Introduction
Cholangiocarcinoma (CCA) is an epithelial cell
malignancy that originates from cholangiocytes, and which can occur
at any level of the biliary tree (1). The overall incidence of CCA has been
increasing worldwide over the course of the last few decades
(1–3). CCAs are usually symptomless in the
early stages of the disease; therefore, they are often not
diagnosed until the disease has progressed to an advanced stage,
which severely compromises the therapeutic options available,
resulting in an unfavorable prognosis (2). Anatomically, CCAs can be classified
into three groups as follows: Intrahepatic, perihilar and distal
(4). Perihilar CCA (pCCA) is the
most common subtype of CCA, representing ~50–60% of all cases of
CCA; ~30–40% of cases are distal carcinomas, whereas intrahepatic
carcinomas represent <10% of CCA cases (5,6).
Surgical resection has been the mainstay of curative treatment for
the three subtypes of CCA. However, patients with pCCA are not able
to receive the full benefits of surgical resection due to the
difficulty of early detection, vascular invasion and lymphatic
metastasis in the advanced stages of the disease (7). Although significant progress has been
made with chemotherapeutics, targeted therapy and immunotherapy as
alternative treatments, to date, these therapies have not
demonstrated any notable benefits in terms of the overall survival
(OS) rates of patients with pCCA (1). Therefore, there is an urgent need for
the identification of novel biomarkers and drug targets for
improving the diagnosis and treatment of patients with pCCA.
Krüppel-like factor 4 (KLF4) is a zinc
finger-containing transcription factor that is able to regulate
diverse cellular processes, including cell growth, proliferation
and differentiation (8). It is
predominantly expressed in terminally differentiated epithelial
tissues, such as the skin, lungs and the gastrointestinal tract
(9). However, a variety of studies
have demonstrated that KLF4 functions via diverse and even opposite
mechanisms in various types of tumors. For example, KLF4 is
downregulated in a number of types of epithelial cancer, including
esophageal, gastric, colorectal and bladder cancer (10–13),
leading to cell proliferation. On the other hand, conflicting
results have demonstrated that KLF4 is overexpressed in primary
breast and prostate cancer (14,15),
where it plays an oncogenic role in tumor development and
progression. However, the underlying mechanisms through which KLF4
exerts its functions in pCCA, and its prognosis value, have yet to
be fully elucidated.
Growth and differentiation factor 15 (GDF15) is a
divergent member of the TGF-β superfamily. Over the course of the
past few years, it has been shown to participate in carcinogenesis
and tumor progression. Previous studies have indicated that GDF15
expression is elevated in certain types of tumor, including
non-small cell lung cancer (16),
cervical cancer (17), liver cancer
(18), head and neck cancer
(19), esophageal cancer (20), and so on (21–24).
However, certain studies have suggested that GDF15 may have a
tumor-suppressive activity in various types of cancer cells, or at
different stages of tumor growth (25). GDF15 has been shown to function as a
tumor activator or suppressor through the Smad, AKT and ERK
signaling pathways (26). However,
to date, the biological functions of GDF15 in pCCA remain
unclear.
KLF4 and KLF5, the closest members of the KLF
family, fulfill key roles in tumor proliferation, differentiation
and carcinogenesis in esophageal cancer and gastrointestinal
carcinoma (27,28). Zhao et al (16) reported that GDF15, as the downstream
target of KLF5, was able to increase the rate of tumor cell
proliferation. However, the association between KLF4 and GDF15 in
pCCA has yet to be elucidated.
In the present study, it was demonstrated that KLF4
was a favorable prognostic factor in pCCA. It has been shown to
inhibit the proliferation and promote the apoptosis of
cholangiocarcinoma cells. The findings presented herein also
demonstrate that KLF4 suppression enhances the transcription of
GDF15 in pCCA, suggesting that the KLF4/GDF15 signaling axis may be
a potential therapeutic target in pCCA.
Materials and methods
Patients and follow-up
The present primary study cohort comprised 242
patients who were diagnosed with pCCA at Linyi People's Hospital
(Linyi, China), and who underwent surgical resection from 2010 to
2020. A validation cohort contained 114 patients who were selected
from the primary cohort according to the following inclusion
criteria: i) Patients who underwent radical resection with a clear
surgical margin; ii) patients with available formalin-fixed tumor
tissues, follow-up information and complete medical records; iii)
patients with a post-surgical survival time of >1 month; and iv)
patients with no history of other malignancies. The primary end
point of follow-up was the OS rate (note that determination of the
OS rate excluded deaths that may have resulted from any other cause
besides cancer-specific death).
Clinical tissue samples
A total of 20 paired tumor specimens and adjacent
normal tissues were obtained from patients with pathologically
verified pCCA who received R0 surgery at Linyi People Hospital. The
present study was approved by the Medical Ethics Committee of Linyi
People's Hospital (approval no. YX200626). Written informed consent
was obtained from each patient prior to inclusion in the study.
Tissue microarray (TMA) and
immunohistochemistry (IHC)
A pCCA TMA containing the aforementioned 114
specimens was immunohistochemically stained for KLF4 (cat. no.
ab215036; Abcam) and GDF15 (cat. no. ab206414; Abcam). The TMA
slides were submerged in EDTA (pH 9.0) buffer for optimal antigen
retrieval. Primary KLF4 antibody (1:500) or GDF15 antibody (1:100)
was applied and incubated with the specimens at 4°C overnight. A
biotin-labeled goat anti-rabbit antibody (OriGene Technologies,
Inc.; formerly ZSGB/ZSGB-BIO) was applied to the specimens for 30
min at room temperature. Subsequently, the slides were incubated
with conjugated horseradish peroxidase-streptavidin. The peroxidase
reaction was developed using 3,3-diaminobenzidine (DAB) solution
(OriGene Technologies, Inc.). The stained TMA was subsequently
scanned using a HistoRX PM-2000 (TM) imaging system and analyzed
using AQU Analysis software (version 2.3.3.2, HistoRx, Inc.), which
was used to generate the IHC scores. The IHC scores were calculated
according to the following formula: IHC score=(percentage of cells
with weak staining intensity ×1) + (percentage of cells with
moderate staining intensity ×2) + percentage of cells with strong
staining intensity ×3). The cohort of specimens was divided into
two subgroups, according to the cut-off value of KLF4 scores, which
were identified as the point with the highest sum of specificity
and sensitivity in the receiver operating characteristic (ROC)
curve. The cut-off value for KLF4 in pCCA was identified to be
71.2. The scores of KLF4 and GDF15 of the TMA were analyzed to find
the associated correlation values using Spearman's correlation
analysis.
Cell lines and cell culture
The human pCCA cell lines, QBC939 (cat. no.
CC-Y1636) and FRH0201 (cat. no. YS1612C), the intrahepatic
cholangiocarcinoma (IHCC) cell line RBE (cat. no. HTX1698), and the
biliary epithelial cell line [human intrahepatic biliary epithelial
cells (HIBEpiC; cat. no. YS2223C)] were obtained from the Cell Bank
of Shanghai Yaji Biotechnology Co., Ltd. The QBC939 and HIBEpic
cell lines were maintained in Gibco® DMEM (Thermo Fisher
Scientific, Inc.), containing 10% Gibco® fetal bovine
serum (FBS; Thermo Fisher Scientific, Inc.) and 100 U/ml
Gibco® penicillin/streptomycin (Thermo Fisher
Scientific, Inc.). The RBE and FRH-0201 were cells maintained in
Gibco® RPMI-1640 medium (Thermo Fisher Scientific, Inc.)
supplemented with 10% Gibco® FBS and 100 U/ml
Gibco® penicillin/streptomycin (Thermo Fisher
Scientific, Inc.). All the cell lines were cultured at 37°C in a
humidified incubator containing 5% CO2.
Transient transfection and lentiviral
transduction
A KLF4 overexpression plasmid and negative control
vector, KLF4-specific shRNA and non-coding shRNA were synthesized
by GenePharma. GDF15 siRNAs were purchased from GeneChem, Inc. The
sequence of the siRNA targeting GDF15 was as follows:
5′-GCTACAATCCCATGGTGCTCA-3′. The sequence of the control siRNA was
as follows: 5′-UUCUCCGAACGUGUCACGUTT-3′. For transient
transfection, the cells were seeded in six-well plates at a density
of 4×105 cells/well. Upon reaching 70% confluency, the
cells were transfected with the siRNAs or plasmids. siRNA (100
pmol) or plasmids (4 µg) were transfected into the cells using
Lipofectamine 2000 reagent (GenePharma Co., Ltd.) at 37°C for 8 h,
according to the manufacturer's protocol. At 8 h post-transfection,
the cell culture medium was discarded, and fresh DMEM containing
10% v/v FBS was added to each well. After 48 h, the transfection
efficiency was further assessed using western blot analysis and
subsequent experimentations were executed. For lentiviral
transduction, the 2nd generation system was used in the lentivirus
transduction experiment. MOI values and optimal infection
conditions were determined by pre-testing. The cells were seeded in
six-well plates at a density of 3×105 cells/well. Upon
reaching 30% confluency, the cells were transfected with the
shRNAs. The shRNAs (MOI, 60) were transduced into cells (GenePharma
Co., Ltd.) at 37°C for 16 h, according to the manufacturer's
protocol. The cell culture medium was then discarded, and fresh
DMEM containing 10% v/v FBS was added into each well. After 72 h,
the transduction efficiency was observed using a fluorescence
microscope (Olympus Corporation). STable cell lines with KLF4
knockdown were selected using 4 µg/ml puromycin for 7 days. Western
blot analysis was conducted to detect the knockdown efficiency of
KLF4.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the pCCA cultured
cells, tumor tissues and adjacent normal tissues using Invitrogen™
TRIzol™ reagent (Thermo Fisher Scientific, Inc.). Subsequently, the
total RNA was reverse transcribed to obtain first-strand cDNA using
an RNA-PCR kit (cat. no. FSQ-101; Toyobo Life Science), following
the manufacturer's protocols. The resulting cDNA was used for
RT-qPCR using an Applied Biosystems® SYBR-Green PCR
Master Mix kit (cat. no. A25742; Thermo Fisher Scientific, Inc.).
The thermocycling conditions were as follows: Pre-denaturation at
93°C for 2 min; 40 cycles of denaturation at 93°C for 1 min;
annealing at 55°C for 1 min; extension at 72°C for 1 min; and final
extension at 72°C for 7 min. Quantification of the relative mRNA
levels was normalized against that of GAPDH and calculated using
the 2−ΔΔCq method (29).
The primers used were as follows: KLF4 forward,
5′-CTGCGAACCCACACAGGTAG-3′ and reverse,
5′-TGGAAGCTAACCTGGGAAGTC-3′; and GDF15 forward,
5′-ACTCACGCCAGAAGTGCG-3′ and reverse, 5′-CACGTCCCACGACCTTGAC-3′;
and GAPDH forward, 5′-GAAAGCCTGCCGGTGACTAA-3′ and reverse,
5′-GCCCAATACGACCAAATCAGAG-3′.
Western blot analysis
Cell lysates and tissues were lysed using RIPA lysis
buffer (Beyotime, Institute of Biotechnology), and the total
protein concentration was determined using a BCA protein detection
kit (Pierce; Thermo Fisher Scientific, Inc.). Target proteins (30
µg/lane) were separated by SDS-PAGE (8–15% gels) and then blotted
onto polyvinylidene fluoride (PVDF) membranes. After blocking the
membranes with 5% bovine serum albumin at room temperature for 1 h,
the PVDF membranes were incubated with primary antibodies at 4°C
overnight. The following antibodies were used: Anti-KLF4 (1:1,000;
cat. no. ab215036; Abcam); anti-GDF15 (1:1,000; cat. no. ab206414;
Abcam); anti-Lamin B (1:1,000; cat. no. AF1408; Beyotime Institute
of Biotechnology); anti-GAPDH (1:1,000; cat. no. AF1186; Beyotime
Institute of Biotechnology); anti-Bcl-2 (1:1,000; cat. no. ab32124;
Abcam); anti-Bax (1:1,000; cat. no. ab32503; Abcam); anti-AKT
(1:500; cat. no. 4691; Cell Signaling Technology, Inc.); and
anti-phosphorylated (p-)AKT (1:500; cat. no. 4060; Cell Signaling
Technology, Inc.). Subsequently, the membranes were incubated with
secondary antibodies (HRP-linked goat anti-rabbit IgG antibodies;
1:500; cat. no. A0208; Beyotime Institute of Biotechnology) for 1 h
at room temperature. Proteins were visualized using enhanced
chemiluminescence detection (Beijing Solarbio Science &
Technology Co., Ltd.). Protein bands were quantified using ImageJ
software (v 1.46r, National Institutes of Health) if necessary.
Cell proliferation assay
Cell proliferation was detected using Cell Counting
Kit-8 (CCK-8) and 5-ethynyl-2′-deoxyuridine (EdU) assays. The
transfected cells were seeded in 96-well plates at a density of
3×103 cells per well, and incubated at 37°C for time
periods of 24, 48, 72 and 96 h. Every 24 h, the cells were mixed
with 10 µl CCK-8 reagent (Dojindo Laboratories, Inc.) per well, and
subsequently incubated further with the CCK-8 reagent at 37°C for 1
h. The absorbance value at 450 nm was then measured using a
microplate reader (Thermo Fisher Scientific, Inc.). The relative
OD450 values were calculated using the formula:
OD450 of the tested well-OD450 of the empty
medium.
The fraction of DNA-replicating cells, which
represents the cell proliferation status, was assessed using an EdU
detection kit (Guangzhou RiboBio Co., Ltd.), in accordance with the
manufacturer's protocol. Briefly, the FRH0201 and QBC939 cells were
cultured in 96-well plates at 6×103 cells per well, and
incubated at 37°C for 24 h. Subsequently, 50 µM EdU labeling medium
was added to the 96-well plates and the cells were incubated at
37°C for 2 h. The cells were then treated with 4% paraformaldehyde
and 0.5% Triton X-100. The cells were stained with Apollo reaction
mixture at 37°C for 30 min. Hoechst 33342 was used to label cell
nuclei at 37°C for 30 min. The EdU incorporation rate was
calculated as the ratio of the number of EdU-incorporated cells to
the number of Hoechst 33342-stained cells. At least 500 cells were
counted for every group.
Apoptosis detection
Cell apoptosis was detected using a PE-Annexin V
Apoptosis Detection Kit (BD Biosciences). Cells contained in the
supernatant were harvested by centrifugation at 500 × g for 5 min
at 4°C. The cells were then washed with PBS and resuspended in a
binding buffer containing PE-Annexin and 7-aminoactinomycin D
(7-AAD), following the manufacturer's instructions. After the cells
were incubated for 15 min at 25°C in the dark, the percentages of
apoptotic cells were analyzed using a flow cytometer (FACSCanto II,
BD Biosciences).
A terminal deoxynucleotidyl transferase
(TdT)-mediated dUTP nick-end labeling (TUNEL) assay was also used
to assess cell apoptosis. The transfected cells were seeded in
30-mm Petri dishes and cultivated to a confluency of not >80%
per Petri dish. Cell fixation, permeabilization and TdT incubation
were performed following the manufacturer's instructions provided
with the TUNEL Apoptosis Detection kit (Alexa Fluor 647; Shanghai
Yeasen Biotechnology Co., Ltd.).
In vivo tumorigenicity assays
All animal experiments were approved by the Medical
Ethics Committee of Linyi People's Hospital. They were executed in
accordance with the guidelines for the use and care of laboratory
animals provided by Linyi People's Hospital. Female BALB/c nude
mice (6–8 weeks of age; weighing 20–24 g; n=12, 6 mice/cage) were
purchased from GenePharm Biotech Corp and housed in a specific
pathogen-free environment (at 25°C, 60% relative humidity and 12-h
light/dark cycle). The mice were provided with food and water ad
libitum in the animal research center. The mice were randomly
divided into two groups, each containing 6 mice. The FRH0201 cells
(4×106) transfected with shKLF4 or a negative control
(NC) were subcutaneously injected into the right flanks of the
mice. Tumor diameters were measured every 3 days, and the tumor
volumes were calculated using the following formula: V=length ×
width2/2 (mm3). The mice were sacrificed by
cervical dislocation following an intraperitoneal injection of
pentobarbital on the 21st day, and the xenograft tumors were
removed, weighed and photographed (the maximum tumor volume
permitted in our study was <2,000 mm3).
Bioinformatics analyses
The microarray expression data of GSE26566 and
GSE89749 were obtained from the Gene Expression Omnibus database
(GEO). The dataset GSE26566 based on the platform of GPL6104
platform (Illumina humanRef-8 v2.0 expression beadchip) including
104 cholangiocarcinoma tumor samples and 6 normal bile duct
samples. The dataset GSE89749 based on the platform of GPL10558
platform (Illumina HumanHT-12 V4.0 expression beadchip) containing
118 cholangiocarcinoma tumor samples and 2 normal bile duct
samples. To screen the differentially expressed genes (DEGs)
between the cholangiocarcinoma tumor samples and normal bile duct
samples, we used the GEO2R online web tool, which allows users to
compare different gene expression data of two or more groups of
samples. An adjusted P-value <0.05 and |log (FC)|≥1.5
were set as the thresholds for identifying DEGs. DEGs with logFC
>0 were considered as upregulated genes, and those with
logFC <0 were classified as downregulated genes. To
identify the intersectional genes between GSE26566 and GSE89249,
the Venny 2.1 online web tool was used to create a Venn
diagram.
Statistical analysis
Statistical analyses were performed using SPSS17.0
(SPSS, Inc.) and GraphPad Prism 6.0 (Dotmatics) software packages.
Data are expressed as the mean ± standard deviation (SD). All
experiments were repeated three times unless otherwise specified.
The associations between KLF4 and the clinicopathological
characteristics of patients with pCCA were analyzed using the
Chi-squared (χ2) test. Univariate and multivariate Cox
regression analyses were used for the survival data. The
correlation between KLF4 and GDF15 was determined using Spearman's
correlation analysis for IHC Scores. Pearson's correlation analysis
was used for the data obtained from RT-qPCR. A paired or unpaired
t-test was used to compare differences between two groups, whereas
one-way ANOVA was used to analyze the differences among three
groups. If there were significant differences, multiple comparisons
were made between the groups (using Tukey's test). P<0.05 was
considered to indicate a statistically significant difference.
Results
KLF4 expression levels in pCCA and
adjacent tissues
To determine the expression of KLF4 in pCCA, the
expression level of KLF4 was first detected in 20 pairs of pCCA
tissues and adjacent normal bile duct tissues using RT-qPCR
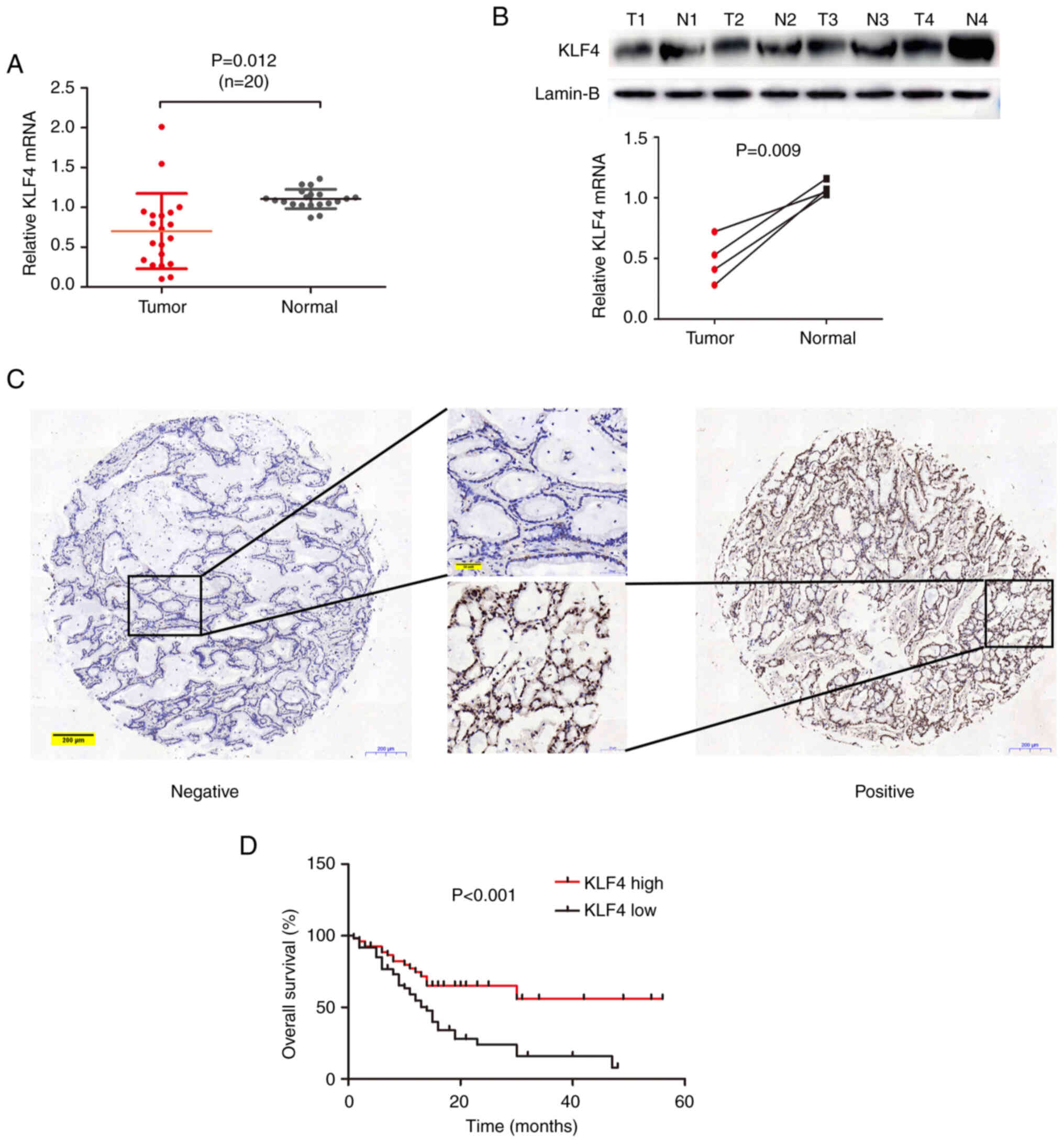
analysis (Fig. 1A). Compared with
the expression level in normal tissues, KLF4 mRNA expression was
found to decreased in the majority of pCCA tissues. KLF4 protein
levels in four pairs of these pCCA tissues were subsequently
detected using western blot analysis, and these experiments
revealed that the expression level of KLF4 protein and the
corresponding mRNA was decreased in pCCA (Fig. 1B). Furthermore, KLF4 expression was
analyzed in human pCCA TMAs; two representative images of KLF4
expression (high and low or positive/negative) obtained from TMAs
are presented in Fig. 1C. As a
transcription factor, KLF4 was shown to be located in the
nucleoplasm and cytosol.
Association between KLF4 and the
pathological features of patients with pCCA
To further determine the level of KLF4 expression,
and to examine the association between KLF4 and the clinical
features of patients with pCCA, pCCA TMAs containing 114 cases of
pCCA were used for IHC. The clinicopathological features of the
patients with pCCA in the TMAs were recorded and analyzed. Cox
survival analysis revealed that a high level of KLF4 expression was
a marker of a favorable prognosis for patients with pCCA
(P<0.001, Fig. 1D).
Subsequently, the associations between KLF4 and the
clinicopathological factors of patients with pCCA was analyzed.
Notably, as shown in Table I, KLF4
expression was found to be negatively associated with histological
grade (P=0.004) and tumor size (P=0.022). However, KLF4 expression
was not found to be associated with age, sex, tumor depth,
lymphatic metastasis and tumor lymph node-metastasis (TNM) stage
(all P>0.05; Table I).
Subsequently, a Cox proportional hazards model was performed in
order to better determine whether KLF4 expression should be
regarded as a valuable biomarker (Table II). Univariate analysis revealed
that histological grade (P=0.001), tumor size (P=0.024), TNM stage
(P=0.003) and KLF4 expression (P=0.002) were significantly
associated with the risk of mortality. Further multivariate
analyses were performed to identify the aforementioned factors, and
the outcomes confirmed that KLF4 expression (P=0.022) was an
independent prognostic indicator for OS in pCCA, as were
histological grade (P=0.017) and TNM stage (P=0.003).
 | Table I.Associations between KLF4 expression
and the clinicopathological characteristics of patients with
pCCA. |
Table I.
Associations between KLF4 expression
and the clinicopathological characteristics of patients with
pCCA.
|
|
| KLF4
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | No. of cases | High (n) | Low (n) | χ2
value | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
>63 | 60 | 25 | 35 | 1.652 | 0.199 |
|
≤63 | 54 | 29 | 25 |
|
|
| Sex |
|
|
|
|
|
|
Male | 81 | 43 | 38 | 3.670 | 0.055 |
|
Female | 33 | 11 | 22 |
|
|
| Histological
grade |
|
|
|
|
|
| Well
and moderately differentiated | 96 | 51 | 45 | 8.082 | 0.004 |
| Poorly
differentiated | 18 | 3 | 15 |
|
|
| Tumor size
(cm) |
|
|
|
|
|
|
>2.5 | 53 | 19 | 34 | 5.272 | 0.022 |
|
≤2.5 | 61 | 35 | 26 |
|
|
| Tumor depth |
|
|
|
|
|
| T1 | 40 | 21 | 19 | 0.651 | 0.420 |
|
T2-T4 | 74 | 33 | 41 |
|
|
| Lymphatic
metastasis |
|
|
|
|
|
|
Absent | 75 | 32 | 43 | 41.944 | 0.163 |
|
Present | 39 | 22 | 17 |
|
|
| TNM stage |
|
|
|
|
|
| I and
II | 73 | 33 | 40 | 0.381 | 0.537 |
| III and
IV | 41 | 21 | 20 |
|
|
 | Table II.Univariate and multivariate analyses
of prognostic factors for the overall survival of patients with
pCCA. |
Table II.
Univariate and multivariate analyses
of prognostic factors for the overall survival of patients with
pCCA.
|
| Univariate
analysis | Multivariate
analysisa |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years (>63
vs. ≤63) | 0.647 | 0.376–1.114 | 0.116 |
|
|
|
| Sex (male vs.
female) | 1.077 | 0.612–1.897 | 0.797 |
|
|
|
| Histological grade
(well and moderately vs. poorly differentiated) | 2.912 | 1.533–5.534 | 0.001 | 2.265 | 1.155–4.443 | 0.017 |
| Tumor size (>2.5
vs. ≤2.5) | 0.538 | 0.314–0.920 | 0.024 |
|
|
|
| Tumor depth (T1 vs,
T2-4) | 1.363 | 0.737–2.520 | 0.323 |
|
|
|
| Lymphatic
metastasis (absent vs. present) | 1.472 | 0.857–2.528 | 0.161 |
|
|
|
| TNM stage (I/II vs.
III/IV) | 2.227 | 1.310–3.784 | 0.003 | 2.305 | 1.341–3.962 | 0.003 |
| KLF4 expression
(high vs. low) | 0.403 | 0.225–0.722 | 0.002 | 0.477 | 0.254–0.897 | 0.022 |
KLF4 suppresses pCCA cell
proliferation in vitro
To examine the effects of an altered KLF4 expression
on the proliferation of pCCA cells, the expression of KLF4 was
detected in a series of cell lines, including the pCCA cell lines,
QBC939 and FRH0201, the IHCC cell line, RBE, and the normal biliary
epithelium cell line, HIBEpic, using RT-qPCR and western blot
analyses. KLF4 was expressed at prominent levels in the FRH0201 and
RBE cells, although its expression was only at a relatively low
level in the QBC939 cells (Fig.
2A). Consequently, the QBC939 cells and FRH0201 cells were used
for further functional analyses. The expression of KLF4 was knocked
down in the FRH0201cells, whereas KLF4 overexpression was induced
in the QBC939 cells. As shown in Fig.
2B and C, the knockdown or overexpression efficiency was
detected using RT-qPCR and western blot analyses. CCK-8 and EdU
assays subsequently revealed that the knockdown of KLF4 expression
promoted cell proliferation (Fig. 2D
and F) in the shKLF4 group, whereas KLF4 overexpression led to
a marked suppression of cell proliferation (Fig. 2E and G) in the KLF4 group compared
with the control and NC groups. Taken together, these results
suggest that KLF4 plays an essential role in the proliferation of
pCCA cells.
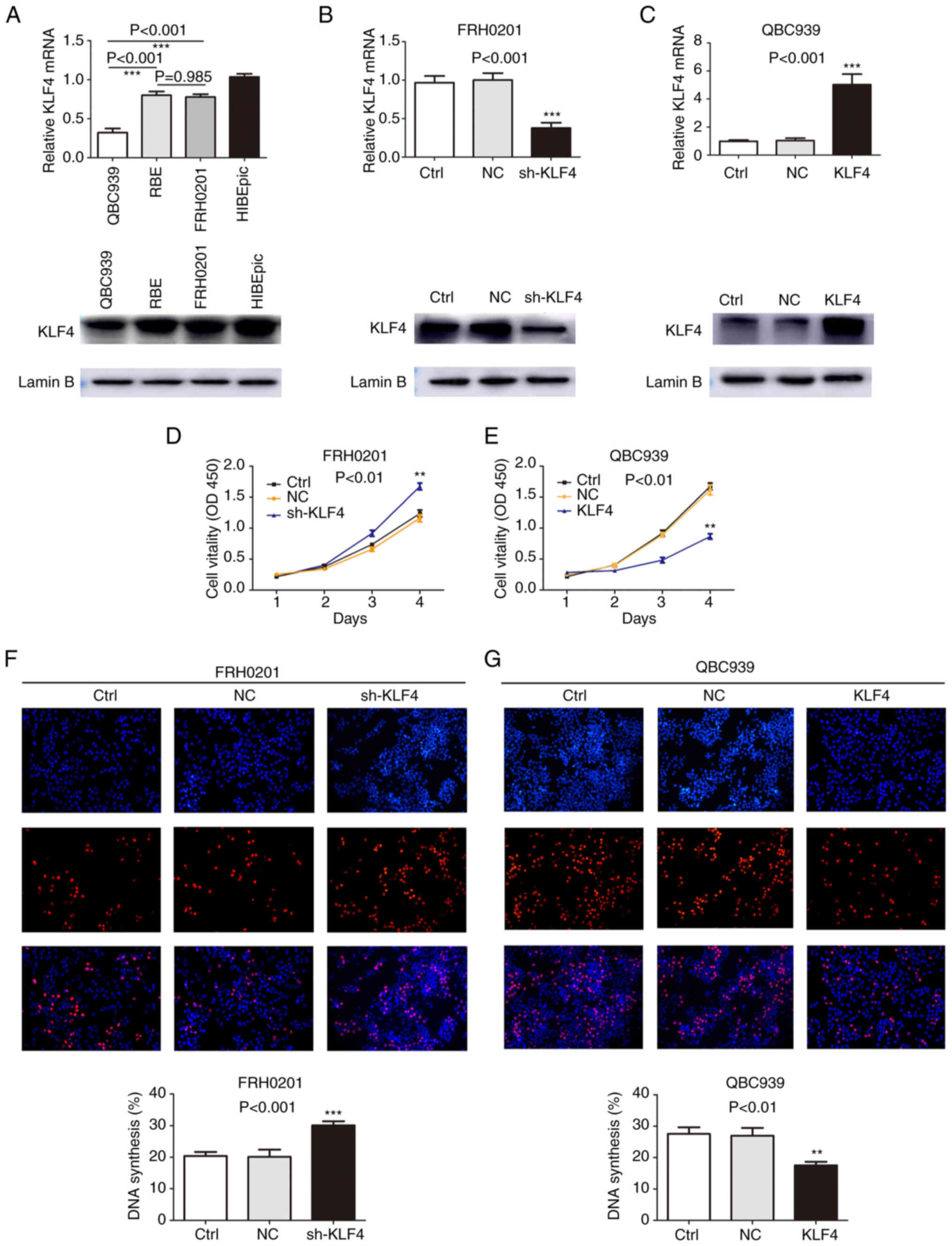 | Figure 2.KLF4 suppresses the proliferation of
pCCA cells. (A) KLF4 expression levels in the human pCCA cell
lines, QBC939, RBE and FRH0201, and the human intrahepatic biliary
epithelial cells (HIBEpiC) were detected using RT-qPCR and western
blot analyses. Lamin B served as the loading control in the western
blot analysis experiments. (B) Knockdown of KLF4 in FRH-0201 cells
with lentivirus infection was verified using RT-qPCR and western
blot analyses. (C) The efficiency of KLF4 overexpression using the
designated plasmid was verified using RT-qPCR and western blot
analyses. (D and F) CCK-8 and EdU assays were used to detect the
proliferation of FRH0201 cells, wherein KLF4 was knocked down. (E
and G) QBC939 cell proliferation was analyzed using CCK-8 and EdU
assays following transfection with the KLF4 overexpression plasmid.
**P<0.01 and ***P<0.001, vs. control group, without
transfection. Ctrl, control; NC group, transfected with a
non-coding shRNA; shKLF4 group, transfected with the KLF4-specific
shRNA; KLF4 group, transfected with the KLF4-over-expression
plasmid. KLF4, Krüppel-like factor 4; pCCA, perihilar
cholangiocarcinoma; RT-qPCR, reverse transcription-quantitative
PCR. |
Knockdown of KLF4 expression
suppresses apoptosis
The inhibition of apoptosis is also crucially
involved in tumor development, in addition to the deregulated
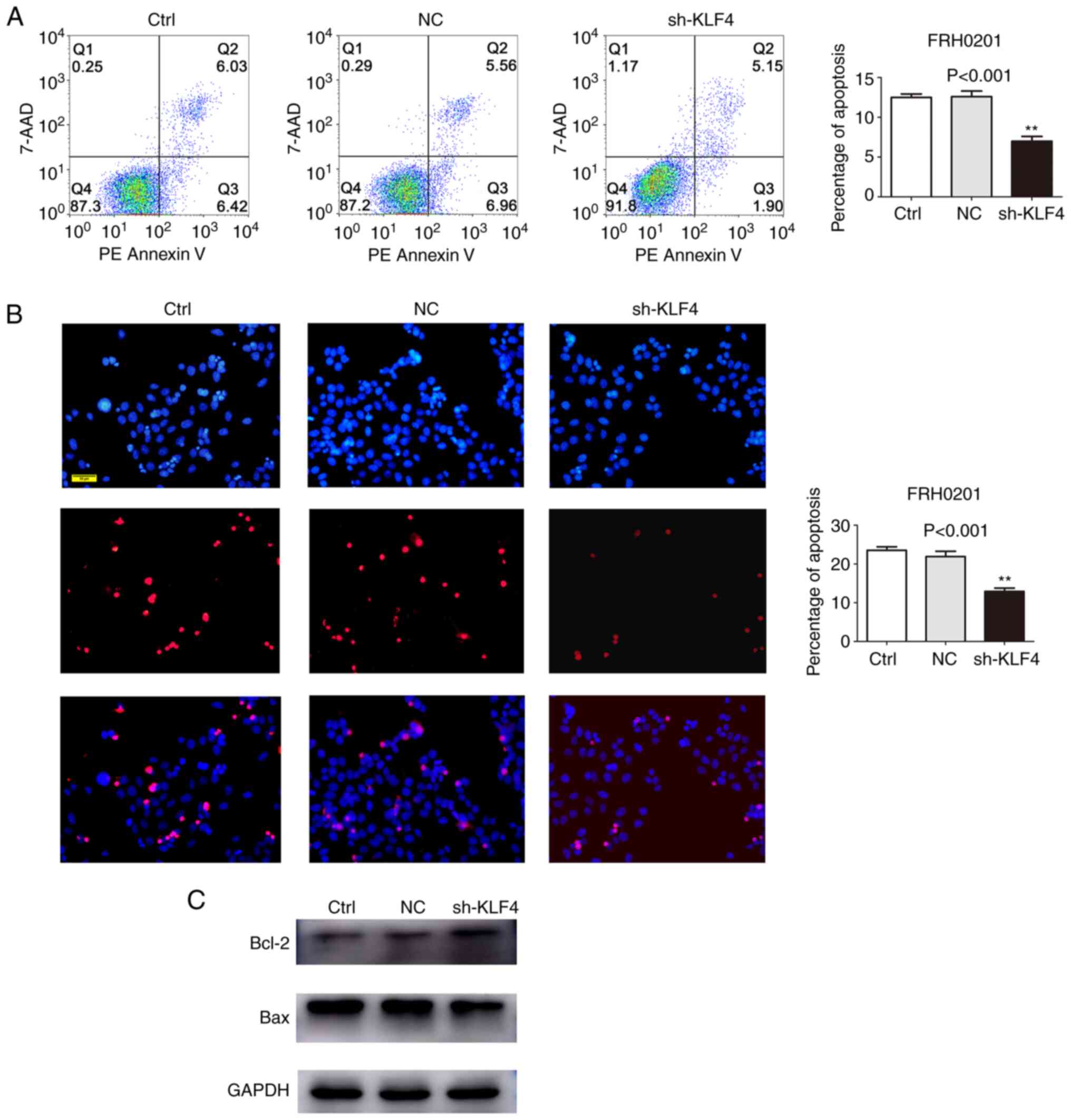
proliferation of cancer cells (30). Therefore, in the present study, flow
cytometric analysis was performed to assess the percentage of
apoptotic FRH0201cells in the shKLF4 group using Annexin V-PE
staining. The silencing of KLF4 was shown to suppress FRH0201 cell
apoptosis (control group, 12.45%; NC group, 12.52%; shKLF4 group,
7.05%; P<0.001; Fig. 3A),
findings that were in accordance with the results of the TUNEL
assays (P<0.001; Fig. 3B). These
findings were additionally supported by the changes in the levels
of biomarker proteins associated with apoptosis. Western blot
analysis demonstrated that the level of Bcl-2 was higher and the
level of Bax was lower, in the shKLF4 group compared with the
control and NC groups (Fig.
3C).
Inverse association between the
expression levels of KLF4 and GDF15 in pCCA
By using the GEO2R online tool from the Gene
Expression Omnibus (GEO), a total of 503 differentially expressed
genes (DEGs) were identified (of which 195 were upregulated and 308
were downregulated DEGs) in the GSE26566 dataset, and 455 DEGs (27
upregulated and 428 downregulated) were found in the GSE89749
dataset, which were differentially expressed between tumor samples
and adjacent normal tissues, as shown by the volcano plots in
Fig. S1A and B (a threshold
defined by a |log2FC|≥1.5 and P<0.05). Further analysis of these
DEGs using a Venn diagram (Fig.
S1C) revealed that there were 61 overlapping DEGs (7
upregulated and 54 downregulated DEGs) comparing between the two
datasets, as shown in Table
SI.
The hTFtarget and TRRUST databases were further
explored to obtain two KLF4 target datasets. A Venn diagram was
constructed to identify one gene at the intersection of two KLF4
target datasets and 61 overlapping DEGs, which was found to be
GDF15 (Fig. S1D).
On the basis of the aforementioned bioinformatics
analyses, it was hypothesized that GDF15 was a downstream target of
KLF4. Therefore, the expression levels of KLF4 and GDF15 were
further evaluated in the pCCA TMAs in order to confirm this
association. As shown in Fig. 4A,
the high expression of GDF15 mainly occurred in the tumor tissue
samples with a low KLF4 expression, whereas the tumor tissue
samples with a high expression of KLF4 had lower levels of GDF15.
Spearman's correlation analysis of the IHC scores demonstrated the
inverse association between KLF4 and GDF15 (Rho=−0.1917, P=0.0411;
Fig. 4B). This result was further
confirmed by the Pearson's correlation analysis of the expression
of KLF4 and GDF15 (data from RT-qPCR analysis of 20 pairs of fresh
tumor tissues) (r=−0.4836, P=0.0308; Fig. 4C).
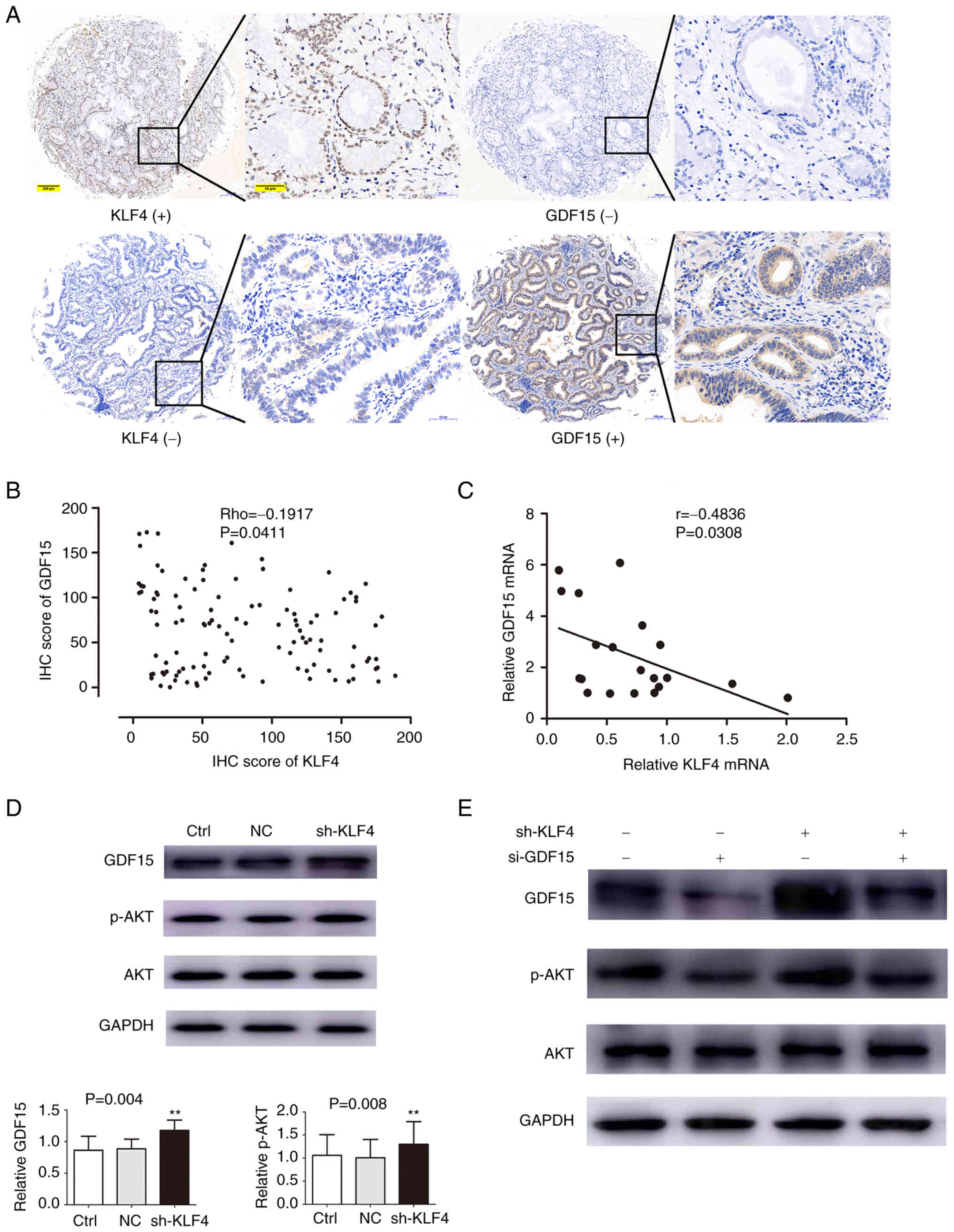 | Figure 4.GDF15 expression is negatively
associated with KLF4 expression in human pCCA tissue. KLF4
suppresses the protein expression of GDF15, thereby regulating the
AKT pathway. (A) GDF15 expression was low in KLF4-positive tissue
samples, and high in KLF4-negative tissue samples. Scale bars: Left
panels, 200 µm; right panels, 50 µm. (B) Correlation of IHC scores
for KLF4 and GDF15 in human pCCA tissues (r=−0.2124, P=0.0233). (C)
Correlation of mRNA expression for KLF4 and GDF15 in human pCCA
tissues (r=−0.4836, P=0.0308). (D) Variations in the levels of
GDF15 and p-AKT were examined using western blot analysis following
the knockdown of KLF4. (E) Western blot analysis revealed that the
increase in p-AKT expression due to knockdown of KLF4 could be
circumvented by si-GDF15. **P<0.01, vs. control group, without
transfection. Ctrl group, control group, without transfection; NC
group, transfected with a non-coding shRNA; shKLF4 group,
transfected with the KLF4-specific shRNA; si-GDF15 group, siRNA
targeting GDF15; KLF4, Krüppel-like factor 4; pCCA, perihilar
cholangiocarcinoma; GDF15, growth/differentiation factor 15; KLF4,
Krüppel-like factor 4. |
Knockdown of KLF4 promotes the
expression of GDF15 and phosphorylation of AKT
To further explore the direct correlation between
KLF4 and GDF15, the effects of KLF4 on GDF15 expression in human
pCCA FRH-0201cells were examined. Furthermore, changes in the
levels of AKT and p-AKT were also detected, since they are the
downstream targets of GDF15 (26).
These experiments revealed that the silencing of KLF4 led to a
significant increase in the levels of GDF15 and p-AKT (Fig. 4D). In the cells in which the
expression of GDF15 was interfered with, the promoting effects of
the silencing of KLF4 on p-AKT were found to be attenuated
(Fig. 4E). Taken together, these
results suggested that KLF4 adjusted and regulated the AKT
signaling pathway by negatively regulating GDF15, whereas the
overexpression of GDF15 mediated by a deficiency of KLF4 may
contribute to pCCA tumor carcinogenesis and development.
Knockdown of KLF4 expression promotes
tumor growth in vivo
As shown in Fig. 5A and
C, the knockdown of KLF4 expression led to a marked promotion
of tumor growth compared with NC transfection in the animal models.
In addition, the average tumor weight was lower in the NC group
compared with the shKLF4 group (242.2±35.94 vs. 548.60±48.61 mg;
Fig. 5B and Table SII). As shown by the results of
western blot analysis and IHC, the knockdown of KLF4 led to an
increase in the expression of GDF15 in the in vivo
experiment (Fig. 5D and E).
Furthermore, the level of p-AKT pertaining to the in vivo
experiments was also measured using western blot analysis; the
results revealed that the level of p-AKT was found to have
increased, along with the augmentation of the GDF15 signal in the
shKLF4group (Fig. 5D). These
results were in accordance with those obtained in the in
vitro experiments, further confirming that the knockdown of
KLF4 expression promoted the progression of pCCA.
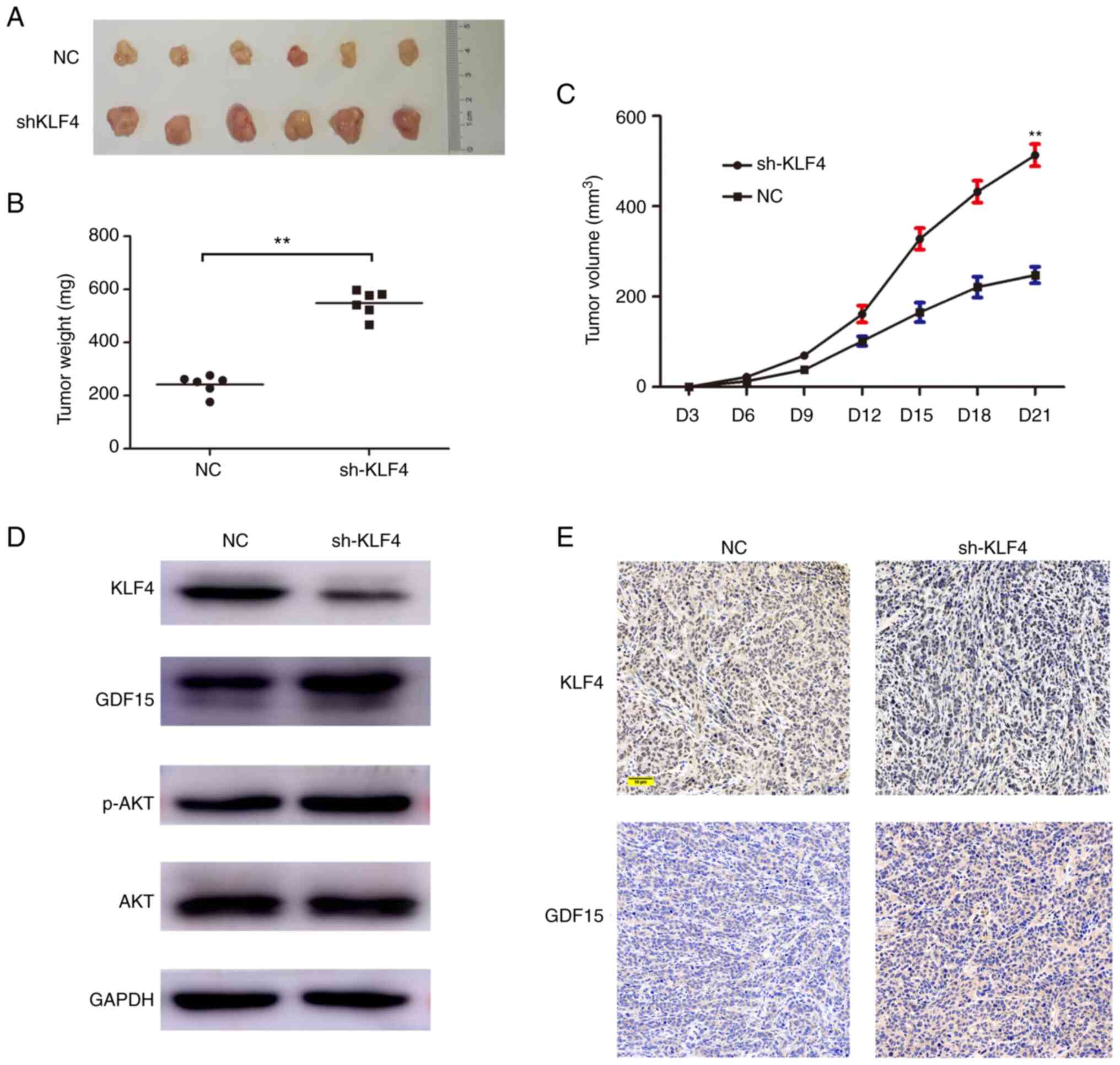 | Figure 5.KLF4 inhibits the growth of HCCA
xenograft tumors. (A) Stable shKLF4 cells and control cells
(transfected with empty vector) were injected subcutaneously into
the right flanks of BALB/c nude mice. At 3 weeks after
implantation, xenograft tumors were observed. (B) Tumor volumes
were measured every 3 days until the mice were euthanized on day
21. (C) Tumor weights were measured on day 21. (D) The expression
levels of KLF4, GDF15, AKT and p-AKT in tumors were measured using
western blot analysis. (E) Immunohistochemical staining revealed
the association between KLF4 and GDF15 in xenograft tumors. Scale
bar, 50 µm. **P<0.01, vs. control. Ctrl, control group without
transfection; NC group, transfected with a non-coding shRNA; shKLF4
group, transfected with the KLF4 specific shRNA; KLF4, Krüppel like
factor 4; pCCA, perihilar cholangiocarcinoma; GDF15,
growth/differentiation factor 15; p, phosphorylated. |
Discussion
Despite the recent developments in early diagnosis
and individual therapy, CCA still remains a highly lethal tumor
(31). Moreover, pCCA is the most
common subtype of CCA, which is associated with the worst prognosis
(32). This is partly due to the
lack of novel biomarkers or molecular profiles and targeted drugs
for pCCA. Therefore, there is an urgent need to unravel the
underlying molecular mechanisms of the oncogenesis and progression
of pCCA.
In the present study, the expression levels and
potential roles of KLF4 and GDF15 in human pCCA were determined. It
was confirmed that the expression level of KLF4 was decreased in
pCCA tumor tissues and cell lines. Further analysis of the
association between the clinicopathological characteristics of
patients with pCCA and the expression levels of KLF4 using TMAs
revealed that the absence of KLF4 was closely associated with a
poor histological grade and a large tumor size, also predicting a
poor OS. Multivariate analysis indicated that KLF4 was an
independent favorable prognosis factor in patients with pCCA. In
addition, the cell proliferative ability was reinforced, and cell
apoptosis was suppressed, when KLF4 was knocked down using shRNA.
Finally, cell proliferation was shown to be suppressed when KLF4
was overexpressed.
KLF4 is a critical member of the KLF family. It
fulfills crucial roles in tumor emergence, progression, invasion
and metastasis (9,33). KLF4 was originally shown to be a
tumor suppressor (12,27); however, numerous studies have
demonstrated that it encodes a transcription factor that is
associated with both tumor suppression and oncogenesis (11,14,34){Rowland, 2005 #296}. Several
previously published studies have demonstrated that KLF4 expression
is decreased in tumor types, such as gastrointestinal cancers
(11), colorectal cancer (12), lung cancer (35) and so on (13,27),
and KLF4 overexpression is a predictor of an improved prognosis
(35,36). In accordance with these findings,
the results of the present study demonstrated that KLF4 expression
was decreased in pCCA compared with adjacent tissues. However,
tentative evidence has indicated that KLF4 may function as an
oncogene in primary breast cancer, prostate cancer and skin cancer
(14,15,37).
The role of KLF4 in terms of regulating tumors is dependent on the
different cellular contexts, the expression patterns of other
genes, and so on (34,38,39).
KLF4 can execute its tumor suppressive functions directly or
indirectly by regulating the cell cycle, as an anti-apoptosis
molecule, and via the Wnt, Notch and TGF-β signaling pathways, and
so on (8,9,40,41).
GDF15 is usually expressed abundantly under conditions of stress,
inflammation, diabetes, cardiovascular disorder, cancer and so on
(42). However, the mechanisms
underlying its activation and regulation remains poorly understood.
It has been shown that AKT signaling is associated with cell
proliferation and survival (anti-apoptosis) (43). A disruption in the balance between
cell proliferation and survival leads to the development and
progression of cancer (26,30). Moreover, it has been confirmed that
GDF15 is able to activate AKT when the balance between cell
proliferation and survival is disrupted (26). In the present study, the expression
of GDF15 in TMAs was further investigated. The results obtained
demonstrated that the pCCA specimens in the TMA analysis, which
lost their KLF4 expression, had a high expression of GDF15.
Spearman's correlation analysis confirmed the negative association
between them. This association was also confirmed using RT-qPCR
analysis. Moreover, bioinformatics analyses revealed that GDF15 was
identified as the only DEG that targeted KLF4 in pCCA. Therefore,
it can be hypothesized that GDF15 is a key downstream target of
KLF4 that functions as a transcription factor in pCCA, as
demonstrated herein. In the present study, further experiments
indicated that the silencing KLF4 increased both the expression of
GDF15 and the phosphorylation of AKT in vitro. Moreover, the
knockdown of GDF15 attenuated the suppressive effects of KLF4 on
the AKT pathway. These findings indicated that KLF4, as a
transcription factor, executes its tumor-suppressive role by
regulating the GDF15/AKT signaling pathway.
In conclusion, the present study demonstrated that
the loss of KLF4 in human pCCA results in the overexpression of
GDF15, thereby leading to the active phosphorylation of AKT and
subsequent tumor progression. The findings of the present study not
only provide a novel molecular mechanism of pCCA tumor progression,
but the KLF4/GDF15/AKT signaling pathway has also been identified
as a potential novel molecular target for controlling pCCA tumor
development. KLF4 itself may prove to be a useful biomarker for
predicting the prognosis of patients with pCCA.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ, CL and HZ were involved in the conception and
design of the study. XZ and CL performed the research. XZ and WW
were involved data analysis and interpretation. All authors were
involved in the writing of the manuscript. XZ, CL and HZ confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
For the use of human samples, the present study was
approved by the Medical Ethics Committee of Linyi People's Hospital
(approval no. YX200626). Written informed consent was obtained from
all patients. All animal experiments were approved by the Medical
Ethics Committee of Linyi People's Hospital. They were executed in
accordance with the guidelines for the use and care of laboratory
animals provided by Linyi People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brindley PJ, Bachini M, Ilyas SI, Khan SA,
Loukas A, Sirica AE, Teh BT, Wongkham S and Gores GJ:
Cholangiocarcinoma. Nat Rev Dis Primers. 7:652021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Banales JM, Marin JJG, Lamarca A,
Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen
JB, Braconi C, et al: Cholangiocarcinoma 2020: The next horizon in
mechanisms and management. Nat Rev Gastroenterol Hepatol.
17:557–588. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krasinskas AM: Cholangiocarcinoma. Surg
Pathol Clin. 11:403–429. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rizvi S and Gores GJ: Pathogenesis,
diagnosis, and management of cholangiocarcinoma. Gastroenterology.
145:1215–1229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khan AS and Dageforde LA:
Cholangiocarcinoma. Surg Clin North Am. 99:315–335. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deoliveira ML, Cunningham SC, Cameron JL,
Kamangar F, Winter JM, Lillemoe KD, Choti MA, Yeo CJ and Schulick
RD: Cholangiocarcinoma: Thirty-one-year experience with 564
patients at a single institution. Ann Surg. 245:755–762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Groot Koerkamp B, Wiggers JK, Gonen M,
Doussot A, Allen PJ, Besselink MG, Blumgart LH, Busch OR,
D'angelica MI, DeMatteo RP, et al: Survival after resection of
perihilar cholangiocarcinoma-development and external validation of
a prognostic nomogram. Ann Oncol. 27:7532016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ghaleb AM and Yang VW: Krüppel-like factor
4 (KLF4): What we currently know. Gene. 611:27–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo X, Zhang Y, Meng Y, Ji M and Wang Y:
Prognostic significance of KLF4 in solid tumours: An updated
meta-analysis. BMC Cancer. 22:1812022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN
and Wu M: Down-regulation of gut-enriched Kruppel-like factor
expression in esophageal cancer. World J Gastroenterol. 8:966–970.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei D, Kanai M, Huang S and Xie K:
Emerging role of KLF4 in human gastrointestinal cancer.
Carcinogenesis. 27:23–31. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao W, Hisamuddin IM, Nandan MO, Babbin
BA, Lamb NE and Yang VW: Identification of Krüppel-like factor 4 as
a potential tumor suppressor gene in colorectal cancer. Oncogene.
23:395–402. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ohnishi S, Ohnami S, Laub F, Aoki K,
Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F and Yoshida T:
Downregulation and growth inhibitory effect of epithelial-type
Krüppel-like transcription factor KLF4, but not KLF5, in bladder
cancer. Biochem Biophys Res Commun. 308:251–256. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pandya AY, Talley LI, Frost AR, Fitzgerald
TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA,
Krontiras H, et al: Nuclear localization of KLF4 is associated with
an aggressive phenotype in early-stage breast cancer. Clin Cancer
Res. 10:2709–2719. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Z, Zhang Y, Chen X, Wu P and Chen D:
Long non-coding RNA LINC00673 silencing inhibits proliferation and
drug resistance of prostate cancer cells via decreasing KLF4
promoter methylation. J Cell Mol Med. 24:1878–1892. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao C, Li Y, Qiu W, He F, Zhang W, Zhao
D, Zhang Z, Zhang E, Ma P, Liu Y, et al: C5a induces A549 cell
proliferation of non-small cell lung cancer via GDF15 gene
activation mediated by GCN5-dependent KLF5 acetylation. Oncogene.
37:4821–4837. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li S, Ma YM, Zheng PS and Zhang P: GDF15
promotes the proliferation of cervical cancer cells by
phosphorylating AKT1 and Erk1/2 through the receptor ErbB2. J Exp
Clin Cancer Res. 37:802018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Q, Xu HX, Li JP, Wang S, Fu Z, Jia J,
Wang L, Zhu ZF, Lu R and Yao Z: Growth differentiation factor 15
induces growth and metastasis of human liver cancer stem-like cells
via AKT/GSK-3β/β-catenin signaling. Oncotarget. 8:16972–16987.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin Y, Jung SN, Lim MA, Oh C, Piao Y, Kim
HJ, Liu L, Kang YE, Chang JW, Won HR, et al: Transcriptional
regulation of GDF15 by EGR1 promotes head and neck cancer
progression through a positive feedback loop. Int J Mol Sci.
22:111512021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dong G, Huang X, Jiang S, Ni L, Ma L, Zhu
C and Chen S: SCAP mediated GDF15-induced invasion and EMT of
esophageal cancer. Front Oncol. 10:5647852020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Joshi JP, Brown NE, Griner SE and Nahta R:
Growth differentiation factor 15 (GDF15)-mediated HER2
phosphorylation reduces trastuzumab sensitivity of
HER2-overexpressing breast cancer cells. Biochem Pharmacol.
82:1090–1099. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang M, Narita S, Koizumi A, Nara T,
Numakura K, Satoh S, Nanjo H and Habuchi T: Macrophage inhibitory
cytokine-1 induced by a high-fat diet promotes prostate cancer
progression by stimulating tumor-promoting cytokine production from
tumor stromal cells. Cancer Commun (Lond). 41:389–403. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan Y, Yue X, Yang S, Zhao Z, Wu P and Liu
H: The expression of growth differentiation factor 15 in
gallbladder carcinoma. J BUON. 26:218–228. 2021.PubMed/NCBI
|
|
24
|
Li C, Wang J, Kong J, Tang J, Wu Y, Xu E,
Zhang H and Lai M: GDF15 promotes EMT and metastasis in colorectal
cancer. Oncotarget. 7:860–872. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Baek SJ and Eling TE: The diverse
roles of nonsteroidal anti-inflammatory drug activated gene
(NAG-1/GDF15) in cancer. Biochem Pharmacol. 85:597–606. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fang L, Li F and Gu C: GDF-15: A
multifunctional modulator and potential therapeutic target in
cancer. Curr Pharm Des. 25:654–662. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Y, Goldstein BG, Chao HH and Katz JP:
KLF4 and KLF5 regulate proliferation, apoptosis and invasion in
esophageal cancer cells. Cancer Biol Ther. 4:1216–1221. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li JC, Chen QH, Jian R, Zhou JR, Xu Y, Lu
F, Li JQ and Zhang H: The partial role of KLF4 and KLF5 in
gastrointestinal tumors. Gastroenterol Res Pract. 2021:24253562021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang XM, Liu ZL, Qiu B, Xu YF, Pan C and
Zhang ZL: Downregulation of EVI1 expression inhibits cell
proliferation and induces apoptosis in hilar cholangiocarcinoma via
the PTEN/AKT signalling pathway. J Cancer. 11:1412–1423. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo K, Cui J, Quan M, Xie D, Jia Z, Wei D,
Wang L, Gao Y, Ma Q and Xie K: The novel KLF4/MSI2 signaling
pathway regulates growth and metastasis of pancreatic cancer. Clin
Cancer Res. 23:687–696. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rowland BD, Bernards R and Peeper DS: The
KLF4 tumour suppressor is a transcriptional repressor of p53 that
acts as a context-dependent oncogene. Nat Cell Biol. 7:1074–1082.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu W, Hofstetter WL, Li H, Zhou Y, He Y,
Pataer A, Wang L, Xie K, Swisher SG and Fang B: Putative
tumor-suppressive function of Kruppel-like factor 4 in primary lung
carcinoma. Clin Cancer Res. 15:5688–5695. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Patel NV, Ghaleb AM, Nandan MO and Yang
VW: Expression of the tumor suppressor Krüppel-like factor 4 as a
prognostic predictor for colon cancer. Cancer Epidemiol Biomarkers
Prev. 19:2631–2638. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC and
Chen CM: Nuclear Krüppel-like factor 4 expression is associated
with human skin squamous cell carcinoma progression and metastasis.
Cancer Biol Ther. 7:777–782. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hu W, Jia Y, Xiao X, Lv K, Chen Y, Wang L,
Luo X, Liu T, Li W, Li Y, et al: KLF4 downregulates hTERT
expression and telomerase activity to inhibit lung carcinoma
growth. Oncotarget. 7:52870–52887. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rowland BD and Peeper DS: KLF4, p21 and
context-dependent opposing forces in cancer. Nat Rev Cancer.
6:11–23. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ghaleb AM, McConnell BB, Kaestner KH and
Yang VW: Altered intestinal epithelial homeostasis in mice with
intestine-specific deletion of the Krüppel-like factor 4 gene. Dev
Biol. 349:310–320. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cui J, Shi M, Quan M and Xie K: Regulation
of EMT by KLF4 in gastrointestinal cancer. Curr Cancer Drug
Targets. 13:986–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Modi A, Purohit P, Roy D, Vishnoi JR,
Pareek P, Elhence P, Singh P, Sharma S, Sharma P and Misra S: FOXM1
mediates GDF-15 dependent stemness and intrinsic drug resistance in
breast cancer. Mol Biol Rep. 49:2877–2888. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|