Introduction
Piwi-like RNA-mediated gene silencing 2
(PIWIL2) gene has been identified as a common mediator for
the development of various types of cancers. It is activated by an
intragenic promoter, resulting in a tumorigenic variant called
PIWIL2-like protein 60 (PL2L60) (1–6). Piwi
proteins, a subfamily of the Argonaute family, are associated with
Piwi-interacting RNAs (piRNAs), a class of small non-coding RNAs
(snRNAs) and are involved in the biogenesis, transport, and use of
these snRNAs (7,8). The Piwil2 gene, alias
Mili in mouse or Hili in human, is inclined to
produce a full length of Piwil2, which binds piRNAs to form
pi-ribonucleoproteins (piRNPs) and initiates transposon silencing
in the germ line (9–11). In adult cells, PIWIL2 gene is
usually silenced but activated upon DNA damage, mediating DNA
repair or promoting cell apoptosis in case that DNA repair is
failed (12).
Alternative gene splicing provides a means by which
cells generate proteins with different properties from a single
mRNA precursor (13,14). The alternative splicing (AS)
produces transcript ‘isoforms’ for most human genes, providing
functional diversity at the level of enzymatic activities and
subcellular localizations, as well as protein-protein, protein-DNA
and protein-ligand physical interactions (15,16).
However, it has been recently found that an isoform of
PIWIL2 gene is not generated from AS of mRNA but derived
from alternative activation of an intragenic promoter (ITP),
resulting in a truncated, alienated product PL2L60 (2,4,5). While
a full length of Piwil2 protein may serve as a tumor barrier
(12), PL2L60 depleted of Piwil2
exon 1–5 is preferentially expressed in various types of human
cancer cells, promoting tumorigenesis and tumor growth (4,12).
However, little is known for the mechanisms that regulate PL2L60
protein expression in cancer cells.
Hypoxia is an intrinsic stress occurring within the
tumor microenvironment due to the rapid growth of cancer cells,
poorly formed neoangiogenic blood vessels, or even radiotherapy and
chemotherapy-induced ischemia (17,18).
The cells deprived of oxygen will initially employ adaptive and
survival strategies (19), but cell
death will eventually occur if hypoxia is sustained (20). The precise mechanisms of
hypoxia-induced cell death remain unclear as apoptosis, necrosis
and autophagy have all been reported in response to hypoxic stress
(21). Since PL2L60 plays a
critical role for the survival and proliferation of cancer cells,
it was hypothesized that hypoxia-induced cell death might be
associated with degradation of PL2L60 proteins mediated by
autophagy.
Autophagy is a catabolic process that maintains
cellular homeostasis through the degradation of cellular
constituents and the generation of basic building blocks for the
synthesis of new macromolecules (22). Autophagy is best characterized to be
induced under stressful conditions, such as organelle damage,
hypoxia or nutrient deprivation, and is followed by the elongation
of the autophagosome membrane around its cargo (23). The ubiquitin-like protein Atg8,
which is conjugated to phosphatidylethanolamine (PE), associates
with outer and inner membranes of autophagic structures (24). Autophagosomes eventually fuse with
lysosomes to degrade their content. The autophagic process requires
a set of evolutionarily conserved proteins, most of which are known
as autophagy-related (ATG) proteins, functioning at different
phases of autophagy formation (24). Initially described as a key survival
feature for cancer cells, autophagy has raised great attention for
its potential ability to promote cell death (25,26).
While autophagosomes were initially identified in dying cells, a
phenomenon that led to the term ‘autophagic cell death’ to describe
a cell death mode is distinct from apoptosis (26). Therefore, autophagy plays a critical
role in maintaining cellular homeostasis and is regarded as a
double-edged sword. While autophagy primarily acts in a
cyto-protective manner, the dysregulated states could result in
compromised cell function and even death (19,20,25–28).
Previously, it was verified by the authors that severe hypoxia
could induce autophagic cell death (28). Since PL2L60 is associated with tumor
cell proliferation (2), it is
possible that decreased PL2L60 might play a key role in the
hypoxia-induced autophagic cell death.
In the present study, the involvement of PL2L60 in
hypoxia-induced autophagic cancer cell death was investigated using
in vitro and in vivo models.
Materials and methods
Cell lines and reagents
Human cancer cell lines including breast [MDA-MB-231
(cat. no. HTB-26™), MDA-MB-468 (cat. no. HTB-132™), MCF-7 (cat. no.
HTB-22™), T-47D (cat. no. HTB-133™) and SKBR3 (cat. no. HTB-30™)],
lung [A549 (cat. no. CCL-185™)], colon [HCT116 (cat. no.
CCL-247™)], cervix [HeLa (cat. no. CCL-2™)], prostate [PC-3 (cat.
no. CRL-1435™)] and liver [HepG2 (cat. no. HB-8065™)] were obtained
from the American Type Culture Collection. The cells were cultured
in Dulbecco's modified Eagle medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum and 100
U/ml penicillin-streptomycin (Gibco; Thermo Fisher Scientific,
Inc.). The cells were maintained in a humidified incubator
containing 5% CO2 and air at 37°C. Cobaltic chloride
(CoCl2; cat. no. C8661; Sigma-Aldrich; Merck KGaA) was
dissolved in water to create 40 mmol/l stock solutions and stored
at −20°C. Rapamycin (cat. no. HY-10219; MedChemExpress) was
dissolved in DMSO to create 10 mmol/l stock solutions and stored at
−20°C.
Transfection of small interfering RNA
(siRNA)
Non-targeting control siRNA and targeting siRNA were
purchased from Shanghai GenePharma Co., Ltd. The siRNA sequences
were as follows: BECN1: 5′-UGUGAAUGGAAUGAGAUUATT-3′;
ATG5: 5′-CCAUCAAUCGGAAACUCAUTT-3′; Piwil2-siE5
(Piwil2 full length): 5′-GCCTGTTAAGCTTCAACAATT-3′;
Piwil2-siE21 (PL2L60): 5′-CUAUGAGAUUCCUCAACUACAGAAG-3′;
HIF-1α: 5′-GGAAAUGAGAGAAAUGCUUTT-3′; microtubule-associated
protein 1 light chain 3 (LC3): 5′-CUCCCUAAGAGGAUCUUUATT-3′,
p62: 5′-GGAGUCGGAUAACUGUUCATT-3′; and negative control:
5′-UUCUCCGAACGUGUCACGUTT-3′. After breast cancer (MDA-MB-231), lung
cancer (A549), or cervix cancer (HeLa) cancer cells being seeded at
2.0–2.4×105 per well in six-well plates overnight, siRNA
duplexes (50 pmol) were transfected into the target cell
populations using Lipofectamine® 2000 Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) for 6 h according to
the manufacturer instructions. After 48 h of transfection, cells
were used for subsequent experiments.
Cell viability assay
The siRNA transfected cells (3×103
cells/well) were initially seeded in 96-well flat-bottomed plates
in quintuplicate for another 24 h. Viable proliferating cells were
detected using the Cell Counting Kit-8 (Dojindo Laboratories,
Inc.), according to manufacturer's instruction. CCK-8 reaction
mixture for each well in one 96-wells plate consisted of 100 µl
DMEM and 10 µl CCK-8 reagent and was incubated for 30 min at 37°C.
The OD value for each well was measured on a Synergy HT microplate
reader (BioTek Instruments, Inc.) at 450 nm wavelength. Cell
viability was calculated with the following formula: Inhibition
percentage (%)=(control siRNA transfected sample-targeted siRNA
transfected sample)⁄(control siRNA transfected sample) ×100%.
Apoptosis assays
Apoptosis was detected by flow cytometric analysis
following PI/Annexin Vdouble staining (cat. no. 640932; Biolegend,
Inc.). The ratio of cells in early apoptosis and late apoptosis
were calculated, and statistical analysis of the data was
performed. All flow cytometric analysis was performed on a BD C6
flow cytometer (BD Biosciences). Data were analyzed using FlowJo
software version 7.6.1 (Tree Star Inc.).
Western blot analysis
Cells were lysed in pre-cold
radioimmunoprecipitation (RIPA) assay buffer (Thermo Fisher
Scientific, Inc.) containing 1X protease inhibitor cocktail (Roche
Diagnostics) on ice. Protein concentration in the soluble lysates
was quantified using a bicinchoninic acid (BCA) protein assay.
Protein samples (20 µg) were separated by 8–12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred to polyvinylidene fluoride membranes (Millipore Sigma).
Membranes were blocked with 5% non-fat dry milk in TBST at room
temperature for 1 h, followed by overnight incubation with primary
antibodies at 4°C in EZ-Buffers N 1X BLOCK BSA in TBS (cat. no.
C500036; Sangon Biotech Co., Ltd.). Western blots were performed
using the following primary antibodies: anti-HIF-1α (1:500; cat.
no. YM0333; Immunoway Biotechnology Company), anti-LC3B (1:1,000;
cat. no. 3868; Cell Signaling Technology, Inc.), anti-Beclin-1
(1:1,000; cat. no. 3495; Cell Signaling Technology, Inc.),
anti-ATG5 (1:1,000; cat. no. 12994; Cell Signaling Technology,
Inc.), anti-p62 (1:2,000; cat. no. 18420-1-AP; Proteintech Group,
Inc.), anti-Piwil2/PL2L60 (1:1,000; generated in the authors' lab)
and anti-β-actin (1:3,000; cat. no. MA5-11869; Invitrogen, Thermo
Fisher Scientific, Inc.). PVDF membranes were washed and incubated
with HRP-conjugated rabbit (1:2,000; cat. no. 7074; Cell Signaling
Technology, Inc.) or mouse (1:3,000; cat. no. 7076; Cell Signaling
Technology, Inc.) secondary antibodies for 1 h at room temperature.
The membranes were visualized with Super Signal® West
Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific,
Inc.), and image acquisition was made with the ChemiDox™
XRS+ system. The differences in protein expression among
various treatments were determined using the image analysis
software ImageJ software version 1.8.0 (National Institutes of
Health), using β-actin as a loading control.
Nuclear and cytoplasmic
fractionations
Nuclear and cytoplasmic extracts were prepared using
NE-PER Nuclear and Cytoplasmic Extraction Reagents (cat. no. 78833;
Pierce, Thermo Fisher Scientific, Inc.). The quality of nuclear and
cytoplasmic extracts was verified by immunoblotting with protein
differentially enriched in the nucleus or the cytoplasm
(β-actin).
Lentiviral transduction
Lentiviral packaging plasmids psPAX2 and pMD2.VSV-G
were co-transfected with the puromycin resistant GFP-LC3 construct
(ChenDu Transvector Biotechnology Co., Ltd.) into 293T cells for
virus production at 37°C under 5% CO2. Briefly, 293T
cells were plated in 10-cm dishes and transfected with GFP-LC3
construct, psPAX2 and pMD2.VSV-G at a ratio of 4:3:1.
Virus-containing media were collected 72 h post-transfection and
used to infect MDA-MB-231 cells in the presence of 5 µg/ml
Polybrene. After 24 h of infection, the infected cells were
selected with puromycin (5 µg/ml) for one week to establish stable
GFP-LC3 expression cells. The cells were further passaged for 1–2
generations before experiments.
Immunofluorescence and imaging
MDA-MB-231 cells stably transfected by LC3-GFP at
50% confluence were fixed with 4% paraformaldehyde (Beijing Dingguo
Changsheng Biotechnology Co., Ltd.) for 15 min at room temperature
followed by permeabilization with 0.25% (w/v) Triton X-100
(MilliporeSigma). Cells were then blocked for 1 h in 3% BSA at room
temperature and incubated overnight with primary antibody (Piwil2;
1:50) at 4°C. Then cells were incubated with secondary antibodies
conjugated with Alexa Fluor® 555 conjugate (Thermo
Fisher Scientific, Inc.) at room temperature for 1 h followed by
three washes with PBS. After washing, the slides were
counterstained with 4′6′-diamidino-2-phenylindole dihydrochloride
(DAPI; 2 µg/ml) (Sigma-Aldrich; Merck KGaA) for 5 min. Images were
acquired by using 20X Ti-S fluorescence scanning microscope (Nikon
Corporation) objective.
Animal experiments
Nude mice (total number, 16; 50% male and 50%
female; weight, ~23 g) were obtained from SLACCAS (Shanghai
Laboratory Animal Center). Mice were bred in the authors' animal
pathogen-free facility (SPF) and maintained under standard
conditions according to the institutional guidelines of Renji
Hospital animal care and ethics review committee (Shanghai, China).
The temperature of SPF animal room was maintained at 20–26°C
(maximum temperature difference <4°C), the relative humidity at
40–70%, the noise was kept at <60 decibels, the ammonia
concentration <14 mg/m3, the ventilation >15 times/h, the
light/dark cycle condition was 12/12-h, and the bottom of the cage
was placed with autoclave corn cob as bedding material. The feeding
density was ≤5 animals per cage. The drinking water was sterilized
by the animal drinking water system. The feed was special sterile
pellets for mice, which were checked twice a day. Feed, drinking
water and cage were changed once a week. Nude mice were used at age
of 6–8 week. All in vivo experiments were approved by the
institutional guidelines of Renji Hospital animal care and ethics
review committee. 1×107 breast cancer MDA-MB-231 or
cervical cancer HeLa cells were suspended in 200 µl of PBS and then
subcutaneously injected into nude mice, and the mice were monitored
for up to 4 weeks. Tumor volume was measured using a caliper every
three days and calculated using the formula: V
(mm3)=0.5× length × width2. Afterwards, the
mice were humanely euthanized via CO2 asphyxiation with
a flow rate displacing 30% of the chamber volume per min when the
tumor diameter reached 2 cm, and each tumor was confirmed by
routine H&E staining of paraffin sections.
Immunohistochemistry (IHC)
Paraffin-embedded tissues were sectioned (5 µm
thickness) and stained with H&E or stored for further paraffin
or fluorescence IHC. Sections were incubated with target retrieval
solution (Dako; Agilent Technologies, Inc.) in a steamer for 45 min
followed by 3% hydrogen peroxide solution for 10 min and protein
block (Dako; Agilent Technologies, Inc.) for 20 min at room
temperature. Sections were incubated overnight in a humid chamber
at 4°C with the following antibodies (all diluted at a ratio of
1:500) against: HIF-1α (cat. no. ab82832; Abcam), LC3, p62, Ki-67
(cat. no. 9027; Cell Signaling Technology, Inc.), cleaved-caspase 3
(cat. no. 9664; Cell Signaling Technology, Inc.) and PL2L60
(generated in the authors' lab) followed by biotinylated secondary
antibody (1:1,000; Vector Laboratories, Inc.) for 30 min and ABC
reagent for 30 min. Immunocomplexes of horseradish peroxidase were
visualized by DAB reaction (Dako; Agilent Technologies, Inc.), and
sections were counterstained with haematoxylin before mounting.
Micrographs of stained sections were captured using a Leica DMIL
LED microscope with an Amscope camera and acquisition software.
Statistical analysis
All data are expressed as the mean ± standard
deviation (SD) and are representative of triplicate samples.
Statistical analysis was performed using GraphPad Prism 5 software
(Dotmatics). Statistical comparisons were made by using two-tailed
unpaired Student t-tests. P≤0.05 was considered to indicate a
statistically significant difference.
Results
PL2L60 is predominantly expressed in
various types of cancer cells but decreased upon oxygen
deprivation
It has been reported by the authors that PL2L60
protein, a variant of PIWIL2, which is alternatively activated by
its ITP, mediates tumorigenesis and tumor growth (2,4). The
PL2L60 expression is significantly inhibited by siRNA against exon
21 (siRNA-E21) but not by siRNA against exon 7 (siRNA-E7) of
PIWIL2 gene (4,5). The expression levels of PIWIL2 and
PL2L60 were therefore further analyzed in a number of human cancer
cell lines. As demonstrated in Fig. 1A
and B, almost all the tumor cell lines examined expressed
PL2L60 in a level markedly higher than PIWIL2. These lines included
cancer cells from breast cancer (MDA-MB-231, MDA-MB-468, MCF-7,
T-47D and SKBR3), lung cancer (A549), colon cancer (HCT116), cervix
cancer (HeLa), prostate cancer (PC3) and liver cancer (HepG2). By
contrast, all lines expressed little full length of PIWIL2,
consistently with the authors' previous observation (2). Since the growth rate of tumor is
always associated with blood or oxygen supply and nutrition supply,
it was investigated whether PL2L60 expression was affected by low
oxygen availability. A total of five different breast cancer cell
lines (MDA-MB-231, MDA-MB-468, MCF-7, T-47D and SKBR3) were exposed
to normoxia (21% O2) and hypoxia (1% O2) for
24 h, respectively, followed by PL2L60 protein expression
measurement by western blot analysis. After 24 h hypoxic exposure,
the PL2L60 protein level was markedly reduced (Fig. 2A). To determine whether the result
was universal or restricted to human breast cancer cells, human
cancer cell lines from other tissues were also examined, including
lung (A549), colon (HCT116), cervix (HeLa), prostate (PC3) and
liver (HepG2). The same results were observed after these cell
lines were exposed to normoxia or hypoxia for 24 h. As expected,
PL2L60 was markedly reduced in the hypoxic cells compared with the
cells exposed to normoxic condition (Fig. 2B). Kinetic analysis revealed that
the levels of PL2L60 began to decrease but slightly as early at 12
h after exposure to hypoxia; at 24 and 48 h, the expression of
PL2L60 was substantially reduced compared with the cells cultured
under normoxic condition (Fig.
2C-F). These data revealed that PL2L60 expression in various
types of cancer cells could be remarkably downregulated at late
stage of hypoxic responses.
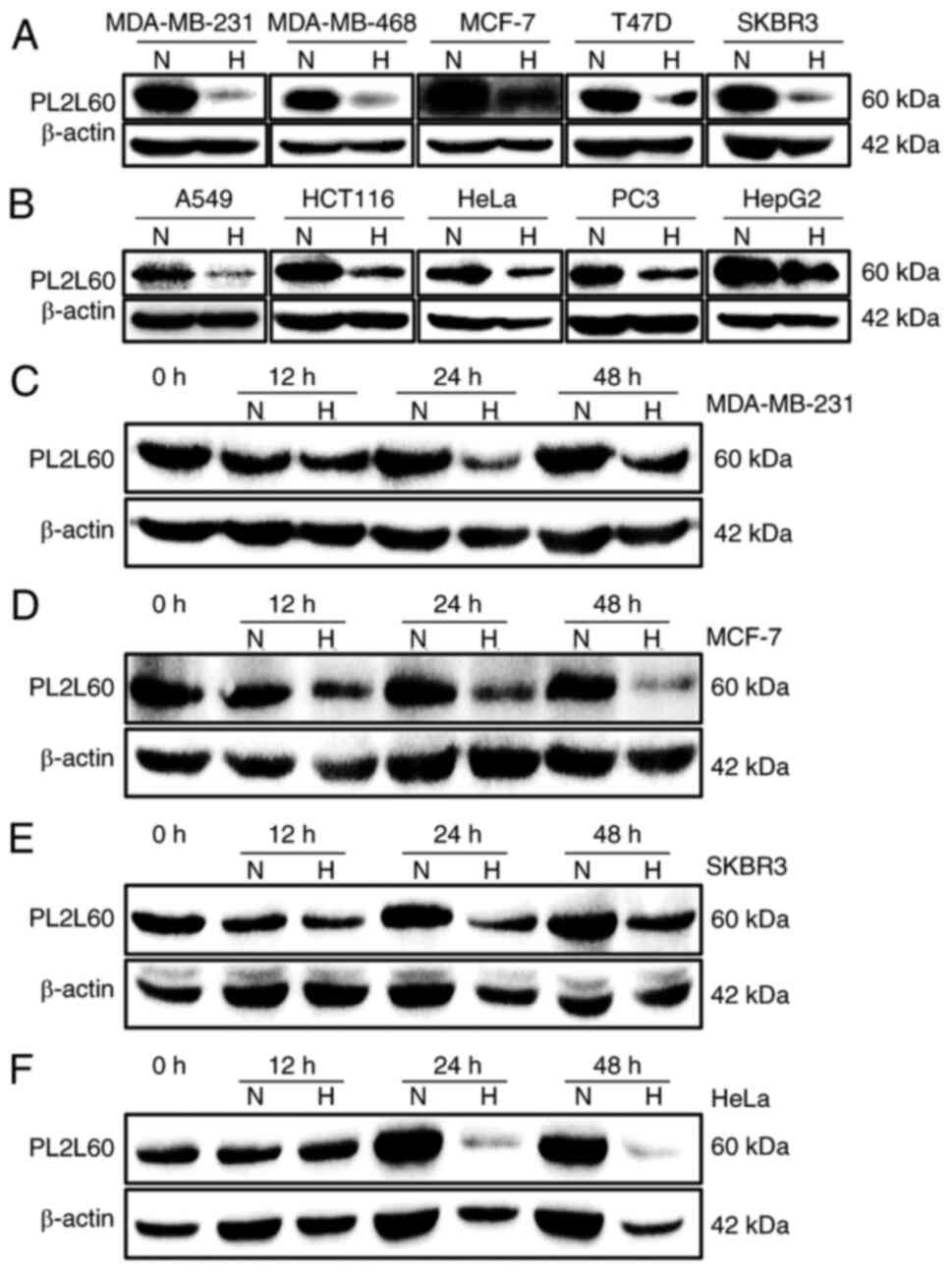 | Figure 2.Hypoxia suppresses PL2L60 protein
expression in various types of cancer cells. (A and B) The effects
of oxygen levels on PL2L60 expression in various types of cancer
cell lines grown in normoxia (N; 21% O2) or hypoxia (H;
1% O2) for 24 h. The cells were harvested and measured
for PL2L60 protein levels by western blotting. (A) Breast cancer
cells including MDA-MB-231, MDA-MB-468, MCF-7, T-47D and SKBR3. (B)
Cancer cell lines from various types of tissues including lung
(A549), colon (HCT116), cervix (HeLa), prostate (PC3) and liver
(HepG2). (C-F) Kinetics of hypoxia-induced decreasing PL2L60
expression in various types of cancer cell lines, including (C)
MDA-MB-231, (D) MCF-7, (E) SKBR3 and (F) HeLa, which were cultured
in hypoxia (H; 1% O2) for 0, 12, 24 and 48 h. It should
be noted that PL2L60 was consistently decreased at 24 h under
hypoxia. PL2L60, Piwil2-like 60 kDa protein. |
Hypoxia-induced downregulation of
PL2L60 is hypoxia- inducible factor 1 alpha subunit
(HIF-1α)-independent
Since the evolutionarily conserved HIF
transcriptional complex is rapidly activated when the O2
tension decreases (29) and the
HIF-1α plays a key role in the regulation of oxygen homeostasis
(30), it was investigated whether
hypoxia-induced HIF-1α was associated with PL2L60 downregulation in
the cancer cells exposed to hypoxic condition. Cobalt chloride
(CoCl2), which stabilizes HIF-1α by inhibiting prolyl
hydroxylases to induce cellular hypoxic responses in vitro
(28,31) was used to treat breast cancer cell
line MDA-MB-231 and cervical cancer cell line HeLa for 24 h in
various concentrations (0, 100, 200 or 400 µM). As demonstrated in
Fig. 3, the expression of PL2L60
protein in both lines of cancer cells was inhibited by CoCl2 in a
dose-dependent manner (Fig. 3A-C),
while HIF-1α was remarkably increased. To determine whether the
reduction of PL2L60 in the hypoxic cells was regulated by HIF-1α,
HIF-1α expression was inhibited in HeLa and MDA-MB-231 cells by
using siRNA (Fig. 3D). As a result,
HIF-1α siRNA did not have any significant effect on the PL2L60
protein expression either in the hypoxic or normoxic condition
(Fig. 3E). Therefore, it was
concluded that hypoxia-induced PL2L60 downregulation was
independent of HIF-1α expression in the hypoxic cancer cells.
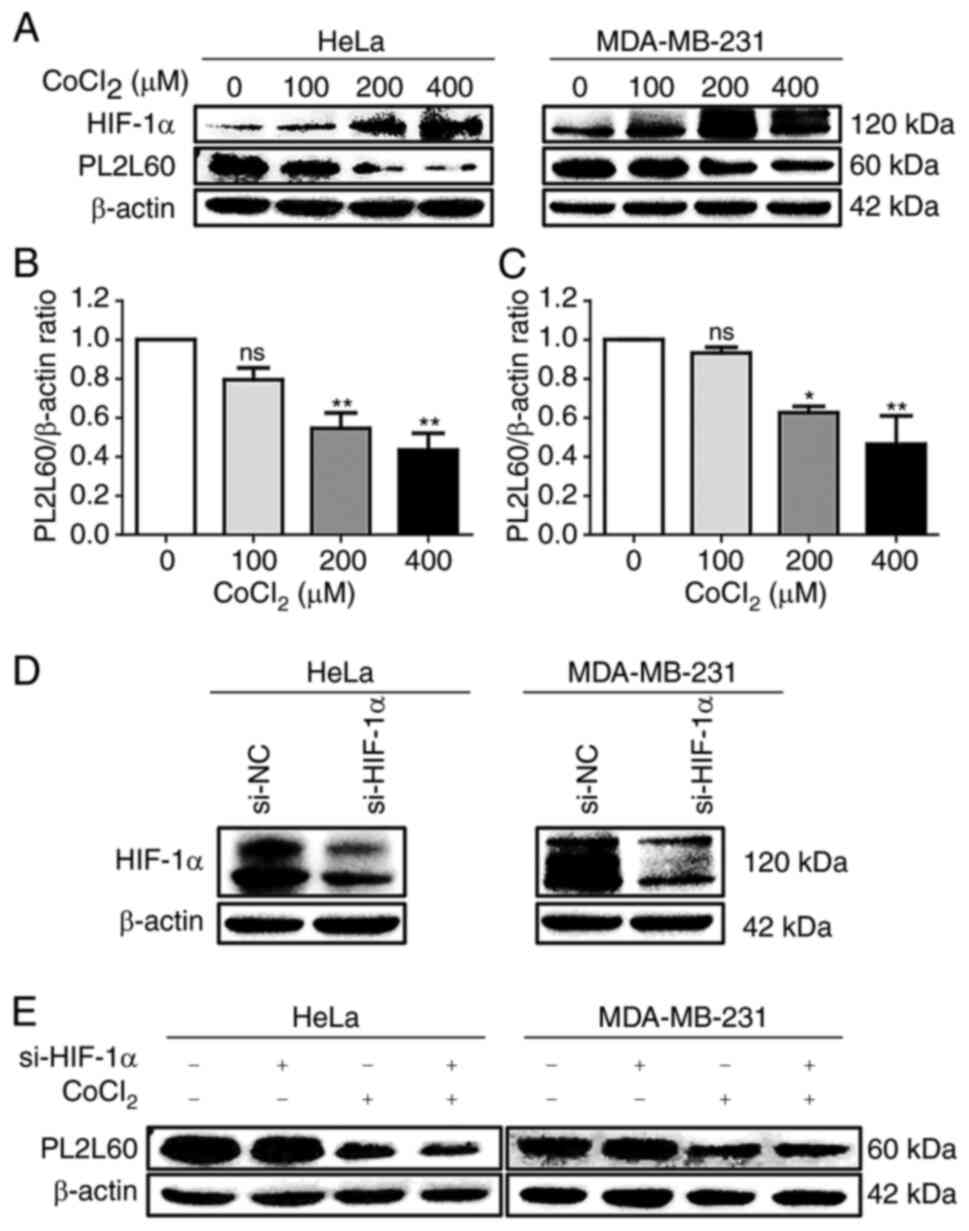 | Figure 3.Hypoxia-induced downregulation of
PL2L60 in cancer cells is HIF-1α-independent. (A-C) Dose-dependent
expression of HIF-1α and PL2L60 in cancer cells treated with cobalt
chloride (CoCl2). (A) The HeLa cells and MDA-MB-231
cells were exposed CoCl2 at the indicated concentration
(0, 100, 200 and 400 µM) for 24 h followed by western blot analysis
of PL2L60, HIF-1α and β-actin. (B and C) The band intensity of
PL2L60 normalized by β-actin was decreased in a dose-dependent
manner of CoCl2. (D and E) Knockdown of HIF-1α had no
effect on PL2L60 expression in the hypoxic cancer cells. (D) The
HeLa and MDA-MB-231 cancer cells were transfected with HIF-1α siRNA
(si-HIF-1α) or control siRNA (si-NC) for 48 h, and HIF-1α protein
expression was analyzed by western blotting. (E) Then, HeLa and
MDA-MB-231 cells transfected with siRNA negative control (si-NC) or
HIF-1α siRNA for 48 h were treated with CoCl2 (400 µM) or vehicles
(PBS) for another 24 h. The results shown were a representative
from 3 reproducible experiments. PL2L60, Piwil2-like 60 kDa
protein; HIF-1α, hypoxia-inducible factor 1 alpha subunit; si-,
small interfering; NC, negative control; ns, no significance.
*P<0.05 and **P<0.01. |
The downregulation of PL2L60
expression in the hypoxic cancer cells is associated with autophagy
activated by hypoxia, rapamycin or nutrient starvation
Since HIF-1α had no effect on PL2L60 protein
expression (Fig. 3), it was
hypothesized that PL2L60 downregulation in the hypoxic cancer cells
might be associated with autophagic activation, because the
autophagy was activated in the human tumor cell lines when exposed
in vitro to hypoxia and/or metabolic stress (28,32,33).
Therefore, the association of LC3 expression with PL2L60
downregulation in the distressed cancer cells was examined, as
autophagy was activated through recruiting LC3 (LC3-I) to
autophagosome membranes, by which LC3-I is conjugated with PE to
form punctate LC3-II (34). Under
the hypoxic condition, HeLa and MDA-MB-231 cancer cells
demonstrated decreased PL2L60 but increased LC3-II (Fig. 4A and B). Simultaneously, the level
of p62 protein was also decreased (Fig.
4A and B), indicating an enhanced autophagic influx. The p62 is
known to make a bridge between polyubiquitinated cargos and
autophagosomes through binding, respectively, to a
ubiquitin-associated domain and a LC3-interacting region. The
results suggested that decreased expression of PL2L60 in the
hypoxic cancer cells was associated with autophagic activation.
Consistently, PL2L60 expression was also decreased in both HeLa and
MDA-MB-231 cancer cells treated by an autophagy activator
rapamycin, an inhibitor of mTOR (Fig.
4C) or deprived of nutrients (Fig.
4D). Collectively, these data verified the hypothesis that
HIF-1α-independent downregulation of PL2L60 in the hypoxic cancer
cells was associated with autophagic activation.
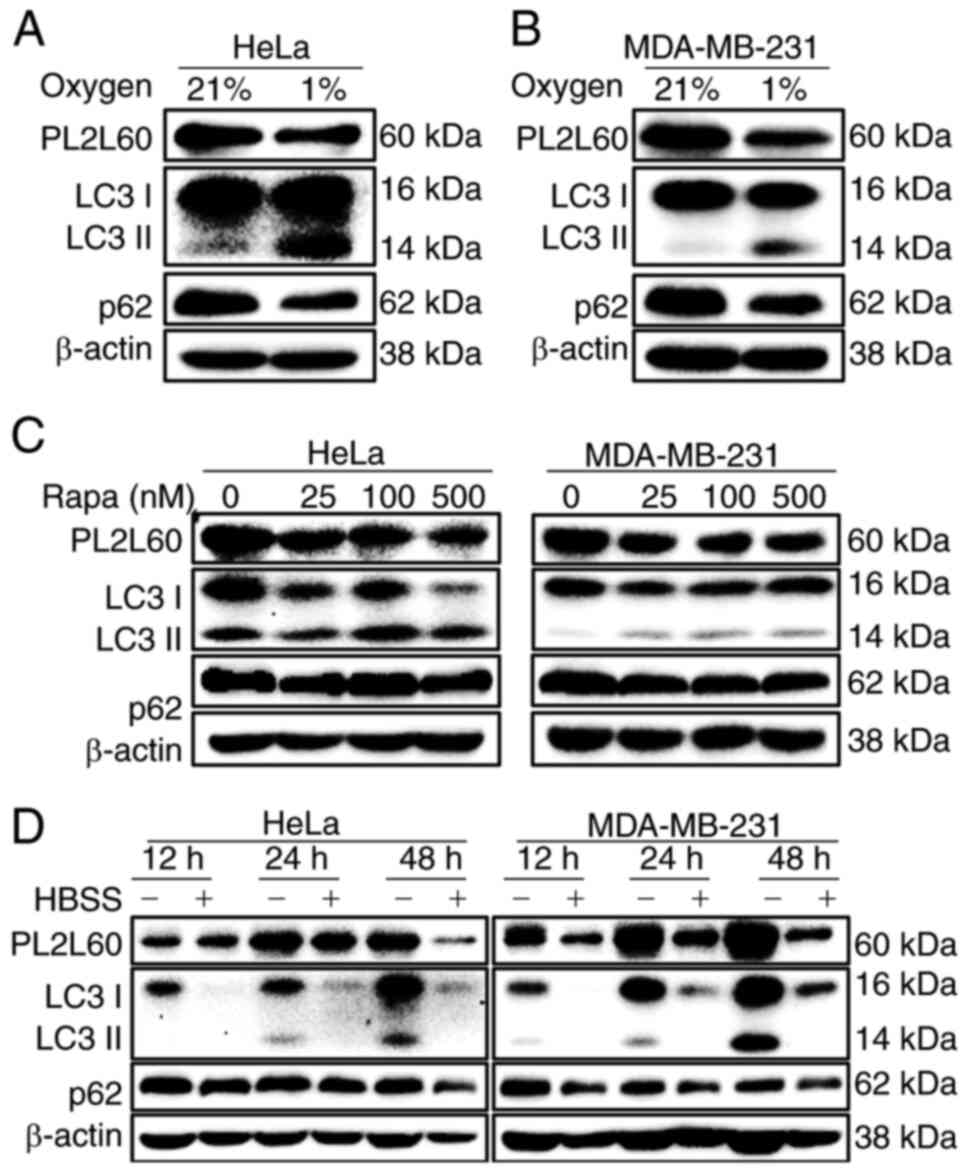 | Figure 4.PL2L60 is synchronically
downregulated with activation of autophagy induced by hypoxia,
rapamycin or nutrient deprivation. (A and B) HeLa and MDA-MB-231
cancer cells were cultured under normoxia (N; 21% O2) or
hypoxia (H; 1% O2) conditions for 24 h, (C) treated with
rapamycin at the indicated concentrations (0, 25, 100 and 500 nM)
for 24 h, or (D) deprived of serum from cultures for the indicated
time intervals (12, 24 and 48 h). The expression levels of PL2L60,
p62, LC3 and β-actin were assessed by immunoblot analysis. The
results shown are representative of 3 reproducible experiments.
PL2L60, Piwil2-like 60 kDa protein; LC3, microtubule-associated
protein 1 light chain 3; HBSS, HBSS, Hank's Balanced Salt
Solution. |
Depletion of autophagic signaling
components blocked PL2L60 degradation in hypoxic cancer cells
To further confirm that autophagic activation was
associated with the decreased expression of PL2L60 in the hypoxic
cancer cells, the effects of depletion of autophagic components on
PL2L60 expression were investigated. The autophagic process
requires a series of evolutionarily conserved proteins, most of
which are known as ATG proteins, functioning at different phases of
autophagy formation. Beclin-1 binds to class III
phosphatidylinositol 3-kinase (PIK3C3 or Vps34), which forms an
initiation complex of autophagy and promotes autophagosomal
membrane nucleation (35,36). Since autophagosomal elongation
requires 2 ubiquitin like conjugation systems, ATG12-ATG5 and
subsequent PE-conjugated form of LC3-II/ATG8-PE (37), Beclin-1, ATG5, p62 and LC3,
respectively, were knocked down in cancer cells, using specific
siRNA, and the levels of PL2L60 in these cells were examined. HeLa
cells were transfected with ATG5 or Beclin-1
(BECN1)-specific siRNA for 48 h and subsequently exposed to
1% O2 (hypoxia) or 21% O2 (normoxia) for
another 24 h. As expected, PL2L60 was not significantly decreased
in the hypoxic HeLa cells when ATG5 or Beclin-1 was
knocked down (Fig. 5A and B).
Similar results were observed when LC3 or p62 was knocked out by
siRNA in the hypoxic HeLa cells (Fig.
5C and D) and MDA-MB-231 cells (Fig. 5E and F). These results verified that
PL2L60 downregulation in the hypoxic cancer cells is mediated by a
mechanism of autophagic degradation.
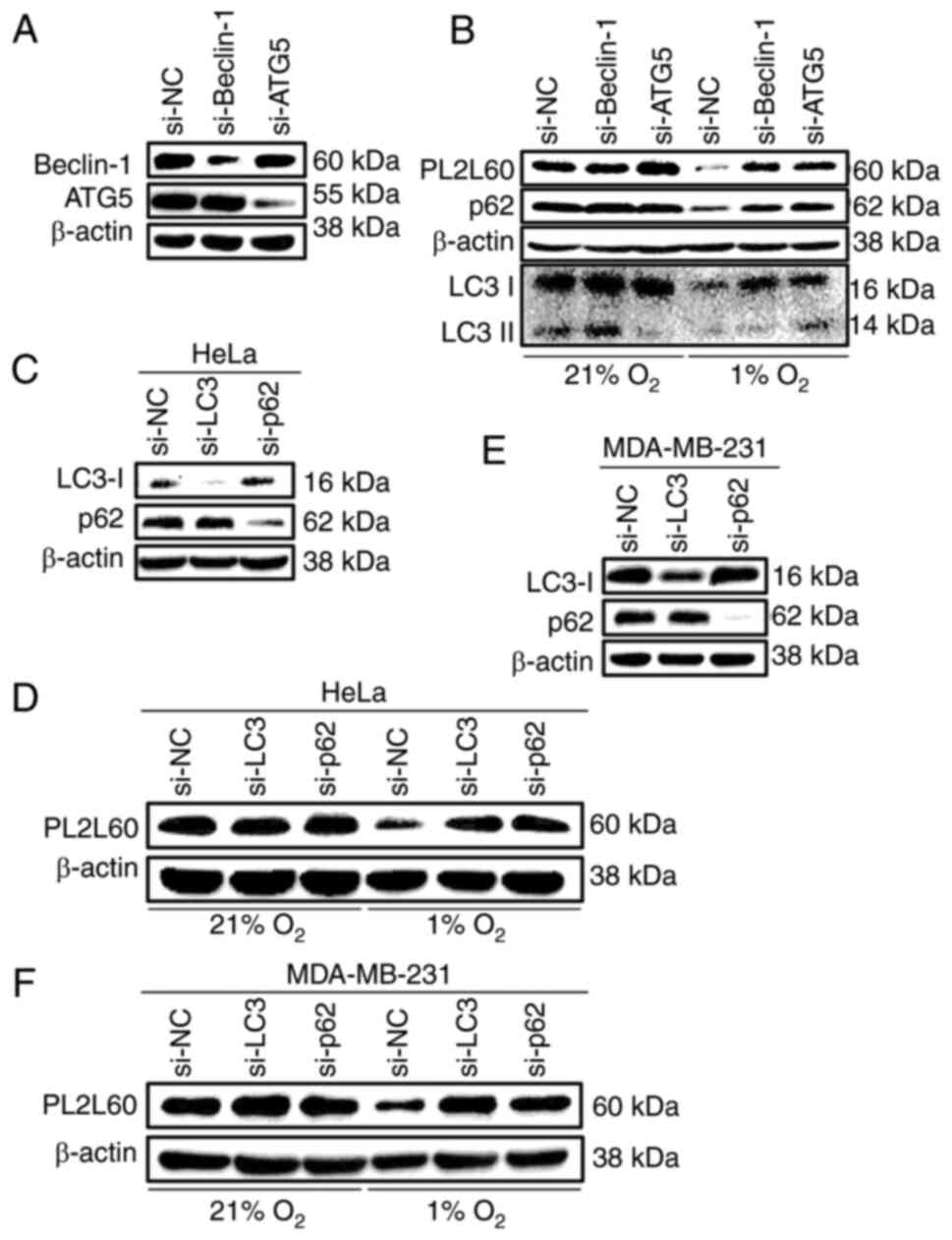 | Figure 5.PL2L60 protein is degraded by
selective autophagy in hypoxia. (A) HeLa cancer cells were
transfected with Beclin-1 siRNA (si-Beclin-1), ATG5 siRNA (si-ATG5)
or control negative control siRNA (si-NC) for 48 h. The levels of
Beclin-1, ATG5 and β-actin were determined by immunoblotting. (B)
HeLa cells were transfected with Beclin-1 siRNA (si-Beclin-1), ATG5
siRNA (si-ATG5) or control siRNA (si-NC) for 48 h and then exposed
to 1% O2 (hypoxia) or 21% O2 (normoxia) for
another 24 h. The cells were harvested and the levels of protein
expression were measured by western blot analysis using antibodies
against PL2L60, p62, LC3 and β-actin. (C) HeLa cancer cells and (E)
MDA-MB-231 cancer cells were transfected with LC3 siRNA (si-LC3),
p62 siRNA (si-p62) or negative control siRNA (si-NC) for 48 h. The
levels of LC3, p62 and β-actin were determined by immunoblotting.
(D) HeLa cancer cells and (F) MDA-MB-231 cancer cells were
transfected with LC3 siRNA (si-LC3), p62 siRNA (si-p62) or control
siRNA (si-NC) for 48 h and then exposed 1% O2 (hypoxia)
or 21% O2 (normoxia) for another 24 h. The cells were
harvested and the levels of protein expression were measured by
western blot analysis using antibodies against PL2L60 and β-actin.
PL2L60, Piwil2-like 60 kDa protein; siRNA, small interfering RNA;
NC, negative control; LC3, microtubule-associated protein 1 light
chain 3. |
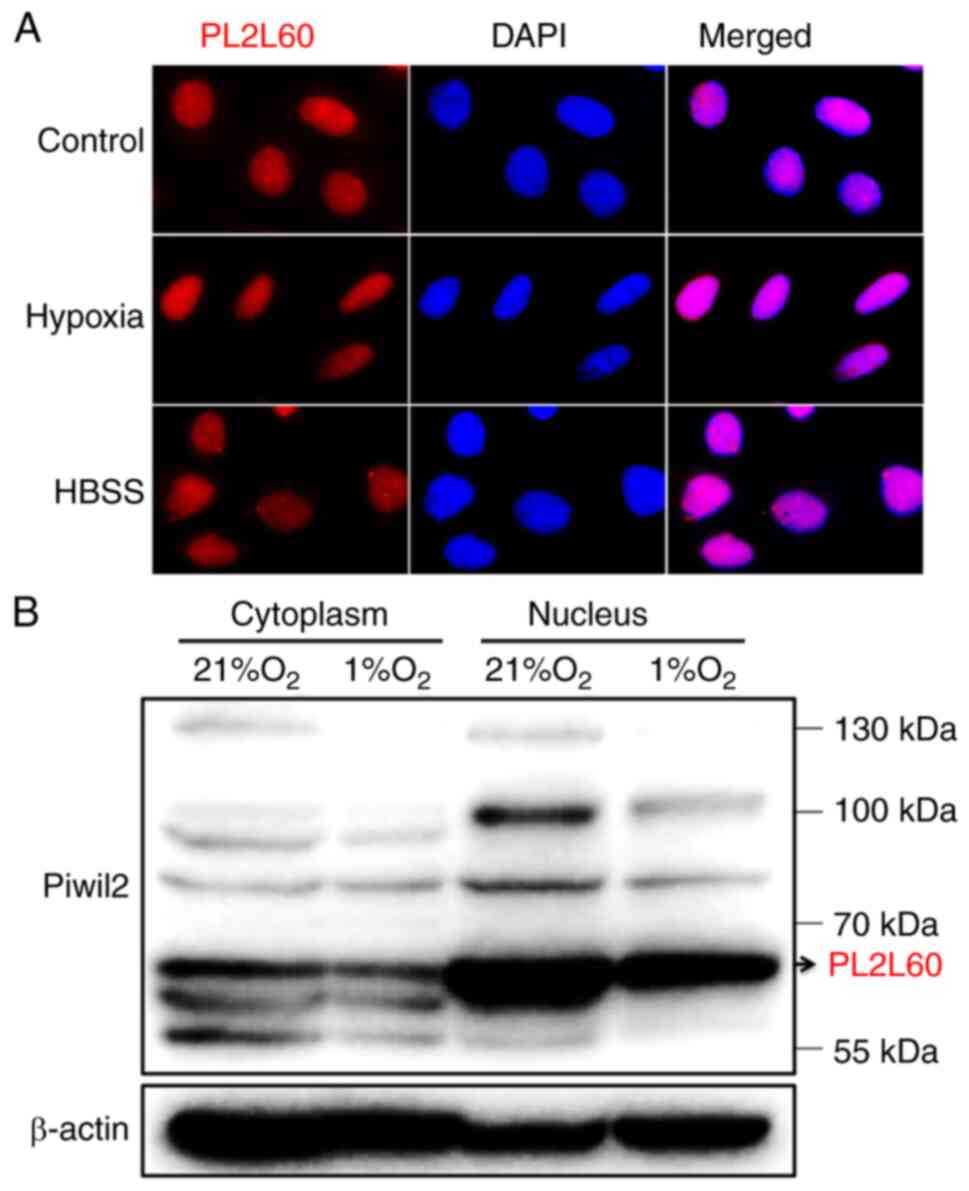
PL2L60 is predominantly expressed in
the nucleus and degraded through forming a complex with
autophagosome in the hypoxic cancer cell lines
PIWI proteins are often detected in the perinuclear
membrane-less organelle called the nuage (also known as Yb bodies,
chromatoid bodies, pi-bodies and piP-bodies); therefore, the nuage
is considered to be the center for piRNA biogenesis and
piRNA-induced silencing complex (piRISC) formation (11,38,39).
However, PL2L60 is a product of alienation-activated PIWIL2
gene by its intragenic promoter (4), and thus is defective in N-terminal,
affecting its subcellular localization and protein functions
(40). Therefore, PL2L60 expression
in the nucleus and cytoplasm was examined using immunofluorescent
microscopy and western blotting. The results revealed that PL2L60
was predominantly expressed in the nucleus of cancer cells
(Fig. 6A and B). Especially, dense
nuage-like granules containing PL2L60 were clearly observed in the
nuclei of cancer cells under normoxic condition. However, the
granules turned out to be obscured in the hypoxic and
nutrients-deprived cancer cells (Fig.
6A), in which autophagy was activated (Fig. 5). In accordance with the
observation, fractionation studies confirmed that PL2L60 was
predominantly expressed in nucleus rather than in the cytoplasm
(Fig. 6B). And the PL2L60 was
significantly reduced in the nuclei of cancer cells under hypoxia
for 24 h (Fig. 6B), suggesting that
the nuclear PL2L60 might be degraded by hypoxia-induced autophagy.
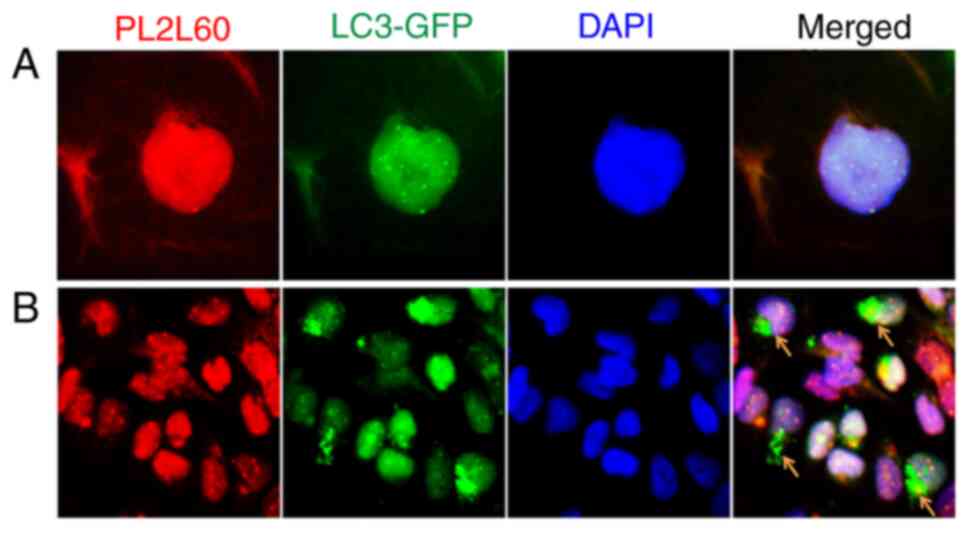
To confirm the hypothesis, breast cancer cell line MDA-MB-231 was
stably transfected with GFP-LC3 and examined for the interaction
between LC3 and PL2L60. As revealed in Fig. 7A, both GFP-LC3 and PL2L60 were
mainly detected in the nuclei. And GFP-LC3 were fused or
colocalized with PL2L60 nuage-like granules in the nucleus.
Interestingly, some perinuclear areas in the cells under normoxic
condition were rich of GFP-LC3 puncta but lacked of PL2L60
(Fig. 7B; arrow heads). Under
normoxic condition, basal autophagy could still be activated in the
cell cytoplasm where the autophagosomes containing LC3 puncta could
be present (23). Therefore, the
GFP-LC3 puncta lack of PL2L60 was likely perinuclear
autophagosomes, in which PL2L60 might have been completely degraded
(Fig. 7B; arrow heads). The results
under basal autophagy condition were similar to the results in
Fig. 6A where autophagy could be
activated by hypoxia or nutrient starvation. The PL2L60 nuage-like
granules in the cells under hypoxia condition were markedly less
dense compared with those in the cells under normoxic condition
(Fig. 6A). The results further
conformed that PL2L60 can be degraded by autophagy in cancer
cells.
Silencing of PL2L60 induces apoptosis
of cancer cells
It has been previously reported by the authors that
PL2L60 mediates tumorigenesis and silencing of PL2L60 using
specific siRNA effectively inhibits tumor cell proliferation
(4). This conclusion is supported
by silencing of PL2L60 using siRNA E21 in cancer cells. MDA-MB-231,
HeLa or A549 cancer cells were transfected with siRNA E21 at the
indicated concentrations for 48 h and then assessed for the cell
growth. As revealed in Fig. 8A and
B, blocking PL2L60 expression potently inhibited growth of
cancer cells including MDA-MB-231, HeLa and A549. This inhibition
was obviously associated with increased apoptosis of cancer cells,
which were transfected with siRNA E21, as demonstrated by flow
cytometric analysis (Fig. 8C and
D). Taken together, PL2L60 silencing could inhibit tumor growth
through inducing apoptosis in cancer cells. To verify the in
vitro results, PL2L60 was examined expression in tumor
cell-derived xenografts.
 | Figure 8.PL2L60 silencing inhibits
proliferation and induces apoptosis in cancer cells. (A and B)
Phase-contrast images of MDA-MB-231, HeLa or A549 cancer cells
transfected with PL2L60 siRNA (si-E21) at indicated concentrations
(250, 500 and 1,000 nM), or negative control siRNA (si-NC) for 48
h. Cell growth inhibition rates are quantified by counting numbers
and expressed as a percentage of si-NC transfected control. The
inhibition rates of si-NC transfected various cancer cells at 24 h
are regarded as 0%. (C and D) MDA-MB-231, HeLa or A549 cancer cells
were transfected with PL2L60 siRNA (si-E21) or negative control
siRNA (si-NC) for 48 h, then cells were stained with PI/Annexin and
detected by flow cytometric analysis. PL2L60, Piwil2-like 60 kDa
protein; siRNA, small interfering RNA; NC, negative control; ns, no
significance. **P<0.01. |
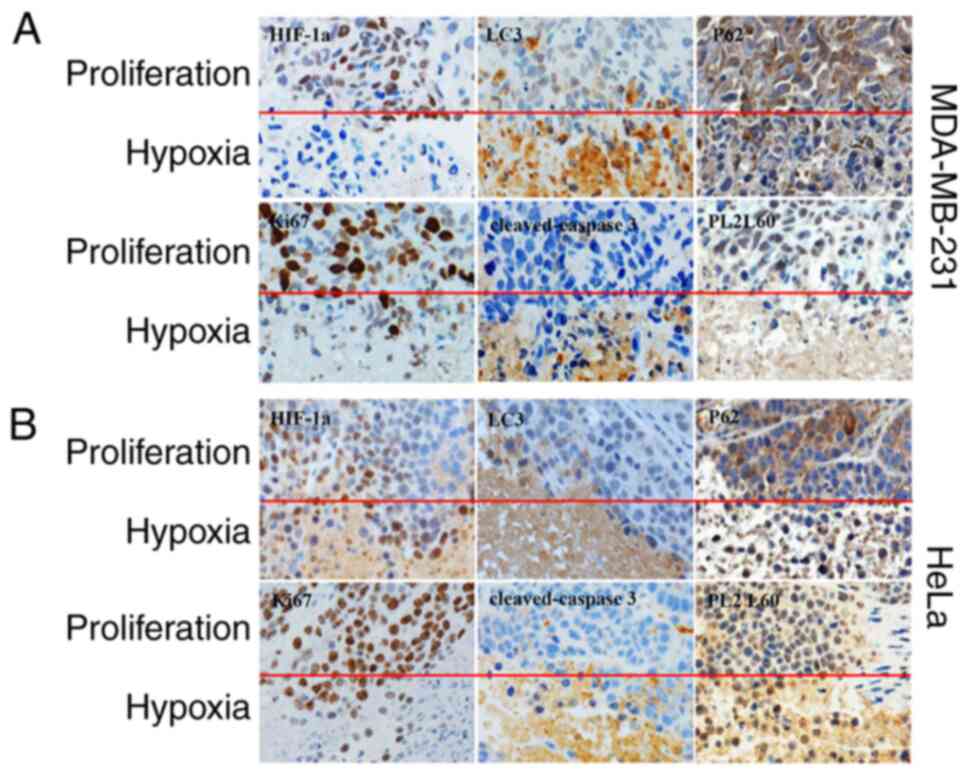
Hypoxia-induced necroptosis of cancers
is a primary site for autophagic cell death and PL2L60
degradation
Since oxygen diffusion distance is no more than 200
microns away from a capillary (41), cancer cells encounter hypoxic stress
at a very early stage during cancer development. Most research have
verified that hypoxia is the primary site for autophagy in tumors
and induces autophagic cell death in cancer cells (20,28,42,43).
Especially, it has been previously reported by the authors that
autophagy can mediate programmed cell death (PCD) of breast and
cervical cancer cells in responding to CoCl2 mimic
hypoxia (28). To determine whether
a similar situation exists in hypoxic tumor regions in vivo,
the prevalence and distribution of HIF-1α, LC3, p62, PL2L60 and
cleaved-caspase 3 was assessed in the MDA-MB-231 and HeLa-derived
xenotransplant tumors. It was found that autophagy, as evaluated by
positive expression of LC3B and downregulated expression of p62,
was more strongly associated with hypoxic tumor regions than
non-hypoxia (proliferating) tumor regions (Fig. 9). Conversely, both HIF-1α and PL2L60
expression decreased significantly in the nested hypoxic tumor area
compared with the normal adjacent proliferating area (Fig. 9). A previous study reported that the
activation of autophagy under hypoxia decreases the amount of
HIF-1α via LC3 dependent manner in the presence of an intact
autophagic machinery (23). This
could explain the downregulation of HIF-1α in the hypoxic tumor
region. In line with the present western blot results (Fig. 2, Fig.
3, Fig. 4, Fig. 5, Fig.
6, Fig. 7), PL2L60 was high in
the normal adjacent proliferating area and low in the nested
hypoxic tumor area (Fig. 9).
Collectively, hypoxia-induced autophagy significantly decreased
PL2L60 expression in vivo or in vitro. In previous
studies, it was confirmed that PL2L60 promotes tumor cell survival
and proliferation (Fig. 9A and B)
and severe hypoxia induces autophagy dependent PCD in cancer cells
by activating caspase-3 (2,28). Autophagy can promote cell death by
selectively reducing the abundance of anti-apoptotic proteins in
the cells (44). Therefore, it was
hypothesized that hypoxia would promote autophagic degradation of
PL2L60 as one cancer cell death mechanism in vivo. IHC using
an antibody against Ki67, a marker of cell proliferation, showed
that Ki67-positive cells were decreased in severe hypoxic areas as
compared with the normal adjacent proliferation area (Fig. 9), demonstrating decreased cell
proliferation induced by hypoxia. H&E staining demonstrated
more dead cells and the evident increase in apoptosis proportion in
severe hypoxic tumor tissues (Fig.
9). IHC also demonstrated the increase in mean areas that
stained positively for cleaved caspase-3 in severe hypoxic tumor
tissues (Fig. 9). Collectively, the
results revealed that hypoxia-induced autophagy inhibits growth and
promotes apoptosis of breast or cervical cancer cells in
vivo with lower levels of PL2L60, which may serve as an
indicator for predicting clinical outcomes of cancer patients.
Discussion
Solid tumor is always confronting the highly
hypoxic, nutrient-poor environment, which may be a major cause of
cancer cell death. However, the mechanisms underlying the metabolic
stresses-induced cancer cell death remain elusive. In the present
study, it was identified for the first time, to the best of our
knowledge, that the cancer cell death induced by hypoxia and
nutrient deprivation was associated with autophagic degradation of
PL2L60 in various types of cancer cells (Fig. 1, Fig.
2, Fig. 3, Fig. 4, Fig.
5, Fig. 6, Fig. 7, Fig.
8, Fig. 9). The hypoxia-induced
PL2L60 downregulation was independent of HIF-1α, which is a key
molecule in the regulation of oxygen homeostasis (Fig. 3). Cells exposed to various stressors
undergo a process of self-digestion known as autophagy, during
which cytoplasmic or nuclear cargo sequestered inside
double-membrane vesicles are delivered to the lysosome for
degradation. Consistently, inhibition of autophagy by targeting
Beclin-1 (BECN1) or ATG5 restored PL2L60 levels in the
hypoxic cancer cells in vitro (Fig. 5A and B). Accordingly, knockdown of
the autophagosome membrane protein Atg8/LC3 or autophagy cargo
protein p62 by siRNA attenuated the hypoxia-induced degradation of
PL2L60 (Fig. 5C-F). Cytological
analysis confirmed existence of LC3/PL2L60 punctate complexes in
the cancer cells (Figs. 6 and
7). These findings indicated that
metabolic stress-induced cancer cell death was associated with the
selective autophagosomal clearance of PL2L60.
In a solid cancer, necroptosis is frequently
observed, which may be caused by reduced oxygen tension (hypoxia)
within the tumor (17,20,30).
The reduced oxygen tensions may induce HIF-1α, which plays an
essential role in homeostatic response to the hypoxia, activating
the transcription of over 40 genes including erythropoietin,
glucose transporters, glycolytic enzymes, vascular endothelial
growth factor and other genes (17,20,30,31).
The protein products of these genes increase oxygen delivery or
facilitate metabolic adaptation to hypoxia. This means that in
addition to protective role of HIF-1α, there should be a pathway
that antagonizes the role of HIF-1α to drive the oxygen-deficient
cancer cells to apoptotic death. In the present study, it was
identified that such a pathway of autophagic degradation of PL2L60
proteins was independent of HIF-1α.
In cancer cells, autophagy is a double sword, which
can be neutral, tumor-suppressive, or tumor-promoting in different
contexts. It has mainly been considered that autophagy can promote
cancer through suppressing p53 and preventing energy crisis, cell
death, senescence, and an antitumor immune response (45). In the present study, however, it was
demonstrated that in the hypoxia or nutrient-deprivation context,
activating autophagy induced apoptosis of cancer cells through
selectively degradation of PL2L60, a key promoter of cancer cell
survival and proliferation (2,4–6). This
finding was consistent with the conclusion that PL2L60 is a
tumorigenic promoter (2–5), and blocking PL2L60 could suppress
tumor growth.
While autophagosomes sequester cytosolic materials
non-specifically in a process called non-selective autophagy, a
process of selective autophagy also occurs in which autophagic
degradation of specific protein aggregates or organelles targeted
for destruction occurs (32,46). A
previous study has reported that LC3 can co-operates with p62 to
deliver mitochondrial proteins to lysosomes for selective
degradation (47). Selective
autophagy degrading specific proteins is often associated with
degradation of p62 or NDP52, a protein complex that binds
ubiquitinated protein aggregates and LC3 family protein to target
them for degradation (47–50). Knockdown of p62 reversed the hypoxic
degradation of PL2L60, suggesting that PL2L60 may be specifically
or selectively degraded by autophagy in the contexts of metabolic
stresses. LC3-PL2L60 interaction alone without p62 was not
sufficient for PL2L60 degradation (Fig.
5C-F), suggesting that PL2L60 was selectively degraded in the
hypoxic environment.
PL2L60 is a truncated protein of PIWIL2, containing
½ PAZ domain and a PIWI domain (2).
Piwi interacts with polycomb group complexes PRC1 and PRC2 in niche
and germline cells to regulate ovarian, germline stem cells and
oogenesis (51). The Mili
(Piwil2)-containing complexes (sometimes named pi-body) can be
isolated from adult mouse testes, which contain some key proteins
such as Tudor domain-containing protein-1 or 12 (Tdrd1/12) and play
a vital role in piRNA biogenesis and maturity in the cytoplasm
(52–54). Indeed, it was observed that PL2L60
speckles accumulated in the cancer nucleus (Figs. 6 and 7). Although both LC3 and p62 plays the key
role in the PL2L60 degradation (Figs.
4 and 5), how PL2L60 as one
autophagic substrate was sequestered in the nucleus and then
transferred into the cytoplasm for autophagic degradation yet
remained unclear. As shown in Fig.
6, PL2L60 is distributed dominantly in the nucleus and fewer
puncta scattered in the cytoplasm. Nuclear autophagy is
evolutionarily conserved in eukaryotes that may target various
nuclear components through a series of processes, including nuclear
sensing, nuclear export, autophagic substrate encapsulation and
autophagic degradation in the cytoplasm (55,56).
For instance, nuclear lamina protein Lamin B1 degradation is
achieved by nucleus-to-cytoplasm transport that delivers Lamin B1
to the lysosome (56). It was also
observed that GFP puncta predominantly distributed in the
perinuclear (Fig. 7B). Importantly,
merging LC3 and PL2L60 fluorescence revealed a nuclear
co-localization for these two proteins while PL2L60 could not be
detected in the GFP-LC3 puncta positive autophagosomes-like
structures (Fig. 7A and B).
Therefore, the predominant perinuclear distribution of lysosomes
(57) may facilitate this cargo and
degradation of PL2L60.
Symmetrical dimethylarginines (sDMA)modification is
required for Piwi protein stability favoring piRNA biogenesis
(58). Most animal Piwi proteins
contain sDMA motifs that are typically clustered close to the amino
terminus (58). Protein
post-translational modification provides a layer of regulation for
the specificity and efficiency of selective autophagy (48,50).
In a previous by the authors, it was confirmed that asymmetric DMA
(aDMA) serves as one specific degradation signal of selective
autophagy (59). Especially, a
previous study found that arginine methylation regulates the
autophagic degradation efficiency of PGL Granules [PGL-1 and PGL-3
(cargo)-SEPA-1 (receptor) complexes] (60). In the present study, some
autophagosomal structures containing PL2L60 were observed to
dominantly express in the nucleus and partly in the perinucleus
(Fig. 6A). Moreover, GFP-LC3 and
PL2L60 accumulated in the nucleus and interacted with each other
(Fig. 7A). Subsequent studies
should evaluate the relative sDMA or aDMA potential of PL2L60 and
assess other regions of their cytoplasmic or nuclear domain
sequences for the involvement in PL2L60 degradation.
The present results indicated that
hypoxia-regulated autophagy induced apoptosis in multiple types of
cancer by suppressing PL2L60. The present study has established an
intimate relationship between autophagy, PL2L60 and apoptosis in
response to hypoxia. The present study may provide rational
strategies for the future clinical evaluation of PL2L60 as one key
cancer immunotherapy antigen in targeted molecular strategy.
Acknowledgements
The authors are grateful to Dr Yanfeng Liu
(Renji-Med X Clinical Stem Cell Research Center, Ren Ji Hospital)
for his stimulating discussion and helpful comments on the
manuscript.
Funding
The present study was supported by the SJTU Interdisciplinary
Research Grant (grant no. YG2015MS56), the National Natural Science
Foundation of China (grant nos. 81402287, 81602700, 81672713,
81372188, and 81371507), the Science and technology support
program, Science and Technology Commission of Shanghai Municipality
(grant no. 1243190074), The Special Fund for Innovation and
Development of Science and Technology and Cultivation Fund for
Major Projects and Innovative Team, the Shanghai Jiao Tong
University, the State Key Laboratory of Oncogenes and Related Genes
in China (grant no. 90-14-06), the Startup Funds from Renji
Hospital and School of Medicine, Shanghai Jiao Tong University and
the Fund for Key Disciplines and Specialties, Shanghai Health and
Family Planning Committee.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
LS, LFL and JXG conceived the project and
supervised the study. LS, FH and GYT initiated the project and
performed the experiments. HLS conducted part of the experiments.
LS, XLC and LFL analyzed and interpreted the data. LS and LFL
designed the experiments and drafted the manuscript. All authors
read and approved the final version of the manuscript. LS, LFL and
JXG confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Nude mice were obtained from SLACCAS (Shanghai
Laboratory Animal Center), bred in our own animal pathogen-free
facility and maintained under standard conditions according to the
institutional guidelines of Renji Hospital animal care and ethics
review committee. All in vivo experiments were approved by
the institutional guidelines of the animal care and ethics review
committee of Renji Hospital (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AS
|
alternative splicing
|
|
PL2L60
|
Piwil2-like 60 kDa protein
|
|
piRNA
|
piwi-interacting RNA
|
|
HIF-1α
|
hypoxia-inducible factor 1 alpha
subunit
|
|
LC3
|
microtubule-associated protein 1
light chain 3 (MAP1LC3)
|
|
ATG
|
autophagy-related gene
|
|
siRNA
|
small interfering RNA
|
|
PCD
|
programmed cell death
|
References
|
1
|
Chen L, Shen R, Ye Y, Pu XA, Liu X, Duan
W, Wen J, Zimmerer J, Wang Y, Liu Y, et al: Precancerous stem cells
have the potential for both benign and malignant differentiation.
PLoS One. 2:e2932007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ye Y, Yin DT, Chen L, Zhou Q, Shen R, He
G, Yan Q, Tong Z, Issekutz AC, Shapiro CL, et al: Identification of
Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS One.
5:e134062010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gainetdinov IV, Skvortsova YV, Stukacheva
EA, Bychenko OS, Kondratieva SA, Zinovieva MV and Azhikina TL:
Expression profiles of PIWIL2 short isoforms differ in testicular
germ cell tumors of various differentiation subtypes. PLoS One.
9:e1125282014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu SS, Liu N, Liu MY, Sun L, Xia WY, Lu
HM, Fu YJ, Yang GL, Bo JJ, Liu XX, et al: An unusual intragenic
promoter of PIWIL2 contributes to aberrant activation of oncogenic
PL2L60. Oncotarget. 8:46104–46120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li LF, Liu N, Liu MY and Gao JX: The
Functions of Piwil2 and Its Prospects in Tumorigenesis. Am J Transl
Med. 1:75–98. 2017. View Article : Google Scholar
|
|
6
|
Gao JX, Liu N and Wu HL: PIWIL2 (piwi-like
RNA-mediated gene silencing 2). Atlas Genet Cytogenet Oncol
Haematol. 18:919–927. 2014.PubMed/NCBI
|
|
7
|
Czech B and Hannon GJ: Small RNA sorting:
Matchmaking for Argonautes. Nat Rev Genet. 12:19–31. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Z, Chen KM, Pandey RR, Homolka D,
Reuter M, Janeiro BK, Sachidanandam R, Fauvarque MO, McCarthy AA
and Pillai RS: PIWI slicing and EXD1 drive biogenesis of nuclear
piRNAs from cytosolic targets of the mouse piRNA pathway. Mol Cell.
61:138–152. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vourekas A and Mourelatos Z: HITS-CLIP
(CLIP-Seq) for mouse Piwi proteins. Methods Mol Biol. 1093:73–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vourekas A, Alexiou P, Vrettos N,
Maragkakis M and Mourelatos Z: Sequence-dependent but not
sequence-specific piRNA adhesion traps mRNAs to the germ plasm.
Nature. 531:390–394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hirakata S and Siomi MC: piRNA biogenesis
in the germline: From transcription of piRNA genomic sources to
piRNA maturation. Biochim Biophys Acta. 1859:82–92. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yin DT, Wang Q, Chen L, Liu MY, Han C, Yan
Q, Shen R, He G, Duan W, Li JJ, et al: Germline stem cell gene
PIWIL2 mediates DNA repair through relaxation of chromatin. PLoS
One. 6:e271542011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim E, Goren A and Ast G: Alternative
splicing: Current perspectives. Bioessays. 30:38–47. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xin D, Hu L and Kong X: Alternative
promoters influence alternative splicing at the genomic level. PLoS
One. 3:e23772008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang X, Coulombe-Huntington J, Kang S,
Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray
RR, et al: Widespread expansion of protein interaction capabilities
by alternative splicing. Cell. 164:805–817. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thadani-Mulero M, Portella L, Sun S, Sung
M, Matov A, Vessella RL, Corey E, Nanus DM, Plymate SR and
Giannakakou P: Androgen receptor splice variants determine taxane
sensitivity in prostate cancer. Cancer Res. 74:2270–2282. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vaupel P, Kelleher DK and Höckel M: Oxygen
status of malignant tumors: Pathogenesis of hypoxia and
significance for tumor therapy. Semin Oncol. 28 (2 Suppl
8):S29–S35. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ueda S, Saeki T, Osaki A, Yamane T and
Kuji I: Bevacizumab induces acute hypoxia and cancer progression in
patients with refractory breast cancer: Multimodal functional
imaging and multiplex cytokine analysis. Clin Cancer Res.
23:5769–5778. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun L, Li T, Wei Q, Zhang Y, Jia X, Wan Z
and Han L: Upregulation of BNIP3 mediated by ERK/HIF-1α pathway
induces autophagy and contributes to anoikis resistance of
hepatocellular carcinoma cells. Future Oncol. 10:1387–1398. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Azad MB, Chen Y, Henson ES, Cizeau J,
McMillan-Ward E, Israels SJ and Gibson SB: Hypoxia induces
autophagic cell death in apoptosis-competent cells through a
mechanism involving BNIP3. Autophagy. 4:195–204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo J, Solimini NL and Elledge SJ:
Principles of cancer therapy: Oncogene and non-oncogene addiction.
Cell. 136:823–837. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bellot G, Garcia-Medina R, Gounon P,
Chiche J, Roux D, Pouysségur J and Mazure NM: Hypoxia-induced
autophagy is mediated through hypoxia-inducible factor induction of
BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol.
29:2570–2581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie Z and Klionsky DJ: Autophagosome
formation: Core machinery and adaptations. Nat Cell Biol.
9:1102–1109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsujimoto Y and Shimizu S: Another way to
die: Autophagic programmed cell death. Cell Death Differ. 12 (Suppl
2):S1528–S1534. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gewirtz DA: The four faces of autophagy:
Implications for cancer therapy. Cancer Res. 74:647–651. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song J, Guo X, Xie X, Zhao X, Li D, Deng
W, Song Y, Shen F, Wu M and Wei L: Autophagy in hypoxia protects
cancer cells against apoptosis induced by nutrient deprivation
through a Beclin1-dependent way in hepatocellular carcinoma. J Cell
Biochem. 112:3406–3420. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun L, Liu N, Liu SS, Xia WY, Liu MY, Li
LF and Gao JX: Beclin-1-independent autophagy mediates programmed
cancer cell death through interplays with endoplasmic reticulum
and/or mitochondria in colbat chloride-induced hypoxia. Am J Cancer
Res. 5:2626–2642. 2015.PubMed/NCBI
|
|
29
|
Philip B, Ito K, Moreno-Sánchez R and
Ralph SJ: HIF expression and the role of hypoxic microenvironments
within primary tumours as protective sites driving cancer stem cell
renewal and metastatic progression. Carcinogenesis. 34:1699–1707.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ivan M, Kondo K, Yang H, Kim W, Valiando
J, Ohh M, Salic A, Asara JM, Lane WS and Kaelin WG Jr: HIFalpha
targeted for VHL-mediated destruction by proline hydroxylation:
Implications for O2 sensing. Science. 292:464–468. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kristensen AR, Schandorff S, Høyer-Hansen
M, Nielsen MO, Jäättelä M, Dengjel J and Andersen JS: Ordered
organelle degradation during starvation-induced autophagy. Mol Cell
Proteomics. 7:2419–2428. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li HY, Zhang J, Sun LL, Li BH, Gao HL, Xie
T, Zhang N and Ye ZM: Celastrol induces apoptosis and autophagy via
the ROS/JNK signaling pathway in human osteosarcoma cells: An in
vitro and in vivo study. Cell Death Dis. 6:e16042015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Maiuri MC, Zalckvar E, Kimchi A and
Kroemer G: Self-eating and self-killing: Crosstalk between
autophagy and apoptosis. Nat Rev Mol Cell Biol. 8:741–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang XH, Jackson S, Seaman M, Brown K,
Kempkes B, Hibshoosh H and Levine B: Induction of autophagy and
inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liang C, Feng P, Ku B, Dotan I, Canaani D,
Oh BH and Jung JU: Autophagic and tumour suppressor activity of a
novel Beclin1-binding protein UVRAG. Nat Cell Biol. 8:688–699.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Johansen T and Lamark T: Selective
autophagy mediated by autophagic adapter proteins. Autophagy.
7:279–296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kobayashi H and Tomari Y: RISC assembly:
Coordination between small RNAs and Argonaute proteins. Biochim
Biophys Acta. 1859:71–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Da Ros M, Lehtiniemi T, Olotu O, Fischer
D, Zhang FP, Vihinen H, Jokitalo E, Sironen A, Toppari J and Kotaja
N: FYCO1 and autophagy control the integrity of the haploid male
germ cell-specific RNP granules. Autophagy. 13:302–321. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Link S, Grund SE and Diederichs S:
Alternative splicing affects the subcellular localization of
Drosha. Nucleic acids research. 44:5330–5343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Olive PL, Vikse C and Trotter MJ:
Measurement of oxygen diffusion distance in tumor cubes using a
fluorescent hypoxia probe. Int J Radiat Oncol Biol Phys.
22:397–402. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shimizu S, Eguchi Y, Kamiike W, Itoh Y,
Hasegawa J, Yamabe K, Otsuki Y, Matsuda H and Tsujimoto Y:
Induction of apoptosis as well as necrosis by hypoxia and
predominant prevention of apoptosis by Bcl-2 and Bcl-XL. Cancer
Res. 56:2161–2166. 1996.PubMed/NCBI
|
|
43
|
Rouschop KM, van den Beucken T, Dubois L,
Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W,
Voncken JW, et al: The unfolded protein response protects human
tumor cells during hypoxia through regulation of the autophagy
genes MAP1LC3B and ATG5. J Clin Invest. 120:127–141. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mariño G, Niso-Santano M, Baehrecke EH and
Kroemer G: Self-consumption: The interplay of autophagy and
apoptosis. Nat Rev Mol Cell Biol. 15:81–94. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Amaravadi R, Kimmelman AC and White E:
Recent insights into the function of autophagy in cancer. Genes
Dev. 30:1913–1930. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Goodwin JM, Dowdle WE, DeJesus R, Wang Z,
Bergman P, Kobylarz M, Lindeman A, Xavier RJ, McAllister G, Nyfeler
B, et al: Autophagy-independent lysosomal targeting regulated by
ULK1/2-FIP200 and ATG9. Cell Rep. 20:2341–2356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Le Guerroué F, Eck F, Jung J, Starzetz T,
Mittelbronn M, Kaulich M and Behrends C: Autophagosomal content
profiling reveals an LC3C-dependent piecemeal mitophagy pathway.
Mol Cell. 68:786–796.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wild P, Farhan H, McEwan DG, Wagner S,
Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et
al: Phosphorylation of the autophagy receptor optineurin restricts
Salmonella growth. Science. 333:228–233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Newman AC, Kemp AJ, Drabsch Y, Behrends C
and Wilkinson S: Autophagy acts through TRAF3 and RELB to regulate
gene expression via antagonism of SMAD proteins. Nat Commun.
8:15372017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jo C, Gundemir S, Pritchard S, Jin YN,
Rahman I and Johnson GVW: Nrf2 reduces levels of phosphorylated tau
protein by inducing autophagy adaptor protein NDP52. Nat Commun.
5:34962014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Peng JC, Valouev A, Liu N and Lin H: Piwi
maintains germline stem cells and oogenesis in Drosophila through
negative regulation of Polycomb group proteins. Nat Genet.
48:283–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reuter M, Chuma S, Tanaka T, Franz T,
Stark A and Pillai RS: Loss of the Mili-interacting Tudor
domain-containing protein-1 activates transposons and alters the
Mili-associated small RNA profile. Nat Struct Mol Biol. 16:639–646.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Aravin AA, van der Heijden GW, Castaneda
J, Vagin VV, Hannon GJ and Bortvin A: Cytoplasmic
compartmentalization of the fetal piRNA pathway in mice. PLoS
Genet. 5:e10007642009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mathioudakis N, Palencia A, Kadlec J,
Round A, Tripsianes K, Sattler M, Pillai RS and Cusack S: The
multiple Tudor domain-containing protein TDRD1 is a molecular
scaffold for mouse Piwi proteins and piRNA biogenesis factors. RNA.
18:2056–2072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Luo M, Zhao X, Song Y, Cheng H and Zhou R:
Nuclear autophagy: An evolutionarily conserved mechanism of nuclear
degradation in the cytoplasm. Autophagy. 12:1973–1983. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dou Z, Xu C, Donahue G, Shimi T, Pan JA,
Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al: Autophagy
mediates degradation of nuclear lamina. Nature. 527:105–109. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tan KP, Ho MY, Cho HC, Yu J, Hung JT and
Yu AL: Fucosylation of LAMP-1 and LAMP-2 by FUT1 correlates with
lysosomal positioning and autophagic flux of breast cancer cells.
Cell Death Dis. 7:e23472016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kirino Y, Kim N, de Planell-Saguer M,
Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA and
Mourelatos Z: Arginine methylation of Piwi proteins catalysed by
dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol.
11:652–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sun L, Xia WY, Zhao SH, Liu N, Liu SS, Xiu
P, Li LF, Cao XL and Gao JX: An asymmetrically dimethylarginated
nuclear 90 kDa protein (p90aDMA) induced by interleukin (IL)-2,
IL-4 or IL-6 in the tumor microenvironment is selectively degraded
by autophagy. Int J Oncol. 48:2461–2471. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li S, Yang P, Tian E and Zhang H: Arginine
methylation modulates autophagic degradation of PGL granules in C.
elegans. Mol Cell. 52:421–433. 2013.Ritio optatur sinvelignis ut
arum que lanturerum il et, quatio. Nam et et escia audi aciis nia
idis eum hilit volor molorat. View Article : Google Scholar : PubMed/NCBI
|