 mRNA expression was
upregulated in the OE group compared with the NC and mock (MOCK)
groups (Fig. 2B). Western blotting
signified that the expression of the FSTL1 protein was upregulated
in the OE group compared with the NC and MOCK groups (Fig. 2C and D). Furthermore, RT-qPCR assay
demonstrated that the expression of FSTL1 mRNA was downregulated in
the KO group compared with the NC and MOCK groups (Fig. 2F). Western blotting assay signified
that the expression of the FSTL1 protein was downregulated in the
KO group compared with the NC and MOCK groups (Fig. 2I and J). Cruiser TM assay showed
that the KO1 and KO3 targets were active, the KO2 target was
inactive, and the KO3 target activity was comparatively improved
(Fig. 2G). The sequencing of the
plasmids containing the three sgRNAs is presented in Fig. 2H. These results indicated that
cervical cancer cells with FSTL1 KO or OE were successfully
created.
OE of FSTL1 inhibits proliferation,
migration and invasion and promotes apoptosis in cervical cancer
cells
To comprehend the biological characteristics of
FSTL1 in vitro, HeLa and SiHa cells were infected with
lentivirus-FSTL1 or lentivirus-NC. The results of the MTT assay
indicated that the proliferation of both HeLa and SiHa cells was
inhibited in the FSTL1 OE group compared with the NC and MOCK
groups (Fig. 3A). Moreover, FSTL1
OE significantly inhibited the proliferation of HeLa and SiHa cells
on the 3rd and 4th day respectively. Subsequently, the colony
formation assay was performed to explore whether FSTL1 affected the
single-cell proliferation of HeLa and SiHa cells. The colony
formation rate of HeLa and SiHa cells in the OE group was lower
than that in the other two control groups (Fig. 3B). Furthermore, the wound-healing
assay demonstrated that FSTL1 OE retarded the migration of HeLa and
SiHa cells when compared with that in the NC and MOCK groups
(Fig. 3C). Furthermore, invasion
and migration in Transwell assay confirmed that the number of
invasive and migratory cells in the FSTL1 OE group was
significantly less than that in the NC and MOCK groups (Fig. 3D and E). Collectively, these data
established that FSTL1 inhibited the migration and invasion
abilities of HeLa and SiHa cells and that it may be a
metastasis-related gene in cervical cancer. Finally, the effect of
FSTL1 on apoptosis was detected using flow cytometric analysis.
Apoptosis assay demonstrated that the number of apoptotic cells in
the FSTL1 OE group was significantly higher than that in the NC and
MOCK groups (Fig. 3F), which proved
that the OE of FSTL1 promoted apoptosis in cervical cancer
cells.
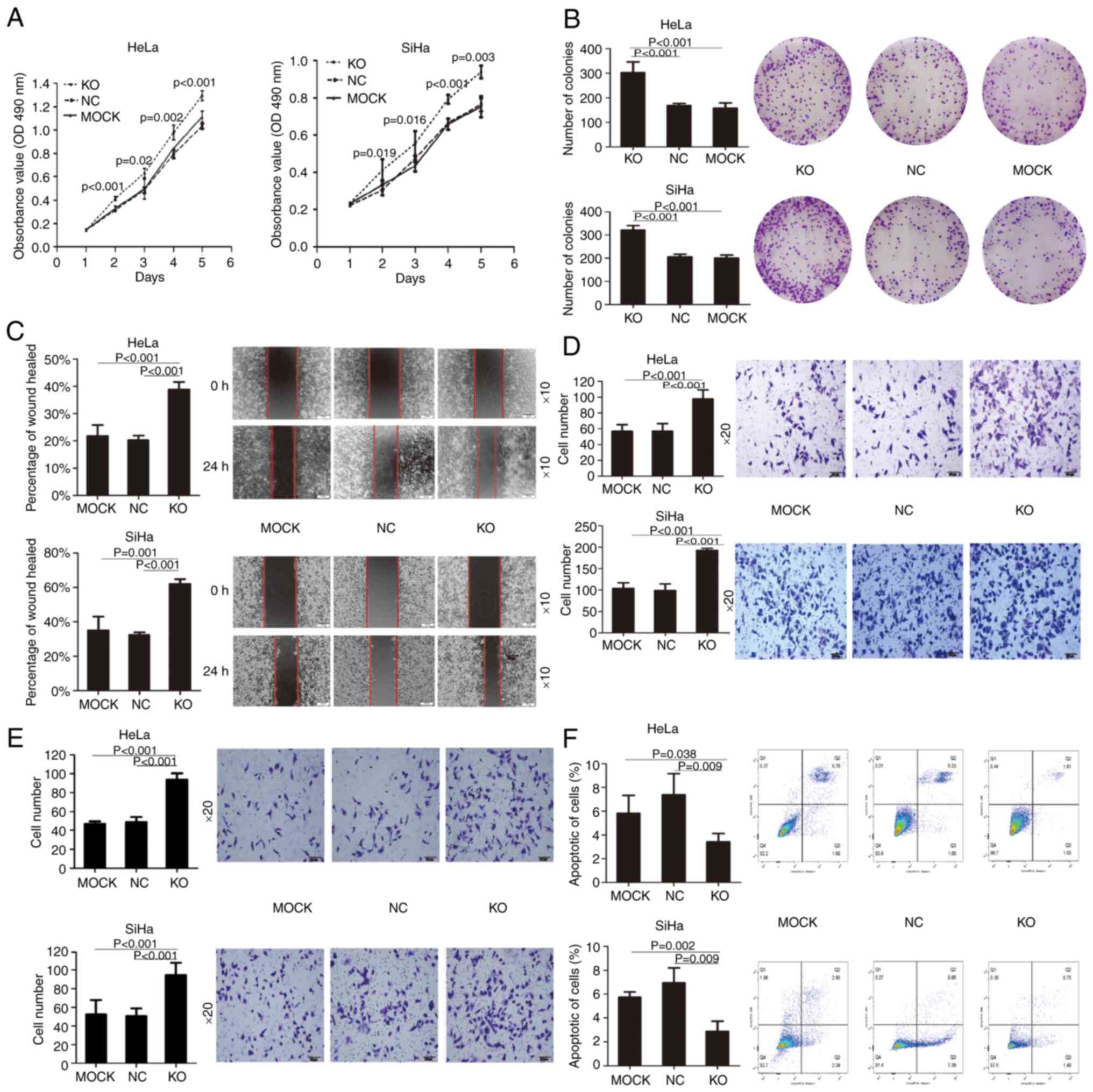
KO of FSTL1 promotes proliferation,
migration and invasion and inhibits apoptosis in cervical cancer
cells
To decipher the effects of FSTL1 depletion on the
biological function of cervical cancer cells, HeLa and SiHa cells
were infected with lentivirus-Cas9-sgRNA-FSTL1 or
lentivirus-Cas9-sgRNA-NC. MTT assay was subsequently performed,
which indicated that the proliferation of both HeLa and SiHa cells
was promoted in the FTL1 KO group compared with the NC and MOCK
groups (Fig. 4A). KO of FSTL1
significantly promoted the proliferation of HeLa and SiHa cells
from the 2nd day. The colony formation assay revealed that the KO
of FSTL1 promoted the single-cell proliferation of HeLa and SiHa
cells (Fig. 4B). Furthermore, the
results of the wound-healing assay demonstrated that the KO of
FSTL1 accelerated the migration of HeLa and SiHa cells compared
with the two control groups (Fig.
4C). Moreover, invasion and migration in the Transwell assay
confirmed that the number of invasive and migratory cells in the KO
group was significantly higher than that in the two control groups
(Fig. 4D and E). Succinctly, these
results confirmed that the KO of FSTL1 augmented the migration and
invasion abilities of HeLa and SiHa cells. Apoptosis assay further
revealed that the number of apoptotic cells in the KO group was
significantly less than that in the two control groups (Fig. 4F). Collectively, these results
confirmed that the KO of FSTL1 inhibited the apoptosis of cervical
cancer cells.
FSTL1 inhibits the growth of cervical
cancer cell line xenografts
To determine the biological function of FSTL1 in
vivo, cervical cancer cells overexpressing FSTL1 were
subcutaneously injected into the axillary region of nude mice.
After 27 days, the average weight and final volume of the tumor in
the FSTL1 OE group were significantly lower than those in the NC
group (Fig. 5A, C and D). RT-qPCR
and IHC assays showed that the expression of FSTL1 at the mRNA and
protein levels was significantly higher compared with the NC group
(Fig. 5B and E). Moreover, cervical
cancer cells with FSTL1 KO were subcutaneously injected into the
axillary region of nude mice (Fig.
6A-E). The findings signified that the biological functions of
the FSTL1 KO group were opposite to those of the OE group in
vivo. Collectively, FSTL1 could inhibit xenograft tumor growth
in nude mice.
FSTL1 possibly regulates the
IGF-1R/PI3K/AKT/BCL-2 signaling pathway
To clarify the regulatory mechanism of FSTL1 in
cervical cancer, gene chip analysis and bioinformatics methods were
used to analyze the interaction and relationship between FSTL1 and
its downstream signaling pathways. The results of gene chip
analysis suggested that compared with the NC group, 881
differentially expressed genes were present in the OE group, of
which 593 were upregulated and 288 were downregulated (Fig. 7B and Table II). Disease and functional
enrichment analysis revealed that the differentially expressed
genes were predominantly involved in regulating biological
processes including cancer development, damage and development of
the body, gastrointestinal diseases and gene expression (Fig. 7A). Furthermore, according to KEGG
and IPA signaling pathway enrichment analysis, the differentially
expressed genes of FSTL1 were chiefly enriched in cancer
transcriptional misregulation, mitogen-activated protein kinase
(MAPK), cancer, P53, and IGF-1 signaling pathways, of which the
IGF-1 signaling pathway was significantly activated in this
experiment (Fig. 7C; Table III). The results of western
blotting analysis suggested that FSTL1 OE downregulated IGF-1R,
PI3K, P-AKT and BCL-2 but upregulated BAX (Fig. 7D-F). On the contrary, FSTL1 KO
restored these changes (Fig. 7D, G and
H). These findings indicated that in cervical cancer, FSTL1 may
be involved in the regulation of the IGF-1R/PI3K/AKT signaling
pathway, thereby leading to the activation of the BCL-2 family.
 | Figure 7.FSTL1 may be involved in the
regulation of the IGF-1R/PI3K/AKT/BCL-2 signaling pathway. (A)
Disease and functional enrichment analyses revealed that
differentially expressed genes of FSTL1 were mainly involved in the
regulation of biological processes such as cancer development,
damage and development of the body, gastrointestinal diseases and
gene expression. (B) Heat map of cluster analysis of upregulated
and downregulated differentially expressed genes in HeLa cells (it
was based on all genes having been sorted using |Fold change |>2
and FDR <0.05). There were 881 differentially expressed genes in
the OE group, of which 593 genes were upregulated and 288 genes
were downregulated. (C) Kyoto Encyclopedia of Genes and Genomes and
Ingenuity Pathway Analysis signaling pathway enrichment analyses
revealed that the differentially expressed genes of FSTL1 were
mainly enriched in the cancer transcriptional misregulation
signaling, mitogen-activated protein kinase (MAPK) signaling,
cancer signaling, P53 signaling and IGF-1 signaling pathways, among
which the IGF-1 signaling pathway was significantly activated in
this experiment. (D) The IGF-1R, PI3K, AKT, P-AKT, BCL-2 and BAX
protein electropherogram. (E-H) The relative protein expression
levels of IGF-1R, PI3K, AKT, P-AKT, BCL-2 and BAX in each group.
Western blotting revealed that the OE of FSTL1 downregulated
IGF-1R, PI3K, AKT, P-AKT and BCL-2, but upregulated BAX. The KO of
FSTL1 restored these changes. Data are expressed as the mean ±
standard deviation and analyzed by one-way analysis of variance
(ANOVA) from three independent experiments. *P<0.05, compared
with the control group. FSTL1, follistatin-like protein 1; OE,
overexpression; KO, knockout; P-, phosphorylated. |
 | Table II.Top 10 upregulated or downregulated
differentially expressed genes in HeLa cells. |
Table II.
Top 10 upregulated or downregulated
differentially expressed genes in HeLa cells.
| Gene symbol | Fold change | P-value | FDR | Regulation |
|---|
| FOS | 32.99038 |
9.83×10−18 |
3.86×10−13 |
Up |
| EGR1 | 19.41814 |
1.77×10−14 |
6.32×10−11 |
Up |
| CTGF | 12.20367 |
4.09×10−14 |
1.15×10−10 |
Up |
| PDZD2 | 10.03935 |
2.04×10−12 |
1.29×10−9 |
Up |
| ZFP36 | 9.669648 |
1.04×10−13 |
1.95×10−10 |
Up |
| ATF3 | 8.939991 |
4.71×10−11 |
9.94×10−9 |
Up |
| NR4A1 | 7.948651 | 2.0
×10−15 |
2.02×10−11 |
Up |
| NR4A2 | 6.967495 |
2.26×10−13 |
2.85×10−10 |
Up |
| MALAT1 | 6.35784 |
2.51×10−11 |
6.66×10−9 |
Up |
| OASL | 5.798434 |
5.50×10−13 |
5.03×10−10 |
Up |
| DAW1 | −23.5035 |
1.89×10−13 |
2.66×10−10 | Down |
| GKN2 | −19.5982 |
9.97×10−13 |
7.63×10−10 | Down |
| CA9 | −9.04366 |
1.62×10−12 |
1.08×10−9 | Down |
| IL7R | −7.51349 |
1.02×10−10 |
1.68×10−8 | Down |
| CDH5 | −6.63583 |
1.04×10−10 |
1.70×10−8 | Down |
| PTGS1 | −6.4919 |
3.74×10−13 |
4.06×10−10 | Down |
| CCL20 | −6.00781 |
1.26×10−9 |
8.09×10−8 | Down |
| LAMA1 | −5.99628 |
1.56×10−13 |
2.37×10−10 | Down |
| STAMBPL1 | −5.8609 |
9.31×10−14 |
1.93×10−10 | Down |
| TSNAX-DISC1 | −5.83368 |
1.12×10−10 |
1.77×10−8 | Down |
 | Table III.Top 5 upregulated or downregulated
KEGG pathway enrichment analysis of differentially-expressed
downstream genes of FSTL1 in HeLa cells. |
Table III.
Top 5 upregulated or downregulated
KEGG pathway enrichment analysis of differentially-expressed
downstream genes of FSTL1 in HeLa cells.
| Gene Ontology
ID | Signaling
pathway | Database | P-value | Genes | Regulation |
|---|
| hsa05202 | Transcriptional
misregulation in cancer | KEGG | <0.001 | 18 |
Up |
| hsa04010 | MAPK signaling
pathway | KEGG | <0.001 | 21 |
Up |
| hsa05200 | Pathways in
cancer | KEGG | 0.003 | 25 |
Up |
| hsa04915 | Estrogen signaling
pathway | KEGG | 0.003 | 10 |
Up |
| hsa04910 | Insulin signaling
pathway | KEGG | 0.01 | 11 |
Up |
| hsa05202 | Transcriptional
misregulation in cancer | KEGG | 0.004 | 9 | Down |
| hsa05222 | Small cell lung
cancer | KEGG | 0.009 | 6 | Down |
| hsa04115 | p53 signaling
pathway | KEGG | 0.018 | 5 | Down |
| hsa05145 | Toxoplasmosis | KEGG | 0.032 | 6 | Down |
| hsa03040 | Spliceosome | KEGG | 0.049 | 6 | Down |
Discussion
In the present study, the expression of FSTL1 in
cervical cancer tissues was detected using RT-qPCR and IHC
analyses. These investigations revealed that the expression of
FSTL1 mRNA and protein was significantly decreased in cervical
cancer tissues compared with the corresponding tumor-adjacent and
normal cervical tissues. These findings were consistent with the
latest relevant studies, affirming that the FSTL1 expression is
reduced in cervical cancer tissues (14). The results also established a
gradual increase in the expression of FSTL1 mRNA in cervical
cancer, tumor-adjacent and normal cervical tissues. Furthermore,
the FSTL1 expression was associated with the clinicopathological
features of patients with cervical cancer. Low FSTL1mRNA expression
was observed in association with the extent of tumor
differentiation and infiltration depth, whereas low FSTL1 protein
expression was associated with the pathological type and
infiltration depth. Thus, infiltration depth was associated with
both mRNA and protein expression levels. Nonetheless, several
discrepancies were observed between the IHC and mRNA expression
levels of FSTL1. This difference may be attributed to multiple
levels of gene expression regulation, the transcription level
regulation being the only one link, and post-transcriptional,
translational and post-translational regulations playing a role in
the final protein expression. Moreover, factors including mRNA
degradation, protein degradation, modification and folding may have
contributed to the inconsistency between mRNA abundance and the
protein expression (18,19). Next, stable strains of cervical
cancer with FSTL1 OE and KO were constructed in HeLa and SiHa
cells. Certain functional assays were performed both in
vitro and in vivo. The findings demonstrated that FSTL1
OE inhibited the proliferation, migration and invasion of HeLa and
SiHa cells and enhanced their apoptosis in vitro. In
addition, FSTL1 OE suppressed xenograft tumor growth in nude mice
in vivo, whereas inhibition of FSTL1 expression resulted in
the opposite effect. These observations support that FSTL1 is a
potential new diagnostic marker for cervical cancer. Therefore, it
is feasible to detect the expression of FSTL1 in sample tissues to
estimate progression in cervical cancer cases. However, there are
several drawbacks to tissue protein detection method, including
that it is time-consuming and expensive and the inaccessibility to
dynamic contrast and follow-up after lesion resection. The
detection of the gene expression in the blood is convenient,
economical and highly accepted by patients. This technique can be
dynamically compared after treatment and is convenient for
follow-up observation. If the expression of FSTL1 mRNA in the blood
is correlated with the clinical pathology of cervical cancer
patients, the FSTL1 level in the blood of cervical cancer patients
could serve as one of the severity indicators to assist in the
judging of the progression of cervical cancer. However, FSTL1
expression in the blood was not examined in the present study.
Previous studies have reported that FSTL1
participates in the occurrence of tumors by regulating multiple
signaling pathways, such as the NF-kB (6) and BMP/Smad signaling pathways
(14). In the present study, it was
revealed that several novel signaling pathways regulated by FSTL1
are closely related to the occurrence and development of cervical
cancer. The results of gene chip analysis indicated the presence of
881 differentially expressed genes after FSTL1 OE. These genes were
chiefly enriched in cancer transcriptional misregulation, MAPK,
cancer, P53 and IGF-1 signaling pathways. Of these, the IGF-1
signaling pathway was markedly altered. Previous studies have
identified that IGF-1 and its receptor (IGF-1R) are aberrantly
expressed in several tumors, stimulating the growth of different
types of cells and blocking their apoptosis (20–23).
IGF-1 promotes cancer development mainly via the PI3K/AKT (24,25)
and Ras/MAPK pathways (26).
Disorders related to the PI3K/AKT signaling pathway occur in
various human diseases, including cancer (27,28),
diabetes (29), cardiovascular
(30) and neurological disease
(31). In cancer, activated AKT
regulates different cellular biological functions in the following
ways: i) regulation of cell growth by activating mTOR and
downstream molecules (25); ii)
regulation of cell proliferation by phosphorylating p27 (32) iii) direct inhibition of cell
apoptosis by inhibiting apoptotic proteins (such as BAX) (33) or transcription factors (such as
FOXO) (34) iv) participation in
cell migration and invasion by regulating E-cadherin and vimentin
(35) and v) regulation of NF-κB
signaling pathway via phosphorylation of IKK (36). Bcl-2 is one of the key effector
molecules downstream of the PI3K/AKT signaling pathway and can
inhibit cell apoptosis and lead to various diseases (37). When PI3K-dependent AKT is activated,
Bcl-2 is phosphorylated, and free Bcl-2 exerts an antiapoptotic
effect (37). Hers et al
(38) reported that the PI3K/AKT
signaling pathway regulates tumor cells by phosphorylating a range
of downstream targets, including Bcl-2 family members and caspase.
Yang et al (13) observed
that FSTL1 inhibited cell apoptosis in hepatocellular carcinoma by
silencing AKT/GSK-3β/Bcl2/BAX/Bim signaling. In the present study,
for the first time to the best of our knowledge, it was
demonstrated that the OE of FSTL1 downregulated IGF-1R, PI3K, P-AKT
and BCL-2 but upregulated BAX. KO of FSTL1 restored these changes.
These findings suggest the involvement of FSTL1 in the regulation
of the IGF-1R/PI3K/AKT/BCL-2 signaling pathway in cervical
cancer.
Collectively, the present study revealed that FSTL1
was downregulated in cervical cancer tissues. FSTL1 inhibited the
proliferation, migration and invasion of cervical cancer cells and
promoted their apoptosis. Furthermore, FSTL1 inhibited xenograft
tumor growth in nude mice. FSTL1 may be involved in the regulation
of the IGF-1R/PI3K/AKT/BCL-2 signaling pathway in cervical cancer.
Hence, it may be exploited as a novel biomarker to evaluate disease
progression in patients with cervical cancer. Prognosis evaluation
and treatment in such patients could therefore be improved.
Acknowledgements
The present study was performed at the Research and
experiment center of the People's Hospital of Guangxi Zhuang
Autonomous Region. The animal experiments were performed at the
Experimental Animal Center of Guangxi Medical University.
Funding
The present study was supported in part by the National Natural
Science Foundation of China (grant no. 81660434), the Natural
Science Foundation of Guangxi (grant no. 2018GXNSFAA050098) and the
Scientific Research Project of Guangxi Health Commission (grant no.
Z-A20220127).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL, XH and HZ conceived and designed the
experiments. ZL and HZ prepared the manuscript. ZL, XH and HZ
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study complied with the guidelines
outlined in the Declaration of Helsinki and was approved (approval
no. KY-LW-2017-5) by the Ethics Committee of the People's Hospital
of Guangxi Zhuang Autonomous Region (Nanning, China). The animal
experiments were approved (approval no. 2201805166) by The Animal
Care & Welfare Committee of Guangxi Medical University
(Nanning, China). Animal ethics review follows the Guiding Opinions
on the Treatment of Laboratory Animals issued by the Ministry of
Science and Technology of China and the Laboratory Animal-Guideline
for Ethical Review of Animal Welfare issued by the National
Standard GB/T35892-2018 of China. Written informed consent was
obtained by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lei J, Ploner A, Elfström KM, Wang J, Roth
A, Fang F, Sundström K, Dillner J and Sparén P: HPV vaccination and
the risk of invasive cervical cancer. N Engl J Med. 383:1340–1348.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Di Domenico M, Giovane G, Kouidhi S, Iorio
R, Romano M, De Francesco F, Feola A, Siciliano C, Califano L and
Giordano A: HPV epigenetic mechanisms related to oropharyngeal and
cervix cancers. Cancer Biol Ther. 19:850–857. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mattiotti A, Prakash S, Barnett P and van
den Hoff MJB: Follistatin-like 1 in development and human diseases.
Cell Mol Life Sci. 75:2339–2354. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bradshaw AD: Diverse biological functions
of the SPARC family of proteins. Int J Biochem Cell Biol.
44:480–488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu YK, Jia YJ, Liu SH and Ma J: FSTL1
increases cisplatin sensitivity in epithelial ovarian cancer cells
by inhibition of NF-κB pathway. Cancer Chemother Pharmacol.
87:405–414. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ni X, Cao X, Wu Y and Wu J: FSTL1
suppresses tumor cell proliferation, invasion and survival in
non-small cell lung cancer. Oncol Rep. 39:13–20. 2018.PubMed/NCBI
|
|
8
|
Liu Y, Han X, Yu Y, Ding Y, Ni C, Liu W,
Hou X, Li Z, Hou J, Shen D, et al: A genetic polymorphism affects
the risk and prognosis of renal cell carcinoma: Association with
follistatin-like protein 1 expression. Sci Rep. 6:266892016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Huang S, Wu S, Yin S, Tang A and
Wen W: Follistatin-like protein-1 upregulates dendritic cell-based
immunity in patients with nasopharyngeal carcinoma. J Interferon
Cytokine Res. 37:494–502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lau MCC, Ng KY, Wong TL, Tong M, Lee TK,
Ming XY, Law S, Lee NP, Cheung AL, Qin YR, et al: FSTL1 promotes
metastasis and chemoresistance in esophageal squamous cell
carcinoma through NFκB-BMP signaling cross-talk. Cancer Res.
77:5886–5899. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu M, Ding Y, Wu N, Jiang J, Huang Y,
Zhang F, Wang H, Zhou Q, Yang Y, Zhuo W and Teng L: FSTL1 promotes
growth and metastasis in gastric cancer by activating AKT related
pathway and predicts poor survival. Am J Cancer Res. 11:712–728.
2021.PubMed/NCBI
|
|
12
|
Su S, Parris AB, Grossman G, Mohler JL,
Wang Z and Wilson EM: Up-regulation of follistatin-like 1 by the
androgen receptor and melanoma antigen-A11 in prostate cancer.
Prostate. 77:505–516. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang W, Wu Y, Wang C, Liu Z, Xu M and
Zheng X: FSTL1 contributes to tumor progression via attenuating
apoptosis in a AKT/GSK-3β-dependent manner in hepatocellular
carcinoma. Cancer Biomark. 20:75–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao C, Chen Z, Zhu L, Miao Y, Guo J, Yuan
Z, Wang P, Li L and Ning W: The BMP inhibitor follistatin-like 1
(FSTL1) suppresses cervical carcinogenesis. Front Oncol.
13:11000452023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu W, Gao G, Hu X, Wang Y, Schwarz JK,
Chen JJ, Grigsby PW and Wang X: Activation of miR-9 by human
papillomavirus in cervical cancer. Oncotarget. 5:11620–11630. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang J, Zhang S, Wu J, Lu Z, Yang J, Wu H,
Chen H, Lin B and Cao T: Clinical significance and prognostic value
of SOX7 expression in liver and pancreatic carcinoma. Mol Med Rep.
16:499–506. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Battle A, Khan Z, Wang SH, Mitrano A, Ford
MJ, Pritchard JK and Gilad Y: Genomic variation. Impact of
regulatory variation from RNA to protein. Science. 347:664–667.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Laurent JM, Vogel C, Kwon T, Craig SA,
Boutz DR, Huse HK, Nozue K, Walia H, Whiteley M, Ronald PC and
Marcotte EM: Protein abundances are more conserved than mRNA
abundances across diverse taxa. Proteomics. 10:4209–4212. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cevenini A, Orrù S, Mancini A, Alfieri A,
Buono P and Imperlini E: Molecular signatures of the insulin-like
growth factor 1-mediated epithelial-mesenchymal transition in
breast, lung and gastric cancers. Int J Mol Sci. 19:24112018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rigiracciolo DC, Nohata N, Lappano R,
Cirillo F, Talia M, Scordamaglia D, Gutkind JS and Maggiolini M:
IGF-1/IGF-1R/FAK/YAP transduction signaling prompts growth effects
in triple-negative breast cancer (TNBC) cells. Cells. 9:10102020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Laron Z and Werner H: Congenital IGF-1
deficiency protects from cancer: Lessons from Laron syndrome.
Endocr Relat Cancer. 30:e2203942023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kwon H, Choi M, Ahn Y, Jang D and Pak Y:
Flotillin-1 palmitoylation turnover by APT-1 and ZDHHC-19 promotes
cervical cancer progression by suppressing IGF-1 receptor
desensitization and proteostasis. Cancer Gene Ther. 30:302–312.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun L, Yuan W, Wen G, Yu B, Xu F, Gan X,
Tang J, Zeng Q, Zhu L, Chen C and Zhang W: Parthenolide inhibits
human lung cancer cell growth by modulating the IGF-1R/PI3K/Akt
signaling pathway. Oncol Rep. 44:1184–1193. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Sun Y, Cong S and Zhang F:
Insulin-like growth factor-1 promotes human uterine leiomyoma cell
proliferation via PI3K/AKT/mTOR pathway. Cells Tissues Organs.
212:194–202. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Solarek W, Koper M, Lewicki S, Szczylik C
and Czarnecka AM: Insulin and insulin-like growth factors act as
renal cell cancer intratumoral regulators. J Cell Commun Signal.
13:381–394. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Zheng P, Jiao T, Zhang M, Wu Y,
Zhang X, Wang S and Zhao Z: Paiteling induces apoptosis of cervical
cancer cells by down-regulation of the E6/E7-Pi3k/Akt pathway: A
network pharmacology. J Ethnopharmacol. 305:1160622023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Chen X, Ding T, Zhang H, Zhang Q,
Dai H, Zhang H, Tang J and Wang X: KAT7 promotes radioresistance
through upregulating PI3K/AKT signaling in breast cancer. J Radiat
Res. 64:448–456. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mao YP, Song YM, Pan SW, Li N, Wang WX,
Feng BB and Zhang JH: Effect of codonopsis radix and polygonati
rhizoma on the regulation of the IRS1/PI3K/AKT signaling pathway in
type 2 diabetic mice. Front Endocrinol (Lausanne). 13:10685552022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou WW, Dai C, Liu WZ, Zhang C, Zhang Y,
Yang GS, Guo QH, Li S, Yang HX and Li AY: Gentianella acuta
improves TAC-induced cardiac remodelling by regulating the Notch
and PI3K/Akt/FOXO1/3 pathways. Biomed Pharmacother. 154:1135642022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tian Q, Guo Y, Feng S, Liu C, He P, Wang
J, Han W, Yang C, Zhang Z and Li M: Inhibition of CCR2 attenuates
neuroinflammation and neuronal apoptosis after subarachnoid
hemorrhage through the PI3K/Akt pathway. J Neuroinflammation.
19:3122022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yoon JH, Shin JW, Pham TH, Choi YJ, Ryu
HW, Oh SR, Oh JW and Yoon DY: Methyl lucidone induces apoptosis and
G2/M phase arrest via the PI3K/Akt/NF-κB pathway in
ovarian cancer cells. Pharm Biol. 58:51–59. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan X, Gong L, Li X, Zhang X, Sun J, Luo
X, Wang Q, Chen J, Xie L and Han S: Promethazine inhibits
proliferation and promotes apoptosis in colorectal cancer cells by
suppressing the PI3K/AKT pathway. Biomed Pharmacother.
143:1121742021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mishra S, Cosentino C, Tamta AK, Khan D,
Srinivasan S, Ravi V, Abbotto E, Arathi BP, Kumar S, Jain A, et al:
Sirtuin 6 inhibition protects against glucocorticoid-induced
skeletal muscle atrophy by regulating IGF/PI3K/AKT signaling. Nat
Commun. 13:54152022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen L, Qing J, Xiao Y, Huang X, Chi Y and
Chen Z: TIM-1 promotes proliferation and metastasis, and inhibits
apoptosis, in cervical cancer through the PI3K/AKT/p53 pathway. BMC
Cancer. 22:3702022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang X, Qu P, Zhao H, Zhao T and Cao N:
COX-2 promotes epithelial-mesenchymal transition and migration in
osteosarcoma MG-63 cells via PI3K/AKT/NF-κB signaling. Mol Med Rep.
20:3811–3819. 2019.PubMed/NCBI
|
|
37
|
Siddiqui WA, Ahad A and Ahsan H: The
mystery of BCL2 family: Bcl-2 proteins and apoptosis: An update.
Arch Toxicol. 89:289–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hers I, Vincent EE and Tavaré JM: Akt
signalling in health and disease. Cell Signal. 23:1515–1527. 2011.
View Article : Google Scholar : PubMed/NCBI
|