Introduction
Colorectal cancer (CRC) is the third most frequent
malignancy and the second leading cause of cancer-associated
mortality worldwide (1). An early
diagnosis is crucial for the survival of patients with CRC. The
5-year survival rate of patients with early-stage CRC can be as
high as 90%, whereas this rate decreases to only ~12% for patients
with distant metastases (2).
Colonoscopy is a well-established method for the diagnosis of CRC;
however, its invasive nature limits its use for certain patients
(3). Therefore, more accurate and
less invasive screening methods are required for the detection of
CRC. Over the past few decades, alongside the rapid development of
tumor biotechnology, including genomics (4), transcriptomics (5), proteomics (6), metabonomics (7) and non-coding RNA omics (8), intensive studies have been conducted
on the mechanisms of CRC tumorigenesis. This has led to the
discovery of novel biomarkers for the diagnosis of CRC and drug
targets for treatment (9,10). CRC is a heterogeneous disease
(11), and factors such as tumor
cell differentiation and proliferation markedly affect disease
development, including metastases, disease outcomes and the
response to anti-tumor drugs (12).
However, the mechanisms through which these tumor heterogeneity
factors are associated with CRC malignancy are not yet well
understood (13); therefore,
identifying novel drivers for CRC progression may shed light on the
tumorigenesis of CRC, and may provide targets for diagnosis and
therapy.
Zinc finger proteins (ZNFs), which contain a wide
variety of zinc finger domains, are one of the largest protein
families in eukaryotes (14). ZNFs
are able to interact with DNA, RNA, poly-ADP-ribose (PAR) and other
proteins, participating in multiple cellular processes, such as
transcriptional regulation, ubiquitin-mediated protein degradation,
signal transduction, DNA repair and cell migration (15). Increasing evidence has demonstrated
that ZNFs play essential roles in the progression of various types
of cancer, including lung, esophageal, colorectal, nasopharyngeal,
thyroid, gastric, breast, ovarian, cervical, prostate and
gallbladder cancer (16–18). ZNF169 is a C2H2-type zinc finger
with Kruppel associated box domain (19). The ZNF169 gene is located at the
D9S196-D9S280 interval on chromosome 9q22.3 (20). The expression of the ZNF169 gene has
been found in multiple organs and tissues, including the alimentary
tract (21). Although the
physiological and pathological functions of ZNF169 are not yet
completely clear, a previous study reported that its dysregulation
may be involved in the progression of skin cancer (22). Via bioinformatic analysis of The
Cancer Genome Atlas (TCGA) database, the clinical relevance and
correlations of ZNF169 with genes in patients with CRC were
revealed. However, the precise function and mechanisms of action of
ZNF169 in CRC remain to be elucidated. Therefore, it is meaningful
to conduct experiments to elucidate the significance of ZNF169
during the development of CRC.
Ankyrin repeat and zinc-finger domain-containing 1
(ANKZF1; also known as ZNF744), another ZNF, is a peptidyl-tRNA
hydrolase and a co-factor of Cdc48 (yeast homolog to p97). The
present study analyzed the genes positively correlating with ZNF169
in CRC tissues based on TCGA database. ANKZF1 was revealed to be
one of the 10 of the most significantly positive genes, Unlike
ZNF169, ANKZF1 is located in the cytoplasm, and is a component of
the ribosome quality control complex that binds to ribosomes and
releases nascent chains from tRNA, resulting in degradation and
subsequent protein synthesis (23).
ANKZF1 plays a role in the regulation of cellular functions,
including cell cycle, apoptosis, autophagy and DNA damage repair
(24). It was recently reported
that ANKZF1 expression was associated with the prognosis of
patients with colon cancer (25,26),
and that it may be a promoting factor in the development of
infantile-onset inflammatory bowel disease (IBD) (27). However, the upstream regulator of
ANKZF1 in CRC remains unclear.
The present study aimed to investigate the clinical
significance, cellular function and underlying mechanisms of ZNF169
in CRC cell growth and proliferation based on the analysis of TCGA
database, the immunohistochemical staining (IHC staining) of CRC
patient tissues and gain-of-function/loss-of-function experiments
on CRC cell lines.
Materials and methods
IHC staining of ZNF169 and ANKZF1
The IHC staining of a human CRC tissue microarray
was conducted by Servicebio Biotechnology Co., Ltd. The paired CRC
tissues and adjacent normal tissues (at a distance of 5 cm from the
tumor margin) were collected from patients with CRC undergoing
surgical treatment between September, 2022 and March, 2023. The
inclusion criteria were patients with primary pathologically
diagnosed CRC. The exclusion criteria were patients with CRC had
been treated with hormonal therapy or chemoradiotherapy, or
patients with chronic inflammatory disease, severe
cerebro-cardiovascular disease, respiratory disease, liver or renal
disease, or female patients who were pregnant. The tissue sections
(5-µm-thick) were subjected to IHC staining for ZNF169 and ANKZF1.
Briefly, the sections were deparaffinized in xylene and hydrated in
a graded alcohol series. Antigen retrieval and endogenous
peroxidase blocking were performed using citrate buffer (pH 6) (cat
no. G1219, Wuhan Servicebio Technology Co., Ltd.) and 3% hydrogen
peroxide (cat no. 88597, MilliporeSigma), respectively. The slides
were incubated with 10% goat serum (BIOSS) for 1 h and with rabbit
polyclonal ZNF169 antibody (1:1,000 dilution; cat no. B-10117,
Weibo Biotechnology Co., Ltd.) and rabbit polyclonal ANKZF1 primary
antibody (1:500 dilution; cat no. 20446-1-AP, Proteintech Group,
Inc.) at 4°C overnight. The antigen-antibody complex was detected
with a biotinylated goat anti-rabbit antibody (1:2,500 dilution;
cat no. BA1003, Boster Biological Technology Co., Ltd.) conjugated
with streptavidin-horseradish peroxidase was used to detect the
antigen-antibody complex. Visualization was conducted using
nickel-enhanced 3,3-diaminobenzidine tetrahydrochloride (Beijing
Solarbio Science & Technology Co., Ltd.). The tissue sections
were then counterstained with hematoxylin (cat no. G1004, Wuhan
Servicebio Technology Co., Ltd.) for 3–5 min at room temperature.
For the mean density analysis of IHC staining, three or more fields
(magnification, ×200) were randomly selected from each section for
imaging. The staining density value is the relative expression
level of the proteins. The high or low expression of ZNF169 and
ANKZF1 was set during the density analysis. The present study was
approved by the Ethics Committee of Beijing Rehabilitation Hospital
of Capital Medical University (Beijing, China). A form of written
informed consent was obtained from each participant in the present
study.
Analysis of ZNF169 and ANKZF1 based on
TCGA
The transcript level of ZNF169 and ANKZF1, the
Spearman's correlation analysis of the correlation between ZNF169
and other genes, as well as the prognostic significance of ZNF169
and ANKZF1 in patients with CRC were analyzed based on TCGA
database (http://cancergenome.nih.gov).
Cells and cell culture
The 293T cells (cat no. CRL-3216) was purchased from
the American Type Culture Collection. The normal human colorectal
cell line NCM460 (cat no. CL0393) was purchased from FENGHUIBIO
(www.fenghbio.cn). The CRC cell lines, RKO (cat
no. CL-0196), HCT-116 (cat no. CL-0096), HT-29 (cat no. CL-0118),
SW620 (cat no. CL-0225) and HCT-8 (cat no. CL-0098), were purchased
from Procell Life Science & Technology Co. Ltd. STR profiling
was used to confirm the authentication of the HT-29 cell line. All
the cells were cultured in RPMI-1640 complete medium (Gibco; Thermo
Fisher Scientific, Inc.), containing 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
solution (Corning, Inc.). The cells were maintained in 6-cm plates
at 37°C in an incubator with 5% CO2.
Knockdown and overexpression of ZNF169
and ANKZF1
For the knockdown assay, the HCT-116, RKO or HT-29
CRC cells were transfected with small interfering RNAs (siRNAs)
against ZNF169, ANKZF1 or negative control siRNA using RNAiMAX
regent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 3
µl of 20 µM siRNAs and 4 µl RNAiMAX were added to 400 µl Opti-MEM
(cat no. 31985070, Gibco; Thermo Fisher Scientific, Inc.) and were
incubated at room temperature for 15 min. The mixture was then
added to 1.6 ml culture medium. After 48 h, the cells were examined
using reverse transcription-quantitative PCR (RT-qPCR), western
blot analysis, cellular function assays and other experiments. The
siRNAs were purchased from HIPPOBIO company (http://www.hippobiotec.com/). The siRNA sequences were
as follows: siCtrl, 5′-UUCUCCGAACGUGUCACGU-3′; siZNF169,
5′-GAAGCUCCAAGAUGCUCUAGU-3′; siANKZF1,
5′-GGUGCUAUAUUUCAAGGAAGA-3′.
For the overexpression assay, the coding sequences
of ZNF169 or ANKZF1 were synthesized and cloned into the pCDH
overexpression plasmids by HIPPOBIO company (http://www.hippobiotec.com/). Subsequently,
lentiviruses were produced by transfecting the 293T cells using
VigoFect (cat no. Vigorous Biotechnology, http://www.vigorousbiol.com/index.htm) with pCDH-Ctrl,
pCDH-ZNF169, or pCDH-ANKZF1 vectors, alongside the packaging
plasmids, including PSPAX2 and PDM2G. After 48 h, the virus
supernatants were harvested, filtered and concentrated.
Subsequently, the lentiviruses were added to the medium, supplied
with polybrene, to infect the HT-29 or HCT-116 cells for 48 h.
Finally, the cell lines with a stable overexpression were
constructed by treating the cells with puromycin (cat no.
HB-PU-1000, HANBIO, http://www.hanbio.net/) for 2 weeks.
RT-qPCR
Total RNA was extracted from the HCT-116, RKO or
HT-29 cells using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. ReverTra Ace™ qPCR RT Master Mix with gDNA Remover
kit (cat no. FSQ-301, Toyobo Life Science) was then applied to
reverse transcribe the RNA into cDNA, according to the
manufacturer's instructions. Subsequently, the quantification of
cDNA was performed using qPCR with SYBR master mixture (Takara Bio,
Inc.) on a Bio-Rad system (Bio-Rad Laboratories, Inc.). The PCR
thermocycling conditions were as follows: i) Holding stage, 95°C
for 3 min; ii) cycling stage, 95°C for 15 sec; iii) Cycling stage,
60°C for 30 sec; iv) cycling stage, 72°C for 30 sec; cycling stage,
2–4; 40 cycles. The qPCR primer sequences were as follows: ANKZF1
forward, 5′-CGGTTTAACCTAAAGCAACGTCT-3′ and reverse,
5′-CCGATCCAGTGTCTGCAAGT-3′; β-actin forward,
5′-CATGTACGTTGCTATCCAGGC-3′ and reverse,
5′-CTCCTTAATGTCACGCACGAT-3′. The quantification of qPCR data was
performed according the 2−ΔΔCq method (28).
Western blot analysis
Total protein was extracted from the NCM460, RKO,
HCT-116, SW620, HT-29 and HCT-8 cells using RIPA buffer (Beyotime
Institute of Biotechnology). The protein concentration was measured
using the BCA kit (Thermo Fisher Scientific, Inc.). A total of
40–70 µg proteins were then separated by SDS-PAGE on 10 or 12%
gels, followed by transfer onto PVDF membranes. The PVDF membranes
were blocked with 5% non-fat milk and incubated with primary
antibodies at 4°C overnight. After washing with PBS (Wuhan
Servicebio Technology Co., Ltd.)-Tween (Beyotime Institute of
Biotechnology), the membranes were incubated with secondary
antibodies for 2 h at room temperature. The protein signal was
detected using the ECL-Plus kit (cat no. RPN2232, Cytiva). The
antibody against ZNF169 (1:1,000; cat. no. ab225924) was purchased
from Abcam, and the antibodies against ANKZF1 (1:800; cat. no.
20447-1-AP) and GAPDH (1:10,000; 10494-1-AP) were obtained from
Proteintech Group, Inc. The mouse anti-rabbit IgG-HRP secondary
antibody (1:8,000; cat. no. sc-2357) was from Santa Cruz
Biotechnology, Inc.
Cell Counting Kit-8 (CCK-8)
The proliferation of the CRC cells was determined
using a CCK-8 kit. Briefly, a total of 3,000 HCT-116, RKO and HT-29
cells were seeded into 96-well plates, which contained 200 µl
culture medium. After 8 h, 20 µl CCK-8 regent (cat no. C0039,
Beyotime Institute of Biotechnology) were added to each well and
incubated at 37°C for 3 h. The OD value was then measured at 450
nm, which represented the relative cell viability of 0 h. At the
time points of 24, 48, 72 and 96 h, the cell viability was
sequentially determined to compare to the OD450 value at 0 h.
Colony formation assay
Following ZNF169 knockdown, 2,000 HCT-116 and RKO
cells from the siCtrl and siZNF169 groups were seeded into six-well
plates, and the cells in completed culture medium were maintained
for 8 days in a cell incubator at 37°C. Following the
overexpression of ZNF169, 500 HT-29 cells from the Ctrl and ZNF169
overexpression groups were cultured in six-well plates for 10 days.
After colonies were formed, they were washed twice with PBS (Wuhan
Servicebio Technology Co., Ltd.). Subsequently, methanol was used
to fix the colonies for 30 min at room temperature and 0.1% crystal
violet (Beyotime Institute of Biotechnology) was used to stain them
for 30 min at room temperature. Finally, the images of the colonies
were captured using a camera (EOS700D, Canon, Inc.).
Measurement of caspase-3/7
activity
Caspase-3/caspase-7 activity was detected using the
Caspase-Glo reagent (Promega Corporation). Briefly, 10,000
suspended HCT-116, RKO or HT-29 cells were seeded into 96-well
plates, to which 100 µl Caspase-Glo was added. After 90 min, the
caspase-3/caspase-7 activity was detected using a microplate reader
(M2009PR, Tecan infinite, Tecan Group, Ltd.).
EdU staining
To assess DNA synthesis, the HCT-116, RKO or HT-29
CRC cells were subjected to EdU staining using the BeyoClick™ EdU
Cell Proliferation kit with Alexa Fluor 555 (Beyotime Institute of
Biotechnology), according to the manufacturer's instructions. A
total of 5×105 HCT-116 and RKO cells transfected with a
negative control siRNA or a siRNA against ZNF169, or a total of
4×105 HT-29 cells infected with an empty control or
ZNF169 overexpression lentivirus, were seeded onto coverslips in
six-well plates. After 24 h, the cells were incubated with EdU
reagent at 37°C for 4–6 h. The cells on the coverslips were then
washed with PBS, fixed with 4% paraformaldehyde (Wuhan Servicebio
Technology Co., Ltd.) for 15 min at room temperature, treated with
0.5% Triton X-100 (Beyotime Institute of Biotechnology) for 15 min
at room temperature, and finally stained with Click Additive
Solution (Beyotime Institute of Biotechnology) for 30 min at room
temperature and DAPI (Beyotime Institute of Biotechnology) for 15
min at room temperature in the dark. Images of the EdU-positive
cells were captured under a microscope and quantified by
Photoshop.
Dual luciferase reporter assay
The promoter of ANKZF1 was cloned into the pGL3
basic vector (Promega Corporation). The HCT-116 and HT-29 cells
were seeded in 24-well plates and co-transfected with the
aforementioned luciferase vectors and the expression vectors
(knockdown or overexpression vector) using VigoFect for 48 h in a
cell incubator at 37°C. The Renilla luciferase vector,
pCMV-RL-TK (Promega Corporation), was used as an internal control
to determine the transfection efficiency. At 48 h following
transfection, the dual luciferase activity was checked by using the
Dual-Luciferase® Reporter Assay System (E1910, Promega
Corporation), according to the manufacturer's protocols.
Chromatin immunoprecipitation
(ChIP)-qPCR
A ChIP assay was performed to determine whether
ZNF169 binds to the promoter of the ANKZF1 gene in the control and
ZNF169-overexpressing cells. The ChIP-qPCR assay was performed
using the SimpleChIP enzymatic chromatin IP kit (cat no. 26156,
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocols. The purified DNA was then subjected to qPCR analysis.
The qPCR primers were designed to amplify the promoter sequence
(2,000 bp upstream of the ANKZF1 transcript) of the ANKZF1 gene.
The qPCR primer sequences were as follows: Forward,
5′-CTCCATGTTCTTCCATCAACT-3′ and reverse
5′-CTCCTTATTGGCAGGTGGTC-3′.
Statistical analysis
GraphPad Prism 8.0 software (Dotmatics) was used for
statistical analyses. The results are presented as the mean ±
standard error of the mean (meean ± SEM). Adobe Illustrator 2021
(Adobe Inc.) was utilized to create the figures. The Chi-squared
test (χ2 value) was performed to assess the relevance of
ZNF169 expression in CRC and normal samples. An unpaired Student's
t-test and one-way ANOVA followed by Tukey's post hoc test were
applied to compare the differences between two groups and among
more than two groups, respectively. P<0.05 was considered to
indicate a statistically significant difference.
Results
ZNF169 expression is upregulated in
patients with CRC
To examine the clinical relevance of ZNF169 in CRC,
the present study first collected data from TCGA and analyzed the
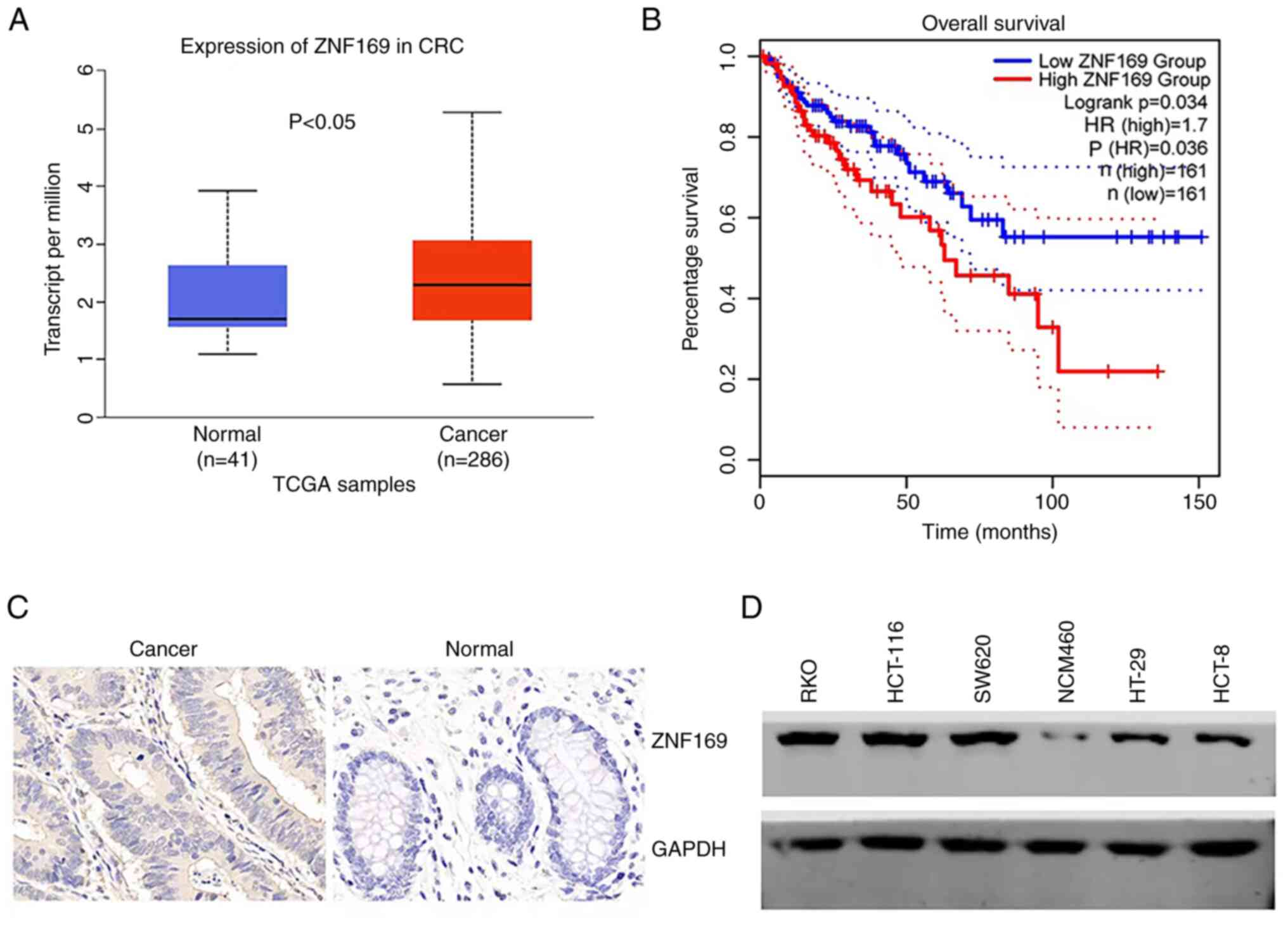
expression of ZNF169 in CRC and normal tissues. It was found that
ZNF169 expression was upregulated in CRC tissues compared with
normal tissues (Fig. 1A). GEPIA is
a newly developed interactive web server which uses the log-rank
test (the Mantel-Cox test) for survival analysis (29). The patients with CRC were
subsequently divided into the ZNF169 high and low expression
groups, and their survival rates were analyzed from GEPIA. The
patients in the high ZNF169 expression group had a shorter survival
time (Fig. 1B), suggesting that
ZNF169 functions as a negative prognostic biomarker for patients
with CRC. In order to validate the protein expression of ZNF169, a
tissue microarray containing cancer and normal tissues from
patients with CRC was subjected to the IHC staining of ZNF169. The
results indicated that the protein expression of ZNF169 was higher
in CRC tissues than in normal tissues (Fig. 1C and Table I). It was also found that ZNF169 was
highly expressed in CRC cell lines, including RKO, HT-29 and
HCT-116, as compared with the NCM460 normal cell line (Fig. 1D).
 | Table I.Expression of ZNF169 in CRC and
normal tissues examined using immunohistochemistry. |
Table I.
Expression of ZNF169 in CRC and
normal tissues examined using immunohistochemistry.
| ZNF169
expression | Cancer tissue (no.
of samples) | Normal tissue (no.
of samples) | χ2
value | P-value |
|---|
| High
expression | 15 | 6 | 6.75 | <0.01 |
| Low expression | 7 | 15 |
|
|
| Total | 22 | 21 |
|
|
ZNF169 promotes the proliferation and
growth of CRC cells
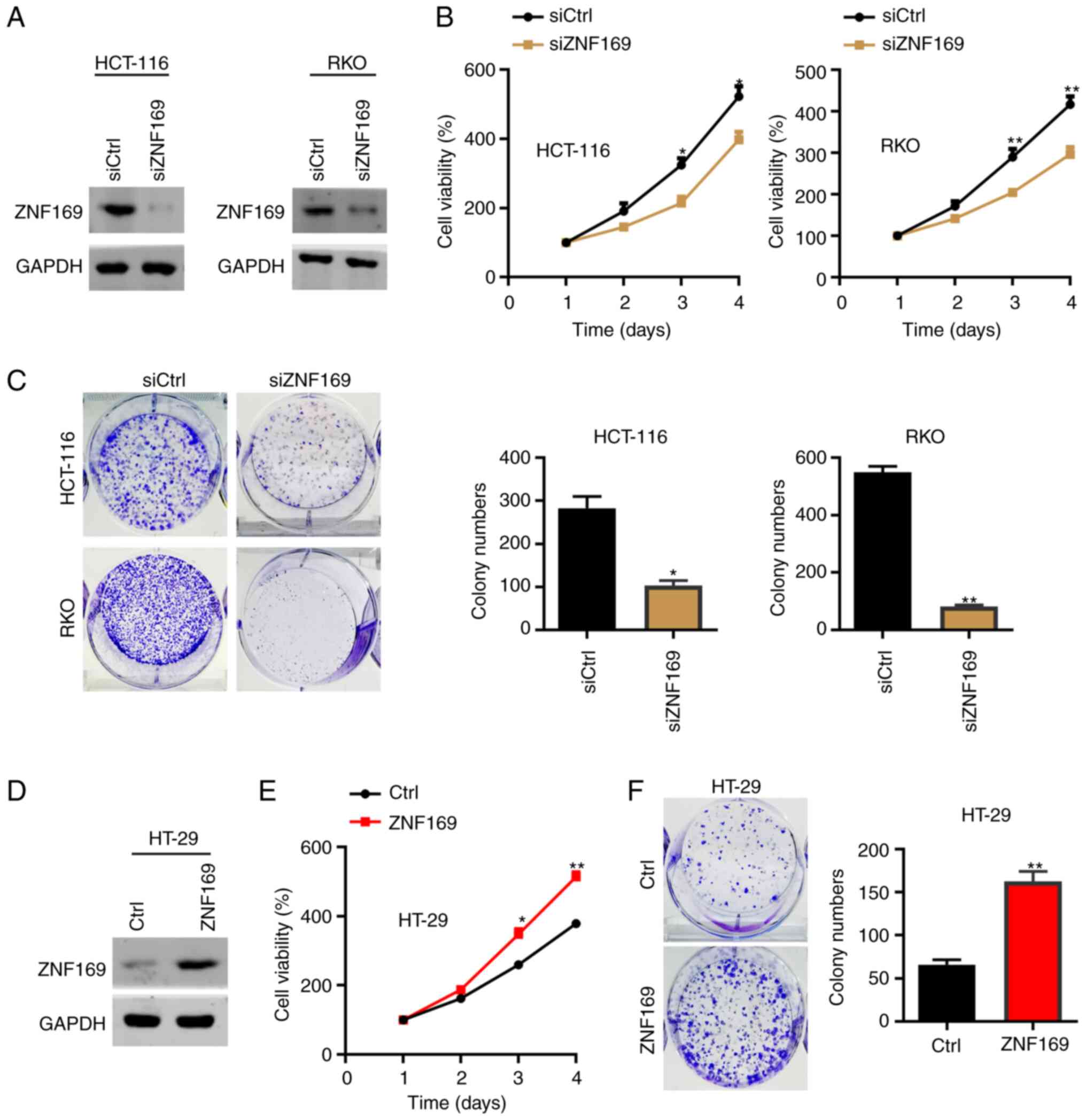
The present study then explored the cellular
function of ZNF169 in CRC cells. As the HCT-116 and RKO cells had a
relatively higher expression of ZNF169, these cells were
transfected with a negative control siRNAs and with siRNAs against
and ZNF169. The results of western blot analysis revealed that
ZNF169 was efficiently silenced in the siZNF169 cells (Fig. 2A). The results of the CCK-8 assay
demonstrated that ZNF169 knockdown suppressed the proliferation of
the HCT-116 and RKO cells (Fig.
2B). Consistently, the colony formation capacity of the HCT-116
and RKO cells was also suppressed following the knockdown of ZNF169
(Fig. 2C). Subsequently, the HT-29
cells were transfected with an empty control or ZNF169
overexpression lentivirus, and the results of western blot analysis
validated the overexpression efficacy (Fig. 2D). The results of CCK-8 and colony
formation assays demonstrated that the ectopic overexpression of
ZNF169 significantly promoted the malignant growth of the HT-29
cells (Fig. 2E and F). These
results suggested that ZNF169 may be critical for the growth and
proliferation of CRC cells.
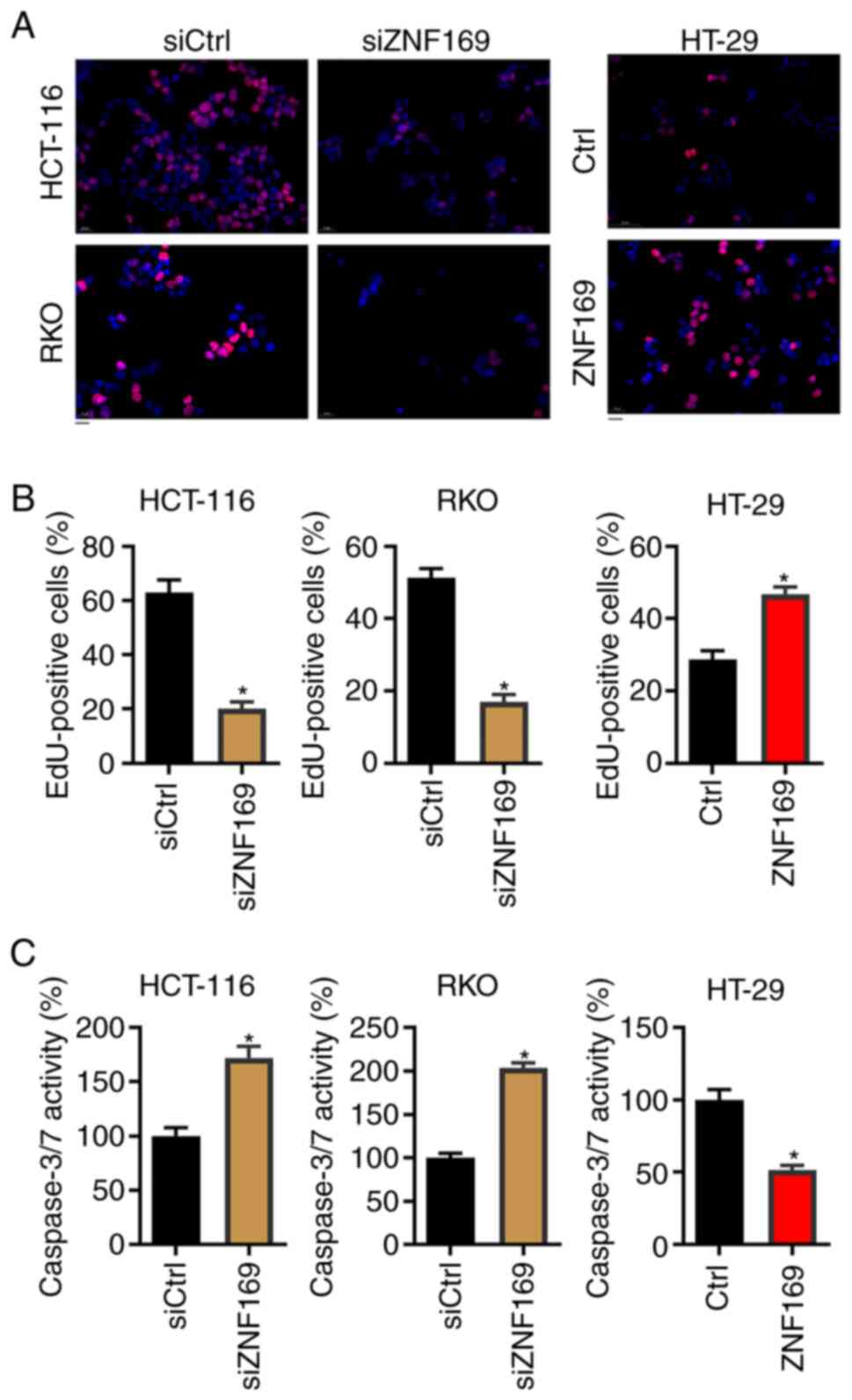
ZNF169 potentiates EdU staining and
inhibits caspase activity in CRC cells
To examine the mechanisms through which ZNF169
contributes to the proliferation of CRC cells, the CRC cells in
which ZNF169 was knocked down or overexpressed were subjected to
EdU staining, which predicts DNA synthesis. It was found that
ZNF169 knockdown significantly suppressed the proportion of
EdU-positive cells in both cell lines (Fig. 3A and B). By contrast, the ectopic
overexpression of ZNF169 enhanced the proportion of EdU-positive
HT-29 cells (Fig. 3A and B). The
present study then analyzed the activity of caspase-3/7, which can
be used to predict apoptosis, following the knockdown or
overexpression of ZNF169 in the cells. As shown in Fig. 3C, ZNF169 knockdown enhanced
caspase-3/7 activity in the HCT-116 and RKO cells. Conversely,
ZNF169 overexpression suppressed caspase-3/7 activity in the HT-29
cells. These results indicated that ZNF169 promotes DNA synthesis
and suppresses the apoptosis of CRC cells.
ZNF169 transcriptionally activates
ANKZF1 in CRC cells
Since ZNF169 serves as a transcription factor in
eukaryotes, there must be downstream effectors that participate in
the ZNF169-induced promotion of CRC cell proliferation. Therefore,
the present study then analyzed the genes positively correlated
with ZNF169 in CRC tissues based on TCGA database. A total of 10 of
the most significantly positively correlated genes, including
ZNF789 and ANKZF1, are illustrated in Fig. 4A. Since the overexpression of ANKZF1
has been reported to be associated with the poor prognosis of
patients with CRC (25,26), the present study aimed to explore
whether ZNF169 regulated ANKZF1 in the present study, and we aim to
explore the significance of other genes positively correlated with
ZNF169, such as ZNF789 and AGAP4, in the subsequent studies.. Based
on the results of RT-qPCR and western blot analysis, it was found
that ZNF169 knockdown markedly suppressed the mRNA and protein
expression levels of ANKZF1 in the HCT-116 and RKO cells (Fig. 4B and C). By contrast, the
overexpression of ZNF169 potentiated the expression of ANKZF1 in
the HT-29 cells (Fig. 4B and C). In
order to examine whether ZNF169 regulates the transcriptional
activity of the ANKZF1 gene, the promoter sequence of the ANKZF1
gene was cloned into a pGL3.basic vector and a dual luciferase
activity assay was performed. It was demonstrated that ZNF169
knockdown inhibited the luciferase activity of the ANKZF1 promoter,
whereas ZNF169 overexpression promoted it (Fig. 4D). In addition, a ChIP-qPCR assay
was carried out to examine the interaction between the ZNF169
protein and the promoter sequence of the ANKZF1 gene. It was found
that the promoter sequence of the ANKZF1 gene was highly enriched
in the ANKZF1-Flag group of CRC cells (Fig. 4E). Collectively, ZNF169 may
potentiate the transcriptional activity of the ANKZF1 gene by
directly interacting with its promoter sequence.
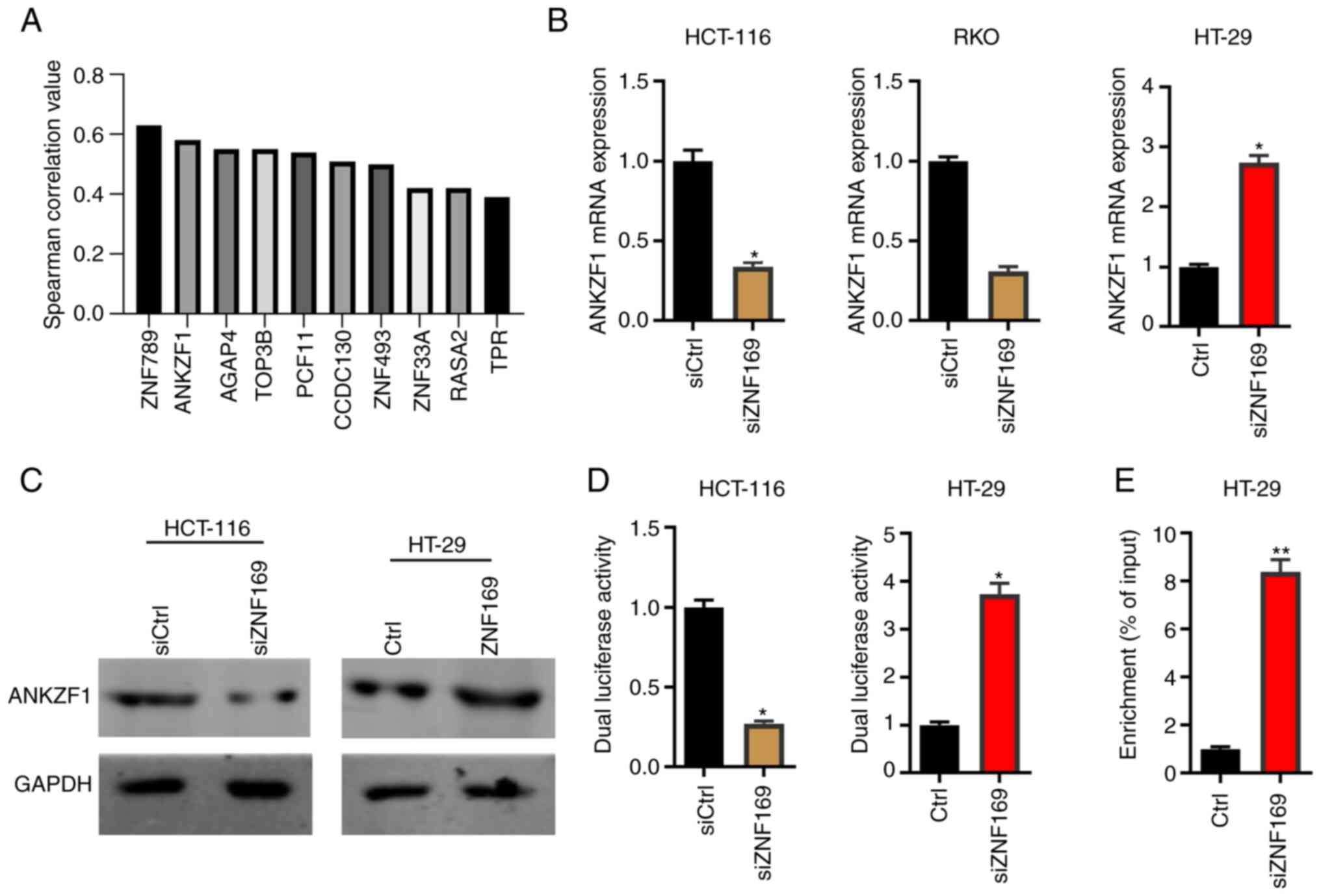 | Figure 4.ZNF169 potentiates the
transcriptional activity of the ANKZF1 gene in CRC cells. (A) The
positively correlated genes of ZNF169 were analyzed in patients
with CRC based on The Cancer Genome Atlas database. (B) Reverse
transcription-quantitative PCR assay was used to detect the mRNA
expression levels of ANKZF1 in siCtrl and siZNF169-transfected
HCT-116 cells and RKO cells, and in Ctrl and ZNF169-overexpressing
HT-29 cells. *P<0.05, vs. the control. (C) Western blot analysis
was used to detect the protein expression levels of ANKZF1 in
siCtrl and siZNF169-transfected HCT-116 cells, and in Ctrl and
ZNF169-overexpressing HT-29 cells. (D) Dual luciferase activity was
detected in siCtrl and siZNF169-transfected HCT-116 cells, and in
Ctrl and ZNF169-overexpressing HT-29 cells. *P<0.05, vs. the
control. (E) Chromatin immunoprecipitation-quantitative PCR was
performed to analyze the interaction between ZNF169 and the
promoter sequence of ANKZF1 in CRC cells. **P<0.01, vs. the
control. During the western blot analysis experiments, the blots
were cut prior to incubation with the antibodies. The results are
presented as the mean ± standard error of the mean. ZNF169, zinc
finger protein 169; CRC, colorectal cancer; ANKZF1, ankyrin repeat
and zinc-finger domain-containing 1. |
ZNF169 contributes to the
proliferation of CRC cells through upregulation of ANKZF1
As ZNF169 was found to promote CRC cell
proliferation and positively regulate the expression of ANKZF1, the
present study then aimed to determine whether ANKZF1 participates
in CRC growth. To this aim, overexpression and knockdown assays of
ANKZF1 were performed in the CRC cells. The results of RT-qPCR
revealed that ANKZF1 could be efficiently silenced by siANKZF1 when
compared with siCtrl in the HT-29 cells (Fig. S1A). Thus, ANKZF1 was knocked down
(Fig. S1B) in
ZNF169-overexpressing CRC cells and the cells were subjected to
CCK-8 and colony formation assays. The results of western blot
analysis revealed that ANKZF1 was efficiently silenced after the
ZNF169-overexpressing cells were transfected with siANKZF1
(Fig. 5A). The results of CCK-8 and
colony formation assays demonstrated that ANKZF1 knockdown
suppressed the growth and proliferation of the CRC cells in which
ZNF169 was overexpressed (Fig. 5B and
C). To validate the function of ANKZF1, ANKZF1 was then
overexpressed in the HCT-116 cells in which ZNF169 was silenced.
ANKZF1 overexpression lentivirus significantly increased the mRNA
level of ANKZF1 in the HCT-116 cells when compared with the cells
transfected with the Ctrl lentivirus (Fig. S1B). Based on the results of western
blot analysis, ANKZF1 was found to be overexpressed in the HCT-116
cells in which ZNF169 was silenced (Fig. 5D). ANKZF1 overexpression restored
the proliferation and growth of the HCT-116 cells in which ZNF169
was knocked down (Fig. 5E and F).
These results suggested that ZNF169 may contribute to the growth
and proliferation of CRC cells through the upregulation of
ANKZF1.
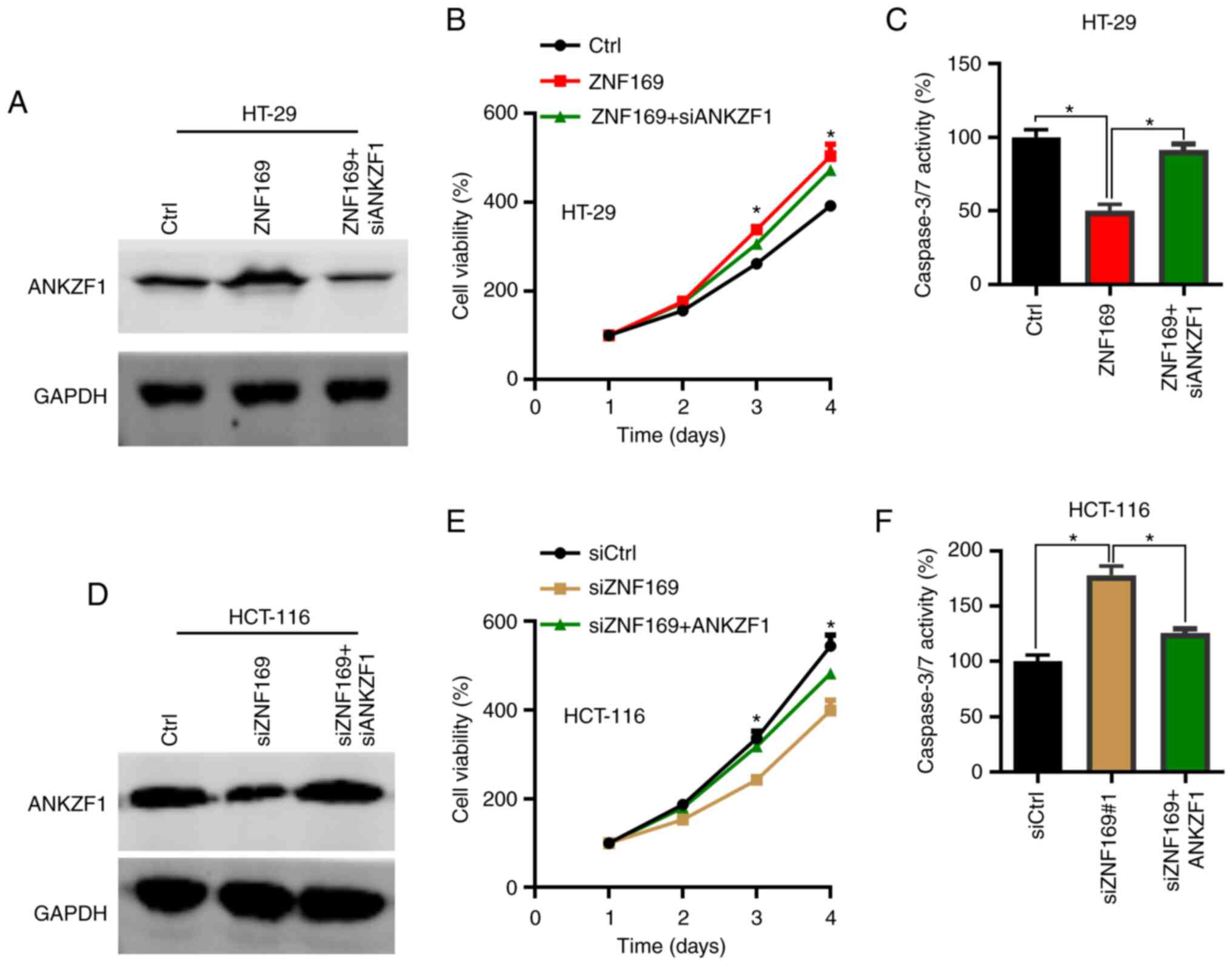 | Figure 5.ANKZF1 is responsible for the
proliferative ability of colorectal cancer cells promoted by
ZNF169. (A) Western blot analysis of ANKZF1 in Ctrl, ZNF169 and
ZNF169 + siANKZF1-transfected HT-29 cells. (B) CCK-8 analysis of
cell proliferation in the Ctrl, ZNF169 and ZNF169 +
siANKZF1-transfected HT-29 cells. *P<0.05, Ctrl vs. ZNF169 and
ZNF169 vs. ZNF169 + siANKZF1 group. (C) The activity of caspase-3/7
was examined in the Ctrl, ZNF169 and ZNF169 + siANKZF1-transfected
HT-29 cells. *P<0.05. (D) Western blot analysis of ANKZF1 in the
siCtrl, siZNF169 and siZNF169 + ANKZF1-transfected HCT-116 cells.
(E) CCK-8 analysis of cell proliferation in the siCtrl, siZNF169
and siZNF169 + ANKZF1-transfected HCT-116 cells. *P<0.05, siCtrl
vs. siZNF169 and siZNF169 vs. siZNF169 + ANKZF1. (F) The activity
of caspase-3/7 was examined in the siCtrl, siZNF169 and siZNF169 +
ANKZF1-transfected HCT-116 cells. *P<0.05. During the western
blot analysis experiments, the blots were cut prior to incubation
with the antibodies. The results are presented as the mean ±
standard error of the mean. ZNF169, zinc finger protein 169;
ANKZF1, ankyrin repeat and zinc-finger domain-containing 1. |
ANKZF1 overexpression is associated
with the poor prognosis of patients with CRC
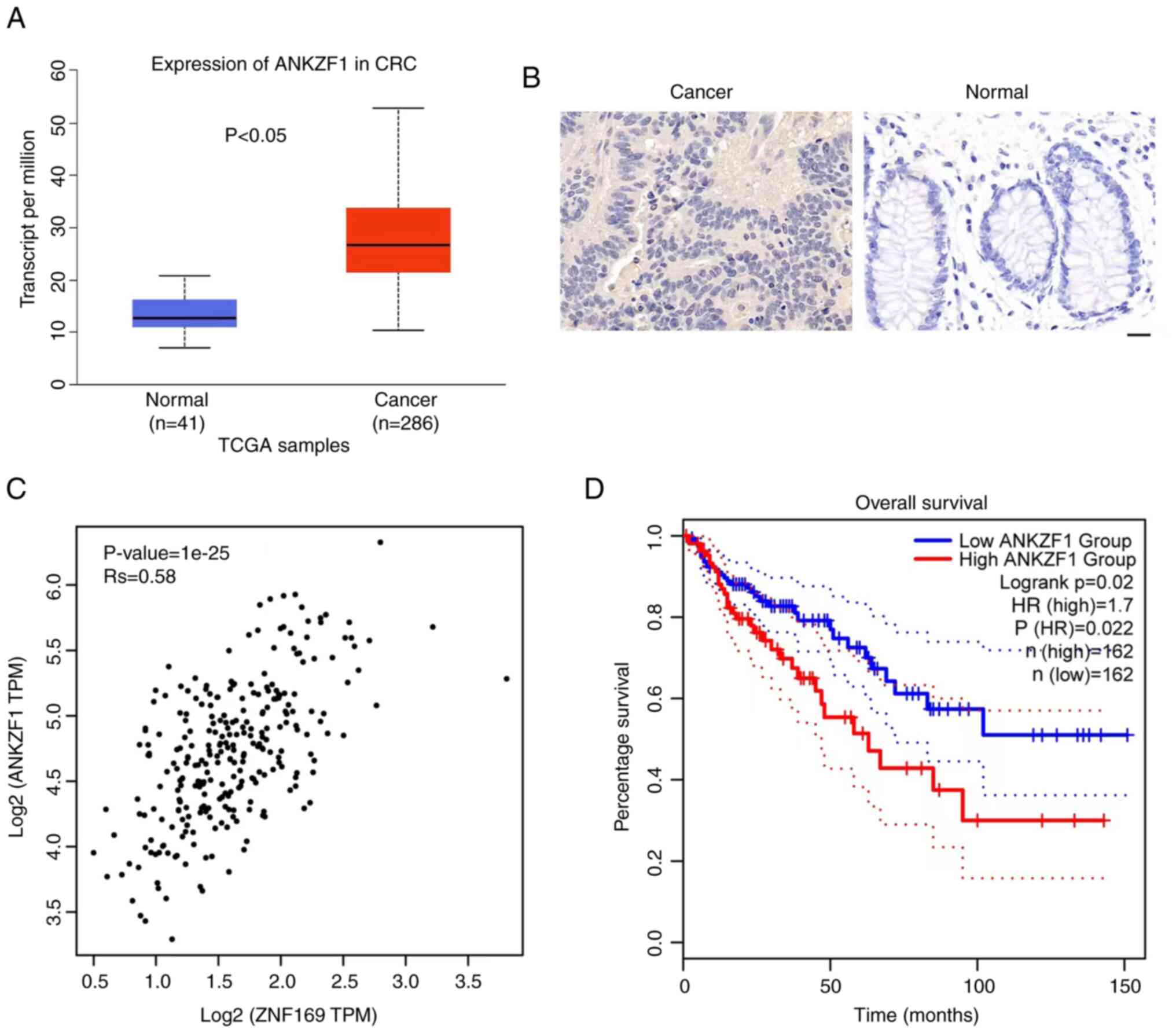
Finally, the clinical relevance of ANKZF1 was
investigated in patients with CRC. Based on TCGA database, ANKZF1
expression was upregulated in the cancer tissues of patients with
CRC compared with normal tissues (Fig.
6A). Consistently, the results of IHC staining revealed that
the protein expression of ANKZF1 was higher in CRC tissues compared
with adjacent normal tissues (Fig.
6B). It was also demonstrated that there was a positive
correlation between ZNF169 and ANKZF1 expression in the tissues of
patients with CRC (Fig. 6C and
Table II). Importantly, a high
expression of ANKZF1 was associated with a poor prognosis of
patients with CRC as compared with those with a low expression of
ANKZF1 (Fig. 6D). Thus, ANKZF1 may
be considered a negative prognostic biomarker for patients with
CRC.
 | Table II.Results of Spearman's correlation
analysis of the correlation between ZNF169 and ANKZF1 expression in
22 CRC tissues examined using immunohistochemistry. |
Table II.
Results of Spearman's correlation
analysis of the correlation between ZNF169 and ANKZF1 expression in
22 CRC tissues examined using immunohistochemistry.
|
| ZNF169 |
|---|
|
|
|
|---|
| Gene | Rs value | P-value |
|---|
| ANKZF1 | 0.621 | <0.001 |
Discussion
Members of the ZNF family can function as either
tumor suppressor genes or oncogenes (16). Recently, the importance of ZNFs in
cancer onset and progression has been actively studied. The
findings have demonstrated that ZNFs are involved in the regulation
of the malignancy of cancer cells through a variety of mechanisms,
including by functioning as transcription factors (15,30).
The results from the present study demonstrated that ZNF169
functioned as an oncogene to promote the development of CRC by
potentiating the expression of ANKZF1. To the best of our
knowledge, this is the first study on the role of ZNF169 in
tumorigenesis. ZNF169 is considered to function as a transcription
factor, and may have functions in DNA-binding transcription
activity, particularly in RNA polymerase II transcription
regulatory region sequence-specific DNA-binding activity (31,32).
RNA polymerase II transcribes all protein-coding genes in
eukaryotic genomes (33);
therefore, ZNF169 could indirectly affect various biological
processes. ZNF169 has been reported to be relevant to obesity,
malignant essential hypertension, speech-language disorder-1,
Fanconi anemia and other human disorders (22,34,35).
However, both the regulation of ZNF169 on target genes and the
regulation of ZNF169 expression are not yet completely clear.
Bhattacharya and Ghosh (36)
demonstrated that herpes virus-associated ubiquitin specific
protease is implicated in the regulation of transcription factors,
including ZNF169. Huttlin et al (37) employed robust affinity
purification-mass spectrometry methodology and demonstrated that
ZNF169 interacts with dozens of other proteins; however, the
precise results of these interactions need to be further
investigated.
The data of the present study demonstrated that
ZNF169 promoted DNA synthesis and reduced caspase-3 and −7 activity
in CRC cell lines. This may be one of the mechanisms underlying the
promoting effects of ZNF169 on CRC malignancy. These results
further indicated the oncogenic role of ZNF169 in the progression
of CRC. The significance of ZNFs has been reported in CRC
development by other studies. Cheng et al (38) found that ZNF277 was overexpressed in
human CRC samples. Xie et al (39) further confirmed that ZNF277 was
uniquely expressed in early colorectal stem cells and
undifferentiated transit-amplifying cells. In addition, it was
reported that the overexpression of ZNF277 is critical for
maintaining CRC growth (39).
ZNF367 is another oncogene in CRC. The depletion of ZNF367 has been
reported to blunt the proliferation and invasion of CRC cells via
the inactivation of the YAP signaling pathway (40). In CRC, ZNF281 also plays a role in
metastasis through the regulation of the epithelial-mesenchymal
transition (EMT). During EMT, c-MYC induces the expression of
ZNF281 in a SNAIL-dependent manner, while microRNA (miR)-34a
decreases the post-transcriptional level of ZNF281. Notably, p53
can promote the expression of miR-34a (41). ZNF281, functioning as a regulator,
has been shown to be associated with an increased
migration/invasion and an enhanced β-catenin activity (42). There are numerous proteins in the
ZNF family, and their functions have a wide range of molecular
effects on several cellular processes (15). It has been reported that ZNFs can
affect cancer malignancy via various mechanisms (16). A single ZNF can function as a
suppressor gene in certain types of tumors, whereas it may function
as an oncogene in others. ZBP89 (also referred to as ZNF148) is
C2H2-type transcription factor. On the one hand, ZBP89 exerts its
oncogenic function by regulating matrix metallopeptidase 3, and
inhibiting ornithine decarboxylase and vimentin, in breast cancer,
melanoma and gastric cancer (43–45).
These three molecules are involved in tumor development, migration,
invasion and metastasis. On the other hand, ZBP89 may function as a
tumor suppressor gene by inhibiting cell proliferation and inducing
apoptosis by regulating the β-catenin pathway in CRC (46,47).
In the present study, it was found that ZNF789 and
ANKZF1 were the top two genes which positively correlated with the
expression of ZNF169 in the tissues of patients with CRC. To the
best of our knowledge, the involvement of ZNF789 has not been
reported in cancer. The present study focused on ANKZF1 and
identified ANKZF1 as a downstream effector in the ZNF169-induced
promotion of CRC cell proliferation. ZNF169 positively regulated
the expression of ANKZF1 by directly interacting with the promoter
of the ANKZF1 gene, and ANKZF1 contributed to the accelerated
proliferation of CRC cells triggered by ZNF169. Apart from the
regulation of ANKZF1 by ZNF169, there may be different pathways
involved in the regulation of ANKZF1 expression, such as the IFN
receptor family (48). Lai and Park
(49) demonstrated that MET, also
known as hepatocyte growth factor receptor, was involved in the
regulation of the transcription of the ANKZF1 gene in gastric
cancer cells, as the expression of the ANKZF1 gene was shown to be
increased when the cells were treated with a MET molecule
inhibitor. Furthermore, the activity of peroxisome
proliferator-activated receptor could inhibit the transcription of
ANKZF1 in monocyte-derived dendritic cells (50). It has been reported that the
increased expression of ANKZF1 may predict the malignant
progression and poor survival of patients with CRC (25,51).
by contrast, the knockdown of ANKZF1 suppresses the growth and
invasion of CRC cells (52). These
results suggest that ANKZF1 may function as an oncogenic protein in
CRC. van Haaften-Visser et al (27) demonstrated that ANKZF1 mutations lad
to a reduction in mitochondrial integrity and respiration, which
may be a pathological factor in the pathogenesis of infantile-onset
IBD.
A number of issues remain unclear, such as the
mechanisms underlying the effects of the upregulation of ZNF169
expression in patients with CRC and the additional mechanisms
upregulating ANKZF1 expression in CRC. These questions need to be
addressed in future studies. DNA methylation of gene promoters and
copy number variants of genes are two key events contributing to
the dysregulation of genes; however, it remains to be determined
whether these events play a role in the expression of ZNF169 and/or
ANKZF1 in CRC. It may be worthwhile to explore the role of the
upstream or downstream factors in the regulation of the
ZNF169/ANKZF1 axis.
In conclusion, the present study demonstrated that
the expression of ZNF169 was markedly increased in CRC, and high
expression pf ZNF169 was associated with the poor overall survival
of patients with CRC. Furthermore, ZNF169 could positively regulate
its downstream factor, ANKZF1, which in turn accelerated CRC cell
progression. The present study demonstrated the biological function
of ZNF169 in CRC, which may provide novel insight into the
diagnosis and treatment of CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific Research Fund
of Beijing Rehabilitation Hospital, Capital Medical University
(2019-043, 2019R-001 2020-056 and 2022-057), and the Scientific
Research Fund of Beijing Anorectal Society (2020ABCP002).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QG, CK and HS designed the study. JZ, YW, SH, MX,
MZ, JW, JS and XC performed the majority of the experiments. QG, CK
and JZ analyzed the data. DD, YW, JW, JS and XC provided assistance
for the experiments and data analysis. QG, CK HS and JZ wrote the
draft of the manuscript. QG revised the manuscript. QG, CK and JZ
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study is approved by the Ethics
Committee of the Beijing Rehabilitation Hospital of Capital Medical
University (Beijing, China) (no. 2022bkkyLW003). A written form of
informed consent was obtained from each participant in the present
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ANKZF1
|
ankyrin repeat and zinc-finger
domain-containing 1
|
|
CCK-8
|
Cell Counting Kit-8
|
|
CRC
|
colorectal cancer
|
|
Chip-qPCR
|
chromatin immunoprecipitation
(Chip)-qPCR
|
|
HIF-1α
|
hypoxia-inducible factor 1α
|
|
IHC staining
|
immunohistochemical staining
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
TCGA
|
The Cancer Genome Atlas
|
|
ZNF169
|
zinc finger protein 169
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu X, Lin H and Li S: Prognoses of
different pathological subtypes of colorectal cancer at different
stages: A population-based retrospective cohort study. BMC
Gastroenterol. 19:1642019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanth P and Inadomi JM: Screening and
prevention of colorectal cancer. BMJ. 374:n18552021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Assis J, Coutinho L, Oyeyemi IT,
Oyeyemi OT and Grenfell RFEQ: Diagnostic and therapeutic biomarkers
in colorectal cancer: A review. Am J Cancer Res. 12:661–680.
2022.PubMed/NCBI
|
|
5
|
Sardo E, Napolitano S, Della Corte CM,
Ciardiello D, Raucci A, Arrichiello G, Troiani T, Ciardiello F,
Martinelli E and Martini G: Multi-Omic approaches in colorectal
cancer beyond genomic data. J Pers Med. 12:1282022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu G, Jin L, Sun W, Wang S and Liu N:
Proteomics of post-translational modifications in colorectal
cancer: Discovery of new biomarkers. Biochim Biophys Acta Rev
Cancer. 1877:1887352022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salita T, Rustam Y, Mouradov D, Sieber OM
and Reid GE: Reprogrammed lipid metabolism and the Lipid-Associated
hallmarks of colorectal cancer. Cancers (Basel). 14:37142022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang M, Yang H, Ji L, Hu X, Tian G, Wang B
and Yang J: A multi-omics machine learning framework in predicting
the survival of colorectal cancer patients. Comput Biol Med.
146:1055162022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Andrei P, Battuello P, Grasso G, Rovera E,
Tesio N and Bardelli A: Integrated approaches for precision
oncology in colorectal cancer: The more you know, the better. Semin
Cancer Biol. 84:199–213. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ashktorab H and Brim H: Colorectal cancer
subtyping. Nat Rev Cancer. 22:68–69. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Ma X, Chakravarti D, Shalapour S and
DePinho RA: Genetic and biological hallmarks of colorectal cancer.
Genes Dev. 35:787–820. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sagaert X, Vanstapel A and Verbeek S:
Tumor heterogeneity in colorectal cancer: What do we know so far?
Pathobiology. 85:72–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bu S, Lv Y, Liu Y, Qiao S and Wang H: Zinc
finger proteins in neuro-related diseases progression. Front
Neurosci. 15:7605672021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cassandri M, Smirnov A, Novelli F, Pitolli
C, Agostini M, Malewicz M, Melino G and Raschellà G: Zinc-finger
proteins in health and disease. Cell Death Discov. 3:170712017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jen J and Wang YC: Zinc finger proteins in
cancer progression. J Biomed Sci. 23:532016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ecco G, Imbeault M and Trono D: KRAB zinc
finger proteins. Development. 144:2719–2729. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Han M, Zhang H, Liu F, Pan Y, Zhu J,
Liao Z, Chen X and Zhang B: Structures and biological functions of
zinc finger proteins and their roles in hepatocellular carcinoma.
Biomark Res. 10:22022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chidambaram A, Gailani M, Gerrard B,
Stewart C, Goldstein A, Chumakov I, Bale AE and Dean M:
Characterization of a YAC contig containing the NBCCS locus and a
novel Kruppel-type zinc finger sequence on chromosome segment
9q22.3. Genes Chromosomes Cancer. 18:212–218. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Levanat S, Chidambaram A, Wicking C,
Bray-Ward P, Pressman C, Toftgard R, Gailani MR, Myers JC,
Wainwright B, Dean M and Bale AE: Pulsed-field gel electrophoresis
and FISH mapping of chromosome 9q22: Placement of a novel zinc
finger gene within the NBCCS and ESS1 region. Cytogenet Cell Genet.
76:208–213. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fagerberg L, Hallström BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
Tissue-specific expression by Genome-Wide integration of
transcriptomics and Antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bose S, Morgan LJ, Booth DR, Goudie DR,
Ferguson-Smith MA and Richards FM: The elusive multiple
self-healing squamous epithelioma (MSSE) gene: Further mapping,
analysis of candidates, and loss of heterozygosity. Oncogene.
25:806–812. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Joazeiro CAP: Ribosomal stalling during
translation: Providing substrates for Ribosome-Associated protein
quality control. Annu Rev Cell Dev Biol. 33:343–368. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stapf C, Cartwright E, Bycroft M, Hofmann
K and Buchberger A: The general definition of the
p97/valosin-containing protein (VCP)-interacting motif (VIM)
delineates a new family of p97 cofactors. J Biol Chem.
286:38670–38678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou X, Shang YN, Lu R, Fan CW and Mo XM:
High ANKZF1 expression is associated with poor overall survival and
recurrence-free survival in colon cancer. Future Oncol.
15:2093–2106. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sajadi M, Fazilti M, Nazem H, Mahdevar M
and Ghaedi K: The expression changes of transcription factors
including ANKZF1, LEF1, CASZ1 and ATOH1 as a predictor of survival
rate in colorectal cancer: A large-scale analysis. Cancer Cell Int.
22:3392022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van Haaften-Visser DY, Harakalova M,
Mocholi E, van Montfrans JM, Elkadri A, Rieter E, Fiedler K, van
Hasselt PM, Triffaux EMM, van Haelst MM, et al: Ankyrin repeat and
zinc-finger domain-containing 1 mutations are associated with
infantile-onset inflammatory bowel disease. J Biol Chem.
292:7904–7920. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye Q, Liu J and Xie K: Zinc finger
proteins and regulation of the hallmarks of cancer. Histol
Histopathol. 34:1097–1109. 2019.PubMed/NCBI
|
|
31
|
Mokry M, Hatzis P, Schuijers J, Lansu N,
Ruzius FP, Clevers H and Cuppen E: Integrated genome-wide analysis
of transcription factor occupancy, RNA polymerase II binding and
steady-state RNA levels identify differentially regulated
functional gene classes. Nucleic Acids Res. 40:148–158. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gaudet P, Livstone MS, Lewis SE and Thomas
PD: Phylogenetic-based propagation of functional annotations within
the Gene Ontology consortium. Brief Bioinform. 12:449–462. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schier AC and Taatjes DJ: Structure and
mechanism of the RNA polymerase II transcription machinery. Gene
Dev. 34:465–488. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Turcot V, Lu YC, Highland HM, Schurmann C,
Justice AE, Fine RS, Bradfield JP, Esko T, Giri A, Graff M, et al:
Protein-altering variants associated with body mass index implicate
pathways that control energy intake and expenditure in obesity. Nat
Genet. 50:766–767. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ewing AD, Cheetham SW, McGill JJ, Sharkey
M, Walker R, West JA, West MJ and Summers KM: Microdeletion of
9q22.3: A patient with minimal deletion size associated with a
severe phenotype. Am J Med Genet A. 185:2070–2083. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bhattacharya S and Ghosh MK: HAUSP
regulates c-MYC expression via de-ubiquitination of TRRAP. Cell
Oncol. 38:265–277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huttlin EL, Bruckner RJ, Paulo JA, Cannon
JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP and Parzen H:
Architecture of the human interactome defines protein communities
and disease networks. Nature. 545:505–509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheng K, Xie G, Khurana S, Heath J,
Drachenberg CB, Timmons J, Shah N and Raufman JP: Divergent effects
of muscarinic receptor subtype gene ablation on murine colon
tumorigenesis reveals association of M3R and zinc finger protein
277 expression in colon neoplasia. Mol Cancer. 13:772014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xie G, Peng Z, Liang J, Larabee SM,
Drachenberg CB, Yfantis H and Raufman JP: Zinc finger protein 277
is an intestinal transit-amplifying cell marker and colon cancer
oncogene. JCI Insight. 7:e1508942022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lei T, Gao Y, Duan Y, Cui C, Zhang L and
Si M: Inhibition of zinc finger protein 367 exerts a tumor
suppressive role in colorectal cancer by affecting the activation
of oncogenic YAP1 signaling. Environ Toxicol. 36:2278–2290. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hahn S, Jackstadt R, Siemens H, Hünten S
and Hermeking H: SNAIL and miR-34a feed-forward regulation of
ZNF281/ZBP99 promotes epithelial-mesenchymal transition. Embo J.
32:3079–3095. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Taank Y and Agnihotri N: Understanding the
regulation of beta-catenin expression and activity in colorectal
cancer carcinogenesis: Beyond destruction complex. Clin Transl
Oncol. 23:2448–2459. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Taniuchi T, Mortensen ER, Ferguson A,
Greenson J and Merchant JL: Overexpression of ZBP-89, a zinc finger
DNA binding protein, in gastric cancer. Biochem Biophys Res Commun.
233:154–160. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rieber MS, Zangemeister-Wittke U and
Rieber M: p53-Independent induc tion-independent induction of
apoptosis in human melanoma cells by a bcl-2/bcl-xL bispecific
antisense oligonucleotide. Clin Cancer Res. 7:1446–1451.
2001.PubMed/NCBI
|
|
45
|
Serova M, Calvo F, Lokiec F, Koeppel F,
Poindessous V, Larsen AK, Laar ES, Waters SJ, Cvitkovic E and
Raymond E: Characterizations of irofulven cytotoxicity in
combination with cisplatin and oxaliplatin in human colon, breast,
and ovarian cancer cells. Cancer Chemoth Pharm. 57:491–499. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bandrés E, Malumbres R, Cubedo E, Honorato
B, Zarate R, Labarga A, Gabisu U, Sola JJ and García-Foncillas J: A
gene signature of 8 genes could identify the risk of recurrence and
progression in Dukes' B colon cancer patients. Oncol Rep.
17:1089–1094. 2007.PubMed/NCBI
|
|
47
|
Essien BE, Sundaresan S, Ocadiz-Ruiz R,
Chavis A, Tsao AC, Tessier AJ, Hayes MM, Photenhauer A,
Saqui-Salces M, Kang AJ, et al: Transcription factor ZBP-89 drives
a feedforward loop of β-Catenin expression in colorectal cancer.
Cancer Res. 76:6877–6887. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mei H, Zhao L, Li W, Zheng Z, Tang D, Lu X
and He Y: Inhibition of ferroptosis protects House Ear
Institute-Organ of Corti 1 cells and cochlear hair cells from
cisplatin-induced ototoxicity. J Cell Mol Med. 24:12065–12081.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lai A and Park M: Time-course
transcriptomic analysis of a panel of MET receptor tyrosine
kinase-inhibited gastric cancer cell lines, This is version 1.0 of
an annotated derivative of the original dataset, which can be found
in GSE54532. Version 1.0. Signaling Pathways Project Datasets.
2014.
|
|
50
|
Szatmari I, Pap A, Rühl R, Ma JX,
Illarionov PA, Besra GS, Rajnavolgyi E, Dezso B and Nagy L:
PPARgamma controls CD1d expression by turning on retinoic acid
synthesis in developing human dendritic cells. J Exp Med.
203:2351–2362. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sajadi M, Fazilti M, Nazem H, Mahdevar M
and Ghaedi K: The expression changes of transcription factors
including ANKZF1, LEF1, CASZ1, and ATOH1 as a predictor of survival
rate in colorectal cancer: A large-scale analysis. Cancer Cell Int.
22:3392022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen P, Li Z, Liang Y, Wei M, Jiang H,
Chen S and Zhao Z: Identification of Hypoxia-Associated signature
in colon cancer to assess tumor immune microenvironment and predict
prognosis based on 14 Hypoxia-Associated genes. Int J Gen Med.
16:2503–2518. 2023. View Article : Google Scholar : PubMed/NCBI
|