In 2020, human digestive system tumors, mainly
esophageal cancer (EC), hepatocellular carcinoma (HCC), pancreatic
cancer (PC), gastric cancer (GC) and colorectal cancer (CRC),
resulted in >5 million new cases and ~4 million cancer deaths
worldwide. These malignancies are associated with personal
suffering, and impose a substantial economic burden on patients,
families and society (1,2). For example, the median medical
expenditure per patient with EC in China increased from 6,851 to
57,554 CNY during 1996–2013, with an average growth rate of 11.89%
(3). From a societal perspective,
the costs associated with CRC in Europe reached ~19 billion EUR in
2018 (4). In the past few decades,
although modern cancer treatments, including surgical treatment,
chemotherapy, radiotherapy, immunotherapy, targeted therapy and
traditional Chinese medicine, have markedly improved the quality of
life of patients, limited effects have been achieved on patients
with advanced or metastatic cancer (5–7). It is
well known that the development processes of cancer are influenced
by genetic and environmental factors, including environmental
agents; lifestyle habits, such as a poor diet; and social behavior,
such as immoderate consumption of alcohol. Various clinical trials
have been conducted on targeted therapy for digestive tract cancer.
Since gene mutations, such as those in the BRCA2 gene, are
prevalent in PC and CRC, the development of corresponding targeted
drugs has been initiated (8–10).
However, these drugs still face the challenge of acquired
resistance, and the efficacy of targeted therapy remains less
pronounced than traditional therapy, such as chemotherapy, in some
tumors. There is therefore a pressing need to identify new
therapeutic targets to provide theoretical support for the clinical
treatment of digestive system cancer.
Long non-coding RNAs (lncRNAs) are single-strand RNA
molecules >200 nucleotides long, which are transcribed by RNA
polymerase II and lack protein-coding ability (11). Extensive research has underscored
the substantial impact of lncRNAs on the development,
proliferation, migration and prognosis of various types of cancer.
These lncRNAs interact with target genes at the transcriptional
level, regulating a series of biological processes, such as histone
modification and chromatin remodeling. They also function as
competing endogenous RNAs (ceRNAs) that interact with microRNAs
(miRNAs), which are ~22 nucleotides long (11–13).
For example, lncRNA BC069792 acts as a ceRNA sponge to interact
with miR-658 and miR-4739, and increases the expression of the
target gene KCNQ4, leading to AKT phosphorylation, and subsequently
to inhibition of the proliferation and invasion of breast cancer
in vitro and in vivo (14).
Small nucleolar RNA host gene 16 (SNHG16) is a
member of the SNHG family that is located on human chromosome
17q25.1 and consists of four exons. SNHG16 was initially identified
as a potent oncogenic factor and has been reported to promote the
progression of neuroblastoma (15).
In addition, SNHG16 has been recognized as non-coding RNA that is
expressed in aggressive neuroblastoma (15). Numerous studies have further
revealed that SNHG16 is extensively involved in the complex
molecular regulatory network of various types of human cancer
(16–18). For example, the knockdown of SNHG16
has been reported to suppress the proliferation and radioresistance
of nasopharyngeal carcinoma cells by regulating the miR-31-5p/SFN
axis (19). Furthermore, SNHG16 has
been implicated as an oncogene, capable of promoting the
proliferation and reducing the apoptosis of bladder cancer cells.
This effect was shown to be achieved by binding and recruiting the
enhancer of zeste homolog 2 (EZH2) to p21 promoter and silencing
the expression of p21 (20). SNHG16
has also been shown to serve a key role in the staging, distant
metastasis and poor prognosis of ovarian cancer by increasing the
expression of MMP9 (21). In oral
squamous cell carcinoma, the expression of SNHG16 is regulated by
the transcription factor c-Myc, which recruits histone
acetyltransferase and induces RNA polymerase II clearance (22). These findings suggested that SNHG16
has an important role in the progression, invasion and
carcinogenesis of human cancer through upstream regulatory and
downstream molecular mechanisms.
The present review begins with a concise summary of
the relationship between risk factors, such as smoking, and human
digestive system tumors. Then, it delves into an examination of
research on the expression, biological function, related mechanisms
and potential clinical significance of SNHG16 for digestive tumors,
indicating a connection between SNHG16 and digestive cancers.
Additionally, it outlines the association of these risk factors
with SNHG16 in all reported publications. These findings
collectively underscore the potential of SNHG16 as both a potential
biomarker and therapeutic target in human digestive system
tumors.
Several factors have been reported to have a
significant import on cancer etiology, including smoking, excessive
alcohol consumption, physical activity, infection, radiation,
living environment, family history, diet, disease and genomic
characteristics. In the subsequent sections, the association of
these risk factors with human digestive system cancers is
described.
Smoking is the leading cause of cancer, and smokers
are at a higher risk of developing digestive system disorders,
including digestive tract cancers (23,24).
In 2019, tobacco smoking was responsible for ~203,000 deaths of
patients with EC worldwide (25).
Increasing evidence has demonstrated that smoking is closely
associated with the development and progression of HCC, with 13% of
HCC cases reported to be caused by smoking worldwide (26,27).
In a statistical analysis of HCC, patients were categorized as
non-smokers, current smokers and ex-smokers, according to smoking
status; notably, non-smokers had higher late survival rates than
current smokers and ex-smokers (28). For PC, tobacco smoking is considered
a major risk factor, with former or current smokers exhibiting a
higher odds ratio of 1.42–1.74 than non-smokers (24,29). A
number of causative factors have been epidemiologically confirmed
to have an association with PC, among which smoking shows the most
positive correlation with the risk of PC and is a recognized risk
factor (30–33). Furthermore, smoking has been shown
to affect the prognosis of patients with PC (34). Similarly, smoking constitutes an
established risk factor for CRC. Compared with 6,866 healthy
individuals, 6,264 patients with CRC had a higher smoking status,
suggesting a strong association between smoking and CRC evident
across early- and late-stage CRC (35).
Notably, research in esophageal squamous cell
carcinoma (ESCC), bladder cancer, non-small lung cancer and CRC has
demonstrated that there is no correlation between SNHG16 expression
and smoking (20,36–38).
Specifically, in lung cancer, SNHG16 expression was not related to
the clinical data of patients, including age and smoking history
(39). Furthermore, a meta-analysis
study indicated that SNHG16 expression was not associated with
smoking (40). By contrast, in a
study on CRC, Zhou et al revealed that smoking influenced
the combination of rs7353, rs8038 and rs15278 sites located in the
SNHG16 gene. In addition, through multifactor dimensionality
reduction analysis, changes in the expression levels of SNHG16 were
revealed to increase or decrease the risk of CRC susceptibility
(41). In the future, the
association between smoking and SNHG16 expression in other types of
cancer requires further research.
Similar to smoking, excessive alcohol consumption is
associated with an increased risk of all digestive system tumors,
including EC (24,42), HCC (43), PC (44), GC (45) and CRC (46). Nevertheless, moderate alcohol
consumption has shown no association with certain types of
digestive cancer, such as HCC, PC, GC and CRC (24,47,48).
An Australian study revealed that the risk of ESCC was
significantly increased with combined tobacco and alcohol use,
surpassing >20-fold higher risk compared with non-smokers and
non-drinkers (49).
Physical activity involves the use of skeletal
muscles and requires energy expenditure. Numerous studies have
demonstrated the association between physical activity and cancers
(50–53). Physical activity can decrease the
risk of EC by 19–51%, GC by 15–19% and colon cancer by 21–27%
(50). In particular, high levels
of physical activity, such as running and jumping rope, may
decrease the risk of PC by 9–25% (50). Moore et al reported that high
levels of physical activity decreased the risk of EC, HCC and GC by
>20% (51). Kasvis and Kilgour
(53) suggested that physical
activity interventions may alleviate malnutrition and muscle
wasting, which are common in PC. In China, a decade-long
prospective study showed that CRC risk was 25% lower in the
highest-level-of-activity group compared with in the
lowest-level-of-activity group (52). In addition, a meta-analysis showed
that a moderate-to-high physical activity level serves as a common
protective factor that can significantly reduce the overall risk of
digestive system cancer (54). To
date, there is an absence of evidence to indicate the relationship
between physical activity and SNHG16 expression, which should be
explored in the future.
Evidence has suggested that bacterial infection
serves a key role in tumor progression, such as in EC (55). Porphyromonas gingivalis is an
important periodontal disease pathogen that has been detected in
61% of ESCC tissues (56). It has
been suggested that EC is moderately positively associated with
chronic hepatitis C virus (HCV) infection, with a combined relative
risk of 1.61 (95% CI, 1.19–2.17) (57). For HCC, the most common risk factor
is chronic hepatitis caused by hepatitis B virus (HBV) and HCV
infection, and long-term chronic hepatitis can lead to cirrhosis
and eventually develop into HCC (58). Furthermore, HBV products and HBV
mutations may disrupt normal cell signaling pathways, leading to
HBV-induced HCC (59).
Fusobacterium has been identified as a potential prognostic
biomarker for PC (60). A
prospective study reported that patients with high concentrations
of P. gingivalis have a higher risk of PC (61). Helicobacter pylori, which is
found only in the human stomach, has been shown to be closely
associated with GC as a separate risk factor (62,63).
Substantial evidence has suggested that H. pylori carrying
CagA and VacA virulence factors is highly associated with distal GC
by promoting GC epithelial-mesenchymal transition (EMT) through
disruption of the gastric tissue microenvironment (64–67).
The Epstein-Barr virus infection can also cause GC, accounting for
~10% of patients with GC (68). In
patients with CRC, Escherichia coli has been reported to
contain a polyketide synthase gene that not only induces
inflammation, epithelial cell damage and cell proliferation, but
also encodes colibactin, which destroys DNA and ultimately leads to
the formation of CRC (69).
Fusobacterium has also been reported to be associated with
the occurrence of CRC (70).
In some clinical samples of HCC, HBV infection
showed no correlation with the expression of SNHG16 (71–75).
In addition, in a meta-analysis by Liu et al, there was no
association detected between SNHG16 expression and HBV infection
(76). To the best of our
knowledge, in other human digestive system tumors, the association
between infection and SNHG16 expression has not been reported. In
some cells, SNHG16 expression was upregulated in
Cryptococcus-treated dendritic cells compared with in
wild-type dendritic cells (77). In
addition, Mycobacterium tuberculosis infection can increase
the expression levels of SNHG16 in a dose- and time-dependent
manner in macrophages (78).
Radiation is a common tool in modern medicine,
including ionizing radiation and radiotherapy, and is one of the
main treatments for cancer (79–81).
However, the disadvantages of radiation cannot be overlooked. In
patients with head and neck tumors, the risk of ESCC is associated
with the dose of radiotherapy (80). In addition, α-radiation emitted by
plutonium is strongly associated with genetic mutations in HCC
(82). Dores et al (83) proposed that both radiotherapy and
chemotherapy can substantially increase the risk of PC in Hodgkin
lymphoma survivors treated previously. In addition, Yusefi et
al (84) showed that ionizing
radiation is a possible risk factor for GC. Low-dose radiation
exposure among uranium miners has been reported to be positively
associated with GC (85).
Computerized tomography radiation slightly increases the risk of
CRC, whereas the benefits of computerized tomography (CT) radiation
far outweigh the risks (86).
Notably, to the best of our knowledge, the association between
radiation and SNHG16 expression has not been reported in
disease.
A study in China revealed that drinking from
untreated water sources can increase the risk of ESCC by 2-fold
(87). The use of polluted water
containing nitrate is considered an essential risk factor for HCC
(88), PC (89), GC (90) and CRC (91). Exposure to external airborne agents,
such as fine particulate matter, may also increase the risk of
digestive system cancer, especially EC and GC (92,93).
Tsai et al (93)
demonstrated that PM2.5 was strongly associated with the mortality
of HCC, which agrees with the results of another study, where a
strong association was detected between PM2.5 and HCC and CRC
(94). A previous study reported
that SNHG16 expression presented no significant differences between
Intensive Care Unit (ICU)-hospitalized and non-ICU hospitalized
patients (95). To the best of our
knowledge, in digestive system cancer, the effect of living
environment on SNHG16 expression is not known.
A number of studies have shown that a family history
of cancer is strongly associated with the incidence rate of certain
types of cancer, such as EC (96),
HCC (97), PC (98), GC (99) and CRC (100). Parents and siblings of a person
with EC and HCC have been reported to exhibit a higher risk of
developing EC and HCC. Research has found that there is a clear
‘dose-response’ relationship with the number of first-degree
relatives of EC (97,101). Furthermore, familial inheritance
is a known cause of GC (102). A
positive first-degree family history of CRC and GC can reduce the
risk of cancer recurrence and death compared with patients without
a family history, based on a well-defined cohort enrolled in a
clinical trial (103,104). Su et al (105) reported that people with a family
history of EC had a 2-fold higher risk of developing the disease
with a poorer prognosis. However, in another study, the HCC
survival rate was higher in the familial cancer group than in the
sporadic cancer group (106).
Furthermore, individuals with a family history of CRC have been
shown to have a slightly increased risk of getting PC (107).
There is a consensus on the strong association
between diet and digestive system tumors. Long-term unbalanced
diets, such as high-calorie hot beverages or food, can lead to
esophageal epithelial cell damage and exacerbate the risk of EC
(108). A number of studies have
demonstrated that reducing vegetable and fruit intake, and
low-fiber diets, may increase the risk of HCC and PC, and
increasing the intake of salted and preserved foods and meat could
increase the risk of HCC and GC (109–114). Similarly, diet also influences
CRC. For example, calcium, fiber, milk, wholegrains and
2′5-hydroxyvitamin D have been shown to inhibit the development of
CRC; however, consumption of a large amount of red or culinary meat
can increase the risk of CRC (115,116). Furthermore, patients with CRC have
been reported to be deficient in vitamin C, vitamin E and folate
(117). To the best of our
knowledge, the association between diet and SNHG16 expression has
not been published.
Existing studies have shown that both infectious
diseases and chronic inflammation may account for ~25% of
carcinogenic factors (118).
Reactive oxygen/nitrogen species are produced under the condition
of chronic inflammation, which can cause DNA damage in various
organs, thereby inducing cellular carcinogenesis (118,119). Furthermore, prolonged acid reflux
can cause reflux esophagitis in the proximal esophagus and expedite
esophageal carcinogenesis (120).
Non-alcoholic fatty liver disease can result in a series of
diseases, including steatosis accumulation, non-alcoholic
steatohepatitis, inflammation, liver fibrosis and cirrhosis, which
may eventually lead to HCC (121–123). Chronic pancreatitis is also a
causative factor in the development of PC. Patients who have had
this disease for >2 years face a 2.71-fold higher risk of
developing PC (124). Chronic
gastritis, one of the most common types of chronic inflammation, is
considered a precursor of GC (125,126). Chronic enteritis and dysbiosis of
the intestinal microflora can increase the risk of CRC (127). Notably, diabetes mellitus (DM) and
obesity are risk factors for digestive system cancer, enhancing the
development of EC (128), HCC
(29), PC (129,130), GC (131,132) and CRC (133). For example, type 2 DM (T2DM) is
often recognized as an independent risk factor for HCC, with a 2-
to 4-fold increased risk in patients with T2DM compared with the
general population (134).
In HCC, liver cirrhosis has been reported to not
necessarily be associated with the expression of SNHG16 (71,74,75,135).
By contrast, investigations into the association between portal
vein tumor thrombus (PVTT) and SNHG16 expression have yielded
dissimilar results. Guo et al (135) revealed a positive correlation
between SNHG16 expression and PVTT, while another study indicated
that high SNHG16 expression was independent of PVTT (136). Patients with sepsis and acute
respiratory distress syndrome (ARDS) have been shown to exhibit a
decline SNHG16 expression compared with those without ARDS,
indicating that SNHG16 may possess a certain ability to
discriminate patients with sepsis and ARDS from those without ARDS,
according to the area under curve (137). Moreover, SNHG16 in patients with
sepsis has been discovered to have a negative correlation with
diabetes and chronic obstructive pulmonary disease history, rather
than other medical history, such as hypertension (137). In addition, some studies have
found that SNHG16 serves an important role in sepsis-induced acute
lung injury and inflammation via an involvement in the pathogenesis
of ARDS (138–140). In patients with acute ischemic
stroke, SNHG16 expression was revealed to be negatively related to
comorbidities, such as hyperlipidemia and disease severity
(141). SNHG16 has been shown to
be upregulated in unilateral ureteral obstruction-induced renal
fibrotic tissues of mice (142).
Cancer is a multi-stage process disease and its
occurrence is not only disturbed by external factors, but also by
intrinsic genetic mutations. The occurrence of genetic mutations
serves an important role in the development of digestive system
tumors. KRAS, a proto-oncogene, affects the cellular proliferation
and differentiation in digestive system cancer, and can influence
the prognosis of these patients (143,144). KRAS mutations have been found in
~85% of patients with PC and ~45% of patients with CRC (145). Similar results have been reported
regarding tumor suppressor genes. Mutations in the TP53 gene
usually occur in the early stages of GC and can accelerate the
progression of GC (146). In HCC,
TP53 mutations have been detected in circulating exosomal DNA and
are associated with the prognosis of patients (147). APC mutations are associated with
tumorigenesis, affecting the overall survival of patients with CRC
(148,149). A previous meta-analysis showed the
neutral function of PIK3CA mutations on the overall survival and
progression-free survival of patients with CRC (144). Specifically, individuals who have
both genetic and lifestyle-related risks have a ~190 times higher
risk of ESCC than those without these risks (150). To the best of our knowledge, the
association between genomic characteristics and SNHG16 expression
has not yet been presented.
Epigenetic influences, such as DNA methylation,
histone modifications and non-coding RNA regulation, are also
important factors (151). The role
of lncRNA in the process of cancer development and progression
cannot be overlooked. A number of studies have shown that SNHG16 is
a key factor in the process of digestive system cancer, including
promoting the proliferation of cancer cells, resisting cancer
therapeutic drugs and enhancing cancer cell invasiveness. The
possible mechanisms and functional characterization of SNHG16 in
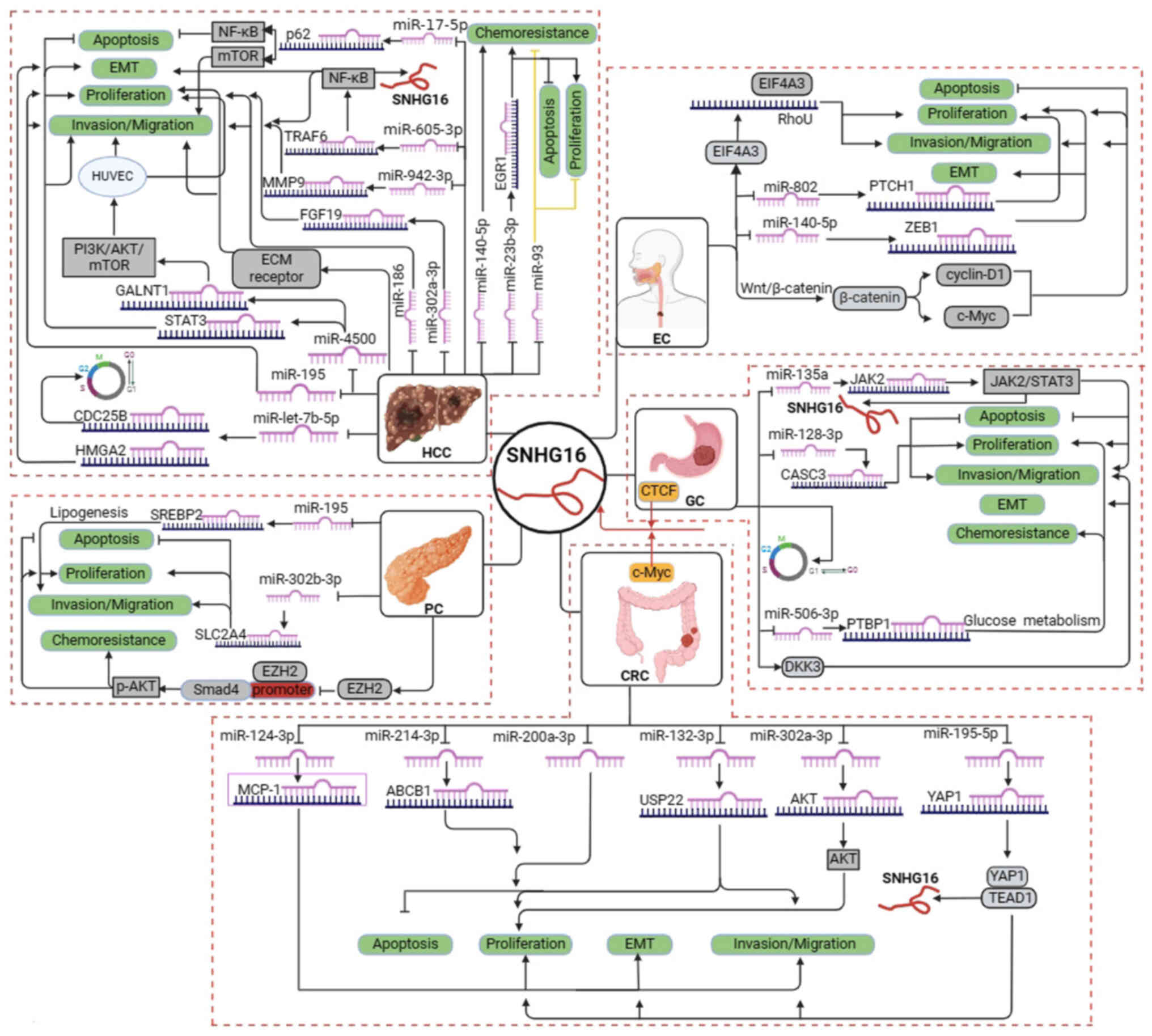
human digestive system cancer are presented in Fig. 1 and Table I, respectively. Furthermore, the
association between SNHG16 expression and clinicopathological
characteristics is summarized in Table
II.
EC is one of the major cancer types worldwide,
ranking 7th (3.1%, 604,100 new cases) and 6th (5.5%, 544, 076
deaths) among all types of cancer in terms of incidence and
mortality rate, respectively (2).
Its incidence and mortality rates vary between geographic regions
(2). For example, due to economic
underdevelopment and dietary habits, the burden of EC is higher in
East Asia with a predominance of patients with ESCC (2). Studies have shown that SNHG16
expression is upregulated in EC, and is closely associated with
tumor stage, lymph node metastasis and clinical stage (36,152–154). The knockdown of SNHG16 has been
reported to suppress the proliferation and invasion, and promote
apoptosis by reducing the expression of β-catenin, cyclin D1
and c-Myc protein in EC-1 and Eca-109 cells (36). In addition, Zhang et al
verified by reverse transcription-quantitative PCR (RT-qPCR) that
the expression levels of SNHG16 were upregulated in EC tissues or
cells compared with those in normal tissues or cells (P<0.01).
This previous study also observed that the disruption of SNHG16
expression suppressed proliferation, promoted apoptosis and
inhibited EMT through the miR-140-5p/ZEB1 axis in vivo and
in vitro (152). In another
study on ESCC, the expression of SNHG16 was revealed to be
associated with tumor differentiation and T stage, and increased
expression of SNHG16 could promote ESCC growth and metastasis. The
underlying mechanism may be that SNHG16 binds to and recruits
EIF4A3 to modulate RhoU expression, thereby enhancing the stability
of RhoU mRNA (154). Zhang et
al (153) demonstrated that
SNHG16 acts as a sponge of miR-802 to upregulate PTCH1 and activate
the Hedgehog pathway, thus facilitating EC proliferation and
self-renewal.
These results show that the upregulation of SNHG16
may be strongly associated with the development of ESCC, suggesting
the potential utility of SNHG16 as a marker for ESCC. These
insights offer novel avenues for the clinical management of
ESCC.
HCC is one of the most common malignancies
worldwide, accounting for 4.7% (906,000 new cases) of all new
cancer cases and 8.3% (830,000 deaths) of all cancer-related
mortalities. HCC is ranked as the 6th most commonly diagnosed
cancer and the 3rd leading cause of cancer-related deaths (2). In most studies, SNHG16 has been
considered a proto-oncogene of HCC, and RT-qPCR has been used to
detect the expression of SNHG16 in HCC tissues and corresponding
non-tumor tissues. The results showed that the expression levels of
SNHG16 in HCC samples were much higher than those in matched
non-tumor samples, and upregulation of SNHG16 expression was highly
associated with poor prognosis and tumor stage of HCC. Furthermore,
the patients with advanced-stage HCC exhibited a significantly
higher SNHG16 expression level than the patients with early-stage
HCC (72,155). Moreover, the high expression of
SNHG16 has been shown to be associated with the tumor size, TNM
stage and vascular infiltration of patients with HCC (74). SNHG16, as a ceRNA, can target STAT3
and GALNT1 through sponging miR-4500 in Huh7 cells and human
umbilical vein endothelial cells (HUVECs), respectively, to promote
proliferation, metastasis and invasion of Huh7 cells, and enhance
angiogenesis of HUVECs (72,156).
In addition, through regulating miR-195, miR-17-5p/P62,
miR-302a-3p/FGF19 and miR-186 expression, SNHG16 can inhibit the
proliferation, migration and invasion of HepG2 and Hep3B cells
(71,73,155,157). Overexpression of SNHG16 may also
affect the G2/M transition of HCC cells by regulating
CDC25B expression through sponging miR-let-7b-5p (158). A previous study reported that
SNHG16 is upregulated in sorafenib-resistant tumor tissues and
cells, and that the overexpression of SNHG16 can enhance sorafenib
resistance in HCC (74). By
contrast, when the expression of SNHG16 is suppressed, sorafenib
resistance disappears (135). Jing
et al (159) also suggested
that SNHG16 may enhance HCC autophagy via the miR-23b-3p/EGR1 axis
and protect HCC from sorafenib resistance. In addition, it has been
reported that SNHG16 can be phagocytized by telocytes and can
mediate telocytes to promote HCC cell metastasis by regulating the
miR-942-3p/MMP9 axis (160).
Furthermore, Hu et al (75)
demonstrated that the overexpression of SNHG16 promotes TRAF6
expression by sponging miR-605-3p, activates NF-κB and exacerbates
the development of HCC. Specifically, the activated NF-κB can
enhance SNHG16 promoter activity, forming a positive SNHG16/NF-κB
feedback loop that further worsens HCC (75). The overexpression of SNHG16 has also
been shown to be associated with tumor recurrence and poor
prognosis after surgery, and mechanistic analyses suggested that
SNHG16 markedly activates the extracellular matrix-receptor
interaction pathway (136).
Studies have demonstrated that SNHG16 regulates a
large lncRNA-miRNA-mRNA network in HCC, and is closely associated
with the infiltration of immune cells, the release of
immunomodulatory factors and the expression of chemokines in tumor
tissues (161–163). Notably, UBE4B and SEMA3F may
promote HCC progression regulated by their upstream
SNHG16/miR-22-3p and SNHG16/let-7c-5p axes, respectively (163,164). Liu et al (76) revealed that SNHG16 can be used as a
potential biomarker for patients with HCC with a poor prognoses. In
summary, SNHG16 may be upregulated in HCC and can promote HCC
development. Notably, a previous study presented the opposite
argument, suggesting that SNHG16 expression may be reduced in HCC
tissue compared with in normal liver tissue, and that
overexpression of SNHG16 could decrease the proliferation of Hep3B
and Huh7 cells, and inhibit HCC development and chemoresistance via
sponging miR-93 (165). This
discrepancy in findings may stem from the diverse dysregulation
patterns of SNHG16 in human cancer, with its expression being
either upregulated or downregulated. Such variations could be
influenced by the specific cancer types, their anatomical locations
and the microenvironments involved (165). This discrepancy prompts further
research into the role of SNHG16 in HCC.
PC is one of the most serious malignancies of the
digestive system. Due to its poor prognosis, PC accounts for almost
as many deaths (466,000) as cases (496,000) in the world, and is
the 7th leading cause of cancer death in both men and women
(2). Similar to EC, the incidence
rate of PC is 4-fold and 5-fold higher in high human development
index (HDI) countries compared with in low HDI countries (2). Studies have shown that the expression
levels of SNHG16 are upregulated in PC tissue compared with those
in normal tissue (166,167). Altering the expression of SNHG16
may inhibit the adipogenesis of AsPC-1 and PANC-1 cells through the
miR-195/SREBP2 axis (168).
Inhibition of SNHG16 expression can result in the release of
miR-302b-3p, which inhibits SLC2A4 expression and promotes
apoptosis in PC cells (166).
Overexpression of SNHG16 has also been reported to be closely
associated with gemcitabine resistance in PC cells. SNHG16 can
interact with EZH2, suppressing SMAD4 expression via EZH2 binding
to the SMAD4 promoter (167).
Downregulation of SMAD4 has a reduced ability to inhibit AKT
phosphorylation, thereby promoting gemcitabine resistance in PC
cells (167). These findings
suggested that SNHG16 may serve a critical role in the development
of PC and that it could be regarded as a marker of poor prognosis
in PC.
GC remains an important type of cancer worldwide,
and is considered the 5th most frequent malignancy (5.6%,
>1,000,000 new cases) and the 4th most common cause of death
(7.7%, ~769,000 deaths) among oncological patients (2). The region with the highest
age-standardized incidence rate is Eastern Asia, followed by
Central and Eastern Europe (2). The
expression of SNHG16 is significantly associated with the depth of
infiltration, lymph node metastasis, TNM stage, histological
differentiation and PTBP1 expression of GC (169,170). Knockdown of SNHG16 can
significantly suppress the migration, invasion and arrest of cells
in the G1 phase, and can decrease c-Myc expression, and
affect the formation of the p27/cyclin D1/CDK6, p53/cyclin E1 and
cyclin A2/CDK2 complexes (169–171). A number of patients with GC
develop 5-fluorouracil (5-FU) resistance, showing a higher
vulnerability than parental GC cells. Notably, blocking the
SNHG16/miR-506-3p/PTBP1 axis may effectively limit 5-FU-resistant
GC cell originated-xenograft tumor growth under 5-FU treatment.
Specifically, PTBP1 stabilizes the mRNA expression of glycolysis
enzymes by directly binding to 3′UTR regions (169). In addition, it has been shown that
SNHG16 can promote EMT by downregulating the WNT signaling pathway
and inhibiting DKK3 expression, and can regulate β-catenin protein
expression without participating in the β-catenin translocation
between the cytoplasm and nucleus (172). In particular, SNHG16 activated by
CCCTC binding factor (CTCF) can modulate gastrointestinal stromal
tumor cell proliferation, migration, invasion and apoptosis through
the miR-128-3p/CASC3 axis (173).
In another study, SNHG16 was also demonstrated to be able to
mediate the upregulation of JAK2 and STAT3 by sponging miR-135a to
influence the proliferation, invasion and apoptosis of GC cells,
with SNHG16 being regulated by phosphorylated-STAT3 directly or
indirectly (174). In summary,
SNHG16 may be closely related to the occurrence and development of
GC, and could be a potential marker of poor GC prognosis.
Notably, >1,900,000 new cases of CRC and 935,000
CRC-related deaths occurred worldwide in 2020; during this year, it
was the third most common cancer, after female breast cancer and
lung cancer, and it exhibited a close mortality rate to lung cancer
(2). Growing evidence has suggested
that the expression levels of SNHG16 are positively associated with
advanced TNM stage, distant metastasis and shorter overall survival
time in CRC (38,175–177). SNHG16 is mainly present in the
cytoplasm, functioning as a ceRNA to regulate multiple miRNAs and
target genes. Li et al (38)
revealed that SNHG16 was associated with malignancy and poor
prognosis in patients with CRC by sponging miR-200a-3p. Tan et
al (177) indicated that
SNHG16 could promote CRC proliferation by upregulating its target
gene ABCB1 through interacting with miR-214-3p. He et al
(178) concluded that SNHG16 could
activate USP22 expression to promote CRC progression via absorbing
miR-132-3p. Ke et al (179)
demonstrated that SNHG16 supported colon cancer cell proliferation
by targeting the miR-302a-3p/AKT axis. Chen et al (176) revealed that the expression of
SNHG16 was higher in cancer tissues from patients than in the
matched normal tissues, and was positively related to CRC grade. It
was also revealed that SNHG16 may serve a contributory role in the
proliferation, migration and EMT of CRC cells through the
miR-124-3p/MCP-1 axis (176). Some
bioinformatics analyses also reached a similar conclusion, in that
SNHG16 may have an important role in CRC (175,180). In particular, SNHG16 has been
reported to be closely associated with autophagy in CRC (175,181).
The expression of SNHG16 may be activated by other
proteins, such as c-Myc. Christensen et al reported that the
expression of SNHG16 is determined by Wnt-regulated transcription
factors such as c-Myc in CRC (182). Specifically, knockdown of
β-catenin could reduce the expression of SNHG16 and c-Myc,
whereas c-Myc knockdown or overexpression could decrease or
increase the SNHG16 expression, respectively (182). In a study by Xiang et al,
the SNHG16/YAP1/TEA domain transcription factor 1 (TEAD1) positive
feedback loop was detected in CRC cells (183). SNHG16 was shown to act as a ceRNA
that can physically bind miR-195-5p, further regulating YAP1
expression and facilitating tumor progression. YAP1 binds to TEAD1
to form a YAP1/TEAD1 complex, which in turn binds to two sites in
the promoter of SNHG16 and activates SNHG16 transcription (183).
In addition, SNHG16 polymorphisms have been shown
to be significantly associated with CRC susceptibility. Research
has revealed that the rs7353 site A>G of the SNHG16 gene is
associated with decreased susceptibility of CRC; however, the
rs8038 site G>A, rs15278 site A>G and rs15278 site G>A
variations may increase CRC susceptibility (41).
SNHG16 has also been studied in other
gastrointestinal tumors. For example, SNHG16 expression has been
shown to be upregulated in cholangiocarcinoma tissues and cell
lines. When SNHG16 expression was suppressed, the proliferation
rate of RBE and HuCCT1 cells was reduced, whereas apoptosis was
activated (184). Wu et al
(184) also revealed that there
was a potential binding site for miR-146a-5p at the 3′UTR end of
GATA6 and that SNHG16 could sponge miR-146a-5p. Interfering with
the expression of miR-146a-5p reversed the SNHG16 knockdown-induced
apoptosis in RBE and HuCCT1 cells, whereas overexpression of GATA6
also achieved the same effect (184). These findings suggested that
SNHG16 is important for the development of cholangiocarcinoma and
it could be a potential target for future drug development against
cholangiocarcinoma.
Neuroendocrine tumors account for a very small
portion of tumors at each site; for example, the incidence of
pancreatic neuroendocrine tumors was <1 case per 100,000
people/year worldwide (1), and the
incidence of gastric neuroendocrine tumors was ~0.4 per 100,000
individuals in America in 2017 (185). However, as the number of patients
with cancer increases, the proportion of neuroendocrine tumors has
also increased, thus highlighting the need for attention to be paid
to neuroendocrine tumors. Although, to the best of our knowledge,
the role of SNHG16 in neuroendocrine tumors in the digestive system
has not been reported, it is a valuable direction for improving the
management of neuroendocrine tumors in the future.
A growing number of studies have demonstrated that
tumorigenesis is caused by a combination of genetics and
environmental factors. At present, environmental factors, such as
diet, require attention to prevent their effects on personal
health. The present review briefly summarized the relationship
between risk factors and human digestive system cancer, identifying
risk factors, such as smoking and diet, which may severely affect
tumorigenesis. Subsequently, this review focused on outlining the
role of SNHG16 in the formation and progression of digestive system
cancer (Fig. 2). Available data
have suggested that SNHG16 is strongly associated with
proliferation, migration, invasion, apoptosis and prognosis in EC,
HCC, PC, GC and CRC.
Upregulation of SNHG16 is clinically important, and
is associated with tumor stage, lymph node metastasis and tumor
size. It may be used as a novel diagnostic or prognostic biomarker
for human digestive system tumors (Table II). For example, SNHG16 was
revealed to be more highly expressed in EC tissues compared with in
normal EC tissues, and similar results were observed in EC cell
lines (36,152,154). In addition, overexpression of
SNHG16 is strongly associated with the tumor stage, lymph node
metastasis and clinical stage of EC (36). A Kaplan-Meier assay demonstrated
that patients with EC and high SNHG16 expression had poorer overall
survival rates than those with low SNHG16 expression (36). Notably, univariate and multivariate
analyses have revealed that SNHG16 expression may be considered an
independent predictor for the overall survival of patients with EC
(36). In a number of studies,
SNHG16 expression has been reported to be higher in HCC tissues or
cells than in normal HCC tissues or cells (71–75,135,156–160); however, a study by Xu et al
demonstrated that SNHG16 was significantly downregulated in both
HCC tissues and cells (165).
Clinical data have revealed that SNHG16 expression may be closely
associated with tumor size, lymph node status and TNM stage, and
enhanced SNHG16 expression could be related to advanced stages and
short overall survival of patients with HCC (72,74,135).
Multivariate analysis indicated that SNHG16 expression was an
independent prognostic factor of HCC. Notably, some studies have
reported that tumor size is not affected by SNHG16 expression,
which may be due to individual differences or the small size of
patients (74,75,136,155). However, reduced expression of
SNHG16 has been reported in mice with smaller tumor sizes (71,155).
In addition to the aforementioned cancer types, similar results
were also reported in GC and CRC (170,183). Generally, the aforementioned
findings suggested that SNHG16 expression is strongly associated
with poor diagnosis or prognosis of human digestive system
tumors.
In recent years, gene therapy has emerged as a
promising avenue for significantly improving the survival rate of
patients with cancer (186). As
aforementioned, the clinical significance and biological functions
of SNHG16 may serve as a promising therapeutic target for human
digestive system cancer (Table I).
However, Xu et al (165)
reported contradictory results, which may be attributed to the
sites and microenvironments specific to certain cancer types. The
present review of publications on SNHG16 and cancer has resulted in
a consistent conclusion that SNHG16 upregulation may promote
proliferation, enhance migration and invasion, inhibit apoptosis,
and affect lipid metabolism and chemoresistance. For example,
silencing the expression of SNHG16 could attenuate the
proliferation, activate the apoptosis, and inhibit the migratory,
invasive and malignant phenotype in GC cell lines. In addition, the
knockdown of SNHG16 could reduce tumor volume and weight in a nude
mouse human-GC xenograft model (173).
The chemoresistance of various types of cancer may
also be associated with the expression of SNHG16. A series of
studies on HCC have demonstrated that downregulation of SNHG16
expression can reverse sorafenib and cisplatin resistance in HCC
cell lines (such as Hep3B) or xenograft models, by acting as an
endogenous sponge for miR-140-5p, miR-23b-3p and let-7b-5p
(74,135,158,159). However, contrasting results
reported by Xu et al showed that upregulation of SNHG16
inhibited 5-FU chemoresistance through competitive linking to
miR-93 in Hep3B and Huh7 cells (165). In PC, SNHG16 may reduce SMAD4 to
induce gemcitabine resistance via EZH2-mediated epigenetic
modifications (167). In GC, the
SNHG16-mediated miR-506-3p/PTBP1 axis effectively led to 5-FU
resistance (169). These findings
suggested that decreasing SNHG16 expression may be a promising
therapy for suppressing GC progression. However, to the best of our
knowledge, there are not currently any clinical trials focused on
SNHG16; the development of inhibitors of SNHG16 have only been
researched in cancer cell lines and animal models. In addition, the
association between SNHG16 expression and chemoresistance in EC and
CRC has not been researched. Therefore, whether SNHG16 will become
a potential target of therapy, requires further study.
In this review, we described the association
between risk factors and SNHG16 expression in ESCC, HCC and CRC.
Notably, smoking may not influence the expression of SNHG16 in
patients with ESCC (36).
Furthermore, both HBV infection and liver cirrhosis were reported
to not affect the expression of SNHG16 in HCC (71–76,135).
The association between PVTT and SNHG16 expression in HCC also
remains controversial and is still under exploration (135,136). In CRC, the expression of SNHG16
could be regulated by risk factors, such as smoking and family
history (41). To date, although
smoking, excessive alcohol consumption, physical activity,
infection, radiation, living environment, family history, diet,
disease and genomic characteristics, are risk factors that can
influence human digestive system cancer, the association between a
number of risk factors and SNHG16 expression remains unclear.
Therefore, further research using large-scale data will be
necessary to thoroughly investigate the relationship between risk
factors and SNHG16 expression.
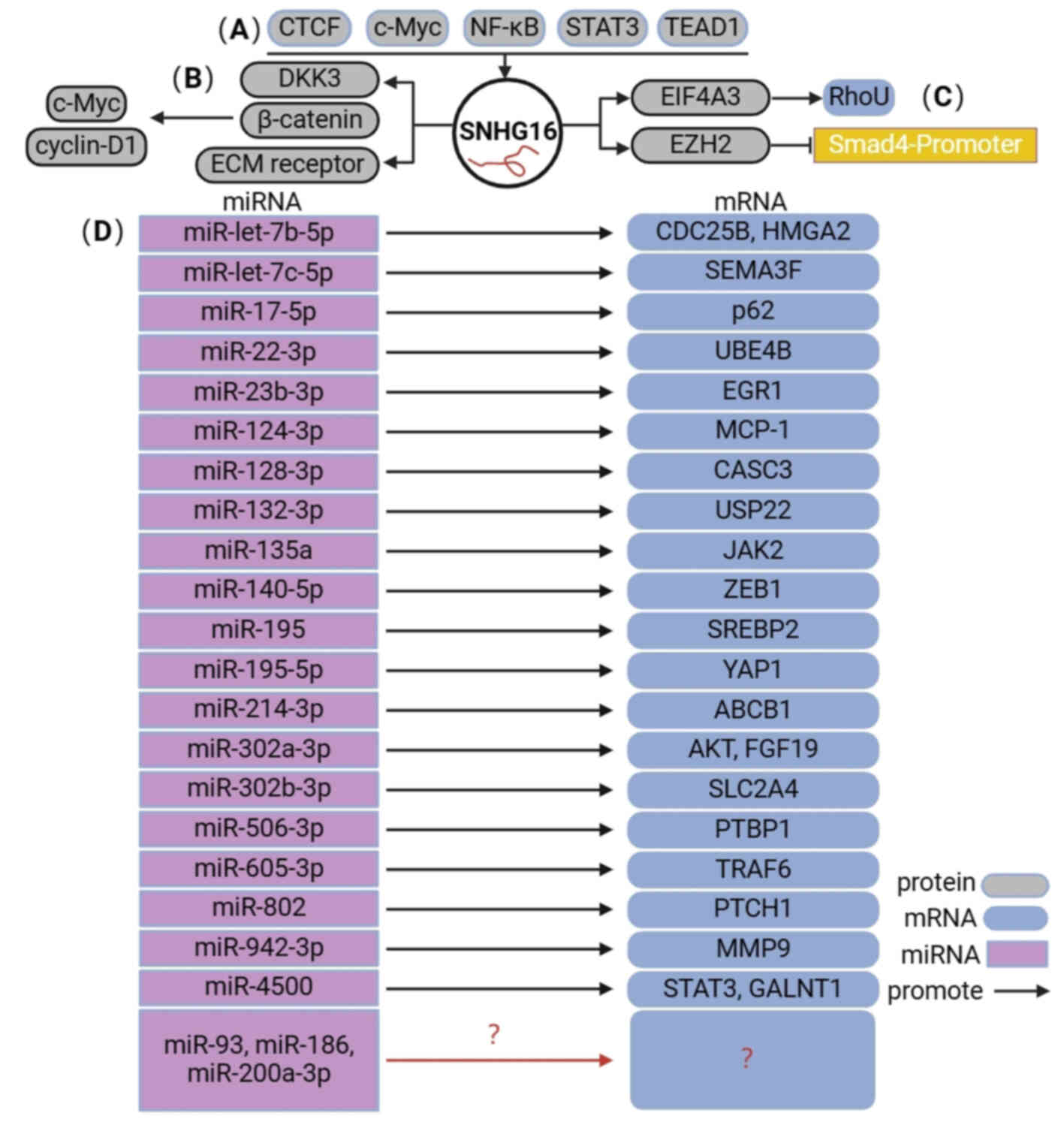
Thus far, the upstream regulatory and downstream
molecular mechanisms of SNHG16 in human digestive system cancer
encompass four primary aspects (Fig.
2). i) Numerous transcription factors, including CTCF, c-Myc,
NF-κB, STAT3 and TEAD1, are positively associated with SNHG16. ii)
SNHG16 directly regulates the expression of downstream target
genes, such as DKK3. iii) SNHG16 can bind to and recruit EIF4A3 to
regulate RhoU expression and enhance RhoU mRNA stability.
Meanwhile, SNHG16 also binds to EZH2 and recruits EZH2 to the SMAD4
promoter, subsequently suppressing SMAD4 expression. iv) SNHG16 can
compete with miRNAs to mediate the expression of downstream target
genes and activate different signaling pathways.
In conclusion, the expression of SNHG16 is
associated with the clinical and pathological characteristics of
patients with cancer, and regulates cell proliferation, apoptosis,
invasion and metastasis through various potential mechanisms. These
findings suggested that SNHG16 may serve an oncogenic role in human
cancer, making it a promising target for cancer diagnosis and
treatment, as well as a potential biomarker for cancer
prognosis.
Not applicable.
This study was supported by the Shandong Youth Natural Science
Foundation of China (grant no. ZR2021QH367); the Scientific and
Technological Innovation Program for Medical Workers in Shandong
Province (grant no. SDYWZGKCJHLH202212); and the Medical and Health
Science and Technology Development Program of Shandong Province
(grant nos. 202103100485 and 202102080115).
Not applicable.
HR, FP and LX conceptualized the study. YL, YK and
LW wrote the manuscript. LW, JP, LZ and HZ searched the literatures
and collected the information from the literature. YL, YK, ZY and
FP designed the figures and tables. LZ and XF revised and submitted
the manuscript. Data authentication is not applicable. All authors
read and approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Assarzadegan N and Montgomery E: What is
New in the 2019 World Health Organization (WHO) classification of
tumors of the digestive system: Review of selected updates on
neuroendocrine neoplasms, appendiceal tumors, and molecular
testing. Arch Pathol Lab Med. 145:664–677. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo LW, Shi CL, Huang HY, Wang L, Yue XP,
Liu SZ, Li J, Su K, Dai M, Sun XB and Shi JF: Economic burden of
esophageal cancer in China from 1996 to 2015: A systematic review.
Zhonghua Liu Xing Bing Xue Za Zhi. 38:102–109. 2017.(In Chinese).
PubMed/NCBI
|
|
4
|
Klimeck L, Heisser T, Hoffmeister M and
Brenner H: Colorectal cancer: A health and economic problem. Best
Pract Res Clin Gastroenterol. 66:1018392023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hojman P, Gehl J, Christensen JF and
Pedersen BK: Molecular mechanisms linking exercise to cancer
prevention and treatment. Cell Metab. 27:10–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu W, Li B, Xu M, Yang T and Hao X:
Traditional Chinese medicine for precancerous lesions of gastric
cancer: A review. Biomed Pharmacother. 146:1125422022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Qiu H, Li C, Cai P and Qi F: The
positive role of traditional Chinese medicine as an adjunctive
therapy for cancer. Biosci Trends. 15:283–298. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Halbrook CJ, Lyssiotis CA, Pasca di
Magliano M and Maitra A: Pancreatic cancer: Advances and
challenges. Cell. 186:1729–1754. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie YH, Chen YX and Fang JY: Comprehensive
review of targeted therapy for colorectal cancer. Signal Transduct
Target Ther. 5:222020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lorenzi L, Avila Cobos F, Decock A,
Everaert C, Helsmoortel H, Lefever S, Verboom K, Volders PJ,
Speleman F, Vandesompele J and Mestdagh P: Long noncoding RNA
expression profiling in cancer: Challenges and opportunities. Genes
Chromosomes Cancer. 58:191–199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Dong X, Guo X, Li C, Fan Y, Liu
P, Yuan D, Ma X, Wang J, Zheng J, et al: LncRNA-BC069792 suppresses
tumor progression by targeting KCNQ4 in breast cancer. Mol Cancer.
22:412023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu
Y, Ozaki T, Isogai E, Nakamura Y, Koda T, et al: High expression of
ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is
associated with poor prognosis in neuroblastoma. Int J Oncol.
34:931–938. 2009.PubMed/NCBI
|
|
16
|
Ghafouri-Fard S, Khoshbakht T, Taheri M
and Shojaei S: A review on the role of small nucleolar RNA host
gene 6 long non-coding RNAs in the carcinogenic processes. Front
Cell Dev Biol. 9:7416842021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong CY, Tang R, Nan W, Zhou KS and Zhang
HH: Role of SNHG16 in human cancer. Clin Chim Acta. 503:175–180.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang M and Wei W: SNHG16: A novel long-non
coding RNA in human cancers. Onco Targets Ther. 12:11679–11690.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang W, Zhou X, Tang Z, Fu L, Zou S and
Tang S: Knockdown of lncRNA SNHG16 attenuates the proliferation and
radioresistance of nasopharyngeal carcinoma cells by mediating
miR-31-5p/SFN axis. Radiat Res. 199:124–131. 2023.PubMed/NCBI
|
|
20
|
Cao X, Xu J and Yue D: LncRNA-SNHG16
predicts poor prognosis and promotes tumor proliferation through
epigenetically silencing p21 in bladder cancer. Cancer Gene Ther.
25:10–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang XS, Wang GX and Luo L: Long
non-coding RNA SNHG16 promotes cell growth and metastasis in
ovarian cancer. Eur Rev Med Pharmacol Sci. 22:616–622.
2018.PubMed/NCBI
|
|
22
|
Li S, Zhang S and Chen J: c-Myc induced
upregulation of long non-coding RNA SNHG16 enhances progression and
carcinogenesis in oral squamous cell carcinoma. Cancer Gene Ther.
26:400–410. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sealock T and Sharma S: Smoking Cessation.
StatPearls [Internet]. StatPearls Publishing; Treasure Island, FL:
2024
|
|
24
|
Yuan S, Chen J, Ruan X, Sun Y, Zhang K,
Wang X, Li X, Gill D, Burgess S, Giovannucci E and Larsson SC:
Smoking, alcohol consumption, and 24 gastrointestinal diseases:
Mendelian randomization analysis. Elife. 12:e840512023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
GBD 2019 Cancer Risk Factors
Collaborators, . The global burden of cancer attributable to risk
factors, 2010-19: A systematic analysis for the global burden of
disease study 2019. Lancet. 400:563–591. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marti-Aguado D, Clemente-Sanchez A and
Bataller R: Cigarette smoking and liver diseases. J Hepatol.
77:191–205. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baecker A, Liu X, La Vecchia C and Zhang
ZF: Worldwide incidence of hepatocellular carcinoma cases
attributable to major risk factors. Eur J Cancer Prev. 27:205–212.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cho WR, Wang CC, Tsai MJ, Lin CC, Yen YH,
Chen CH, Kuo YH, Yao CC, Hung CH, Huang PY, et al: Smoking as a
risk factor for very late recurrence in surgically resected
early-stage primary hepatocellular carcinoma. Clin Med Insights
Oncol. 18:117955492412282322024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Klein AP: Pancreatic cancer epidemiology:
Understanding the role of lifestyle and inherited risk factors. Nat
Rev Gastroenterol Hepatol. 18:493–502. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lynch SM, Vrieling A, Lubin JH, Kraft P,
Mendelsohn JB, Hartge P, Canzian F, Steplowski E, Arslan AA, Gross
M, et al: Cigarette smoking and pancreatic cancer: A pooled
analysis from the pancreatic cancer cohort consortium. Am J
Epidemiol. 170:403–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bosetti C, Lucenteforte E, Silverman DT,
Petersen G, Bracci PM, Ji BT, Negri E, Li D, Risch HA, Olson SH, et
al: Cigarette smoking and pancreatic cancer: An analysis from the
international pancreatic cancer case-control consortium (Panc4).
Ann Oncol. 23:1880–1888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ghadirian P, Lynch HT and Krewski D:
Epidemiology of pancreatic cancer: An overview. Cancer Detect Prev.
27:87–93. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iodice S, Gandini S, Maisonneuve P and
Lowenfels AB: Tobacco and the risk of pancreatic cancer: A review
and meta-analysis. Langenbecks Arch Surg. 393:535–545. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weissman S, Takakura K, Eibl G, Pandol SJ
and Saruta M: The diverse involvement of cigarette smoking in
pancreatic cancer development and prognosis. Pancreas. 49:612–620.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li H, Chen X, Hoffmeister M and Brenner H:
Associations of smoking with early- and late-onset colorectal
cancer. JNCI Cancer Spectr. 7:pkad0042023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han GH, Lu KJ, Wang P, Ye J, Ye YY and
Huang JX: LncRNA SNHG16 predicts poor prognosis in ESCC and
promotes cell proliferation and invasion by regulating
Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci.
22:3795–3803. 2018.PubMed/NCBI
|
|
37
|
Han W, Du X, Liu M, Wang J, Sun L and Li
Y: Increased expression of long non-coding RNA SNHG16 correlates
with tumor progression and poor prognosis in non-small cell lung
cancer. Int J Biol Macromol. 121:270–278. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Lu Y and Chen Y: Long non-coding RNA
SNHG16 affects cell proliferation and predicts a poor prognosis in
patients with colorectal cancer via sponging miR-200a-3p. Biosci
Rep. 39:BSR201824982019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gheliji T, Oskooei VK, Ashrafi Hafez A,
Taheri M and Ghafouri-Fard S: Evaluation of expression of vitamin D
receptor related lncRNAs in lung cancer. Noncoding RNA Res.
5:83–87. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiao R, Jiang W, Wei X, Zhang M, Zhao S
and Huang G: Clinicopathological significance and prognosis of long
noncoding RNA SNHG16 expression in human cancers: A meta-analysis.
BMC Cancer. 20:6622020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou L, Zhang Y, Jin J and Gu X:
Correlation between lncRNA SNHG16 gene polymorphism and its
interaction with environmental factors and susceptibility to
colorectal cancer. Medicine (Baltimore). 99:e233722020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Uhlenhopp DJ, Then EO, Sunkara T and
Gaduputi V: Epidemiology of esophageal cancer: Update in global
trends, etiology and risk factors. Clin J Gastroenterol.
13:1010–1021. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McGee EE, Jackson SS, Petrick JL, Van Dyke
AL, Adami HO, Albanes D, Andreotti G, Beane-Freeman LE, Berrington
de Gonzalez A, Buring JE, et al: Smoking, alcohol, and biliary
tract cancer risk: A pooling project of 26 prospective studies. J
Natl Cancer Inst. 111:1263–1278. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wood LD, Canto MI, Jaffee EM and Simeone
DM: Pancreatic cancer: Pathogenesis, screening, diagnosis, and
treatment. Gastroenterology. 163:386–402.e1. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Laszkowska M, Rodriguez S, Kim J and Hur
C: Heavy alcohol use is associated with gastric cancer: Analysis of
the national health and nutrition examination survey from 1999 to
2010. Am J Gastroenterol. 116:1083–1086. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
McNabb S, Harrison TA, Albanes D, Berndt
SI, Brenner H, Caan BJ, Campbell PT, Cao Y, Chang-Claude J, Chan A,
et al: Meta-analysis of 16 studies of the association of alcohol
with colorectal cancer. Int J Cancer. 146:861–873. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sheikh M, Roshandel G, McCormack V and
Malekzadeh R: Current status and future prospects for esophageal
cancer. Cancers (Basel). 15:7652023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Larsson SC, Carter P, Kar S, Vithayathil
M, Mason AM, Michaëlsson K and Burgess S: Smoking, alcohol
consumption, and cancer: A mendelian randomisation study in UK
Biobank and international genetic consortia participants. PLoS Med.
17:e10031782020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dong J and Thrift AP: Alcohol, smoking and
risk of oesophago-gastric cancer. Best Pract Res Clin
Gastroenterol. 31:509–517. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Friedenreich CM, Ryder-Burbidge C and
McNeil J: Physical activity, obesity and sedentary behavior in
cancer etiology: Epidemiologic evidence and biologic mechanisms.
Mol Oncol. 15:790–800. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Moore SC, Lee IM, Weiderpass E, Campbell
PT, Sampson JN, Kitahara CM, Keadle SK, Arem H, Berrington de
Gonzalez A, Hartge P, et al: Association of leisure-time physical
activity with risk of 26 types of cancer in 1.44 million adults.
JAMA Intern Med. 176:816–825. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Su J, Jiang Y, Fan X, Tao R, Wu M, Lu Y,
Hua Y, Jin J, Guo Y, Lv J, et al: Association between physical
activity and cancer risk among Chinese adults: A 10-year
prospective study. Int J Behav Nutr Phys Act. 19:1502022.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kasvis P and Kilgour RD: Diet and exercise
interventions in patients with pancreatic cancer: A scoping review.
Pancreas. 50:657–666. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xie F, You Y, Huang J, Guan C, Chen Z,
Fang M, Yao F and Han J: Association between physical activity and
digestive-system cancer: An updated systematic review and
meta-analysis. J Sport Health Sci. 10:4–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Garrett WS: Cancer and the microbiota.
Science. 348:80–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nasrollahzadeh D, Malekzadeh R, Ploner A,
Shakeri R, Sotoudeh M, Fahimi S, Nasseri-Moghaddam S, Kamangar F,
Abnet CC, Winckler B, et al: Variations of gastric corpus
microbiota are associated with early esophageal squamous cell
carcinoma and squamous dysplasia. Sci Rep. 5:88202015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ponvilawan B, Rittiphairoj T, Charoenngam
N, Rujirachun P, Wattanachayakul P, Tornsatitkul S and Ungprasert
P: Association between chronic hepatitis C virus infection and
esophageal cancer: A systematic review and meta-analysis. J Clin
Gastroenterol. 56:55–63. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chidambaranathan-Reghupaty S, Fisher PB
and Sarkar D: Hepatocellular carcinoma (HCC): Epidemiology,
etiology and molecular classification. Adv Cancer Res. 149:1–61.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Singh AK, Kumar R and Pandey AK:
Hepatocellular carcinoma: Causes, mechanism of progression and
biomarkers. Curr Chem Genom Transl Med. 12:9–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga
Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, et
al: Association of Fusobacterium species in pancreatic
cancer tissues with molecular features and prognosis. Oncotarget.
6:7209–7220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fan X, Alekseyenko AV, Wu J, Peters BA,
Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R,
Miller G, et al: Human oral microbiome and prospective risk for
pancreatic cancer: A population-based nested case-control study.
Gut. 67:120–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang YC: Medicinal plant activity on
Helicobacter pylori related diseases. World J Gastroenterol.
20:10368–10382. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ailloud F, Didelot X, Woltemate S,
Pfaffinger G, Overmann J, Bader RC, Schulz C, Malfertheiner P and
Suerbaum S: Within-host evolution of Helicobacter pylori
shaped by niche-specific adaptation, intragastric migrations and
selective sweeps. Nat Commun. 10:22732019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Espinoza JL, Matsumoto A, Tanaka H and
Matsumura I: Gastric microbiota: An emerging player in
Helicobacter pylori-induced gastric malignancies. Cancer
Lett. 414:147–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Baj J, Korona-Glowniak I, Forma A, Maani
A, Sitarz E, Rahnama-Hezavah M, Radzikowska E and Portincasa P:
Mechanisms of the epithelial-mesenchymal transition and tumor
microenvironment in Helicobacter pylori-induced gastric
cancer. Cells. 9:10552020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xia R, Zhang B, Wang X and Jia Q:
Pathogenic interactions between Helicobacter pylori adhesion
protein HopQ and human cell surface adhesion molecules CEACAMs in
gastric epithelial cells. Iran J Basic Med Sci. 22:710–715.
2019.PubMed/NCBI
|
|
67
|
Roesler BM, Rabelo-Gonçalves EMA and
Zeitune JMR: Virulence factors of Helicobacter pylori: A
review. Clin Med Insights Gastroenterol. 7:9–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sasaki S, Nishikawa J, Sakai K, Iizasa H,
Yoshiyama H, Yanagihara M, Shuto T, Shimokuri K, Kanda T, Suehiro
Y, et al: EBV-associated gastric cancer evades T-cell immunity by
PD-1/PD-L1 interactions. Gastric Cancer. 22:486–496. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cuevas-Ramos G, Petit CR, Marcq I, Boury
M, Oswald E and Nougayrède JP: Escherichia coli induces DNA
damage in vivo and triggers genomic instability in mammalian cells.
Proc Natl Acad Sci USA. 107:11537–11542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Flemer B, Warren RD, Barrett MP, Cisek K,
Das A, Jeffery IB, Hurley E, O'Riordain M, Shanahan F and O'Toole
PW: The oral microbiota in colorectal cancer is distinctive and
predictive. Gut. 67:1454–1463. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhong JH, Xiang X, Wang YY, Liu X, Qi LN,
Luo CP, Wei WE, You XM, Ma L, Xiang BD and Li LQ: The lncRNA SNHG16
affects prognosis in hepatocellular carcinoma by regulating p62
expression. J Cell Physiol. 235:1090–1102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lin Q, Zheng H, Xu J, Zhang F and Pan H:
LncRNA SNHG16 aggravates tumorigenesis and development of
hepatocellular carcinoma by sponging miR-4500 and targeting STAT3.
J Cell Biochem. 120:11604–11615. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chen H, Li M and Huang P: LncRNA SNHG16
promotes hepatocellular carcinoma proliferation, migration and
invasion by regulating miR-186 expression. J Cancer. 10:3571–3581.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ye J, Zhang R, Du X, Chai W and Zhou Q:
Long noncoding RNA SNHG16 induces sorafenib resistance in
hepatocellular carcinoma cells through sponging miR-140-5p. Onco
Targets Ther. 12:415–422. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Hu YL, Feng Y, Chen YY, Liu JZ, Su Y, Li
P, Huang H, Mao QS and Xue WJ: SNHG16/miR-605-3p/TRAF6/NF-κB
feedback loop regulates hepatocellular carcinoma metastasis. J Cell
Mol Med. 24:7637–7651. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Liu Q, Gao P, Li Q, Xu C, Qu K and Zhang
J: Long non-coding RNA SNHG16 as a potential biomarker in
hepatocellular carcinoma: A meta-analysis. Medicine (Baltimore).
100:e271782021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Teng Y, Li M, Tao X, Huang Y, Ding X, Xu
D, Fan Y and Shen Z: Cryptococcosis inhibits the immune response of
dendritic cells through the snhg1-miR-145a-3p-Bcl2 axis. Exp Clin
Transplant. 21:441–450. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Sun W, Zhang X, He X, Zhang J, Wang X, Lin
W, Wang X and Wu X: Long non-coding RNA SNHG16 silencing inhibits
proliferation and inflammation in Mycobacterium
tuberculosis-infected macrophages by targeting miR-140-5p
expression. Infect Genet Evol. 103:1053252022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Allen C, Her S and Jaffray DA:
Radiotherapy for cancer: Present and future. Adv Drug Deliv Rev.
109:1–2. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nobel TB, Barbetta A, Hsu M, Tan KS,
Pinchinat T, Schlottmann F, Bains MS, Ku GY, Wu AJ, Patti MG, et
al: Outcomes of radiation-associated esophageal squamous cell
carcinoma: The MSKCC experience. J Gastrointest Surg. 23:11–22.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Thierry-Chef I, Cardis E, Damilakis J,
Frija GA, Hierath M and Hoeschen C: Medical applications of
ionizing radiation and radiation protection for European patients,
population and environment. EPJ Nuclear Sci Technol. 8:442022.
View Article : Google Scholar
|
|
82
|
Goerlitz DS, Blancato J, Ramesh A, Islam
M, Graham GT, Revina V, Kallakury B, Zeck J, Kirillova E and
Loffredo CA: Somatic mutation signatures in primary liver tumors of
workers exposed to ionizing radiation. Sci Rep. 9:181992019.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Dores GM, Curtis RE, van Leeuwen FE,
Stovall M, Hall P, Lynch CF, Smith SA, Weathers RE, Storm HH,
Hodgson DC, et al: Pancreatic cancer risk after treatment of
Hodgkin lymphoma. Ann Oncol. 25:2073–2079. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yusefi AR, Bagheri Lankarani K, Bastani P,
Radinmanesh M and Kavosi Z: Risk factors for gastric cancer: A
systematic review. Asian Pac J Cancer Prev. 19:591–603.
2018.PubMed/NCBI
|
|
85
|
Kreuzer M, Straif K, Marsh JW, Dufey F,
Grosche B, Nosske D and Sogl M: Occupational dust and radiation
exposure and mortality from stomach cancer among German uranium
miners, 1946–2003. Occup Environ Med. 69:217–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Albert JM: Radiation risk from CT:
Implications for cancer screening. AJR Am J Roentgenol.
201:W81–W87. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sun Y, Zhang T, Wu W, Zhao D, Zhang N, Cui
Y, Liu Y, Gu J, Lu P, Xue F, et al: Risk factors associated with
precancerous lesions of esophageal squamous cell carcinoma: A
screening study in a high risk Chinese population. J Cancer.
10:3284–3290. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang L, Wan X, Shi R, Gong P and Si Y:
Comparing spatial patterns of 11 common cancers in Mainland China.
BMC Public Health. 22:15512022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yang CY, Tsai SS and Chiu HF: Nitrate in
drinking water and risk of death from pancreatic cancer in Taiwan.
J Toxicol Environ Health A. 72:397–401. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Picetti R, Deeney M, Pastorino S, Miller
MR, Shah A, Leon DA, Dangour AD and Green R: Nitrate and nitrite
contamination in drinking water and cancer risk: A systematic
review with meta-analysis. Environ Res. 210:1129882022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Nasseri Maleki G, Bayati Khatibi M,
Khamnian Z, Jalali Z, Dastgiri S and Ghodrati Aroogh H: Association
between nitrate concentration in drinking water and rate of
colorectal cancer: A case study in northwestern Iran. Int J Environ
Health Res. 32:1791–1800. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lee W, Kim J, Lim SS, Kim Y, Ahn YS and
Yoon JH: External airborne-agent exposure increase risk of
digestive tract cancer. Sci Rep. 10:86172020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tsai SS, Hsu CT and Yang C: Risk of death
from liver cancer in relation to long-term exposure to fine
particulate air pollution in Taiwan. J Toxicol Environ Health A.
86:135–143. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Pritchett N, Spangler EC, Gray GM,
Livinski AA, Sampson JN, Dawsey SM and Jones RR: Exposure to
outdoor particulate matter air pollution and risk of
gastrointestinal cancers in adults: A systematic review and
meta-analysis of epidemiologic evidence. Environ Health Perspect.
130:360012022. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Taheri M, Rad LM, Hussen BM, Nicknafs F,
Sayad A and Ghafouri-Fard S: Evaluation of expression of
VDR-associated lncRNAs in COVID-19 patients. BMC Infect Dis.
21:5882021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhang S, Chen J, Li B, Cai X, Wang K, Tan
Z, Zheng Y and Liu Q: Family history of cancer is a prognostic
factor for better survival in operable esophageal squamous cell
carcinoma: A propensity score matching analysis. Front Oncol.
12:9459372022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Yang Y, Wu QJ, Xie L, Chow WH, Rothman N,
Li HL, Gao YT, Zheng W, Shu XO and Xiang YB: Prospective cohort
studies of association between family history of liver cancer and
risk of liver cancer. Int J Cancer. 135:1605–1614. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Abe K, Kitago M, Kitagawa Y and Hirasawa
A: Hereditary pancreatic cancer. Int J Clin Oncol. 26:1784–1792.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Choi IJ, Kim CG, Lee JY, Kim YI, Kook MC,
Park B and Joo J: Family history of gastric cancer and
Helicobacter pylori treatment. N Engl J Med. 382:427–436.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Sninsky JA, Shore BM, Lupu GV and Crockett
SD: Risk factors for colorectal polyps and cancer. Gastrointest
Endosc Clin N Am. 32:195–213. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Chen T, Cheng H, Chen X, Yuan Z, Yang X,
Zhuang M, Lu M, Jin L and Ye W: Family history of esophageal cancer
increases the risk of esophageal squamous cell carcinoma. Sci Rep.
5:160382015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Setia N, Clark JW, Duda DG, Hong TS, Kwak
EL, Mullen JT and Lauwers GY: Familial gastric cancers. Oncologist.
20:1365–1377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Chan JA, Meyerhardt JA, Niedzwiecki D,
Hollis D, Saltz LB, Mayer RJ, Thomas J, Schaefer P, Whittom R,
Hantel A, et al: Association of family history with cancer
recurrence and survival among patients with stage III colon cancer.
JAMA. 299:2515–2523. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Han MA, Oh MG, Choi IJ, Park SR, Ryu KW,
Nam BH, Cho SJ, Kim CG, Lee JH and Kim YW: Association of family
history with cancer recurrence and survival in patients with
gastric cancer. J Clin Oncol. 30:701–708. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Su Z, Zou GR, Mao YP, OuYang PY, Cao XL,
Xie FY and Li Q: Prognostic impact of family history of cancer in
Southern Chinese patients with esophageal squamous cell cancer. J
Cancer. 10:1349–1357. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Dai WC, Fan ST, Cheung TT, Chok KS, Chan
AC, Tsang SH, Poon RT and Lo CM: The impact of family history of
hepatocellular carcinoma on its patients' survival. Hepatobiliary
Pancreat Dis Int. 11:160–164. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Hamada T, Yuan C, Yurgelun MB, Perez K,
Khalaf N, Morales-Oyarvide V, Babic A, Nowak JA, Rubinson DA,
Giannakis M, et al: Family history of cancer, Ashkenazi Jewish
ancestry, and pancreatic cancer risk. Br J Cancer. 120:848–854.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Chen Y, Tong Y, Yang C, Gan Y, Sun H, Bi
H, Cao S, Yin X and Lu Z: Consumption of hot beverages and foods
and the risk of esophageal cancer: A meta-analysis of observational
studies. BMC Cancer. 15:4492015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Keszei AP, Goldbohm RA, Schouten LJ,
Jakszyn P and van den Brandt PA: Dietary N-nitroso compounds,
endogenous nitrosation, and the risk of esophageal and gastric
cancer subtypes in the Netherlands cohort study. Am J Clin Nutr.
97:135–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ibiebele TI, Hughes MC, Whiteman DC and
Webb PM; Australian Cancer Study, : Dietary patterns and risk of
oesophageal cancers: A population-based case-control study. Br J
Nutr. 107:1207–1216. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Kim J, Cho YA, Choi WJ and Jeong SH:
Gene-diet interactions in gastric cancer risk: A systematic review.
World J Gastroenterol. 20:9600–9610. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Cheng YK, Yao SM, Xu YR and Niu RG:
Life-style habits in a high-risk area for upper gastrointestinal
cancers: A population-based study from Shanxi, China. Asian Pac J
Cancer Prev. 17:4301–4306. 2016.PubMed/NCBI
|
|
113
|
Berretta M, Lleshi A, Fisichella R,
Berretta S, Basile F, Li Volti G, Bolognese A, Biondi A, De Paoli
P, Tirelli U and Cappellani A: The role of nutrition in the
development of esophageal cancer: What do we know? Front Biosci
(Elite Ed). 4:351–357. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zhang Z and Zhang X: Salt taste
preference, sodium intake and gastric cancer in China. Asian Pac J
Cancer Prev. 12:1207–1210. 2011.PubMed/NCBI
|
|
115
|
Thanikachalam K and Khan G: Colorectal
cancer and nutrition. Nutrients. 11:1642019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Song M, Garrett WS and Chan AT: Nutrients,
foods, and colorectal cancer prevention. Gastroenterology.
148:1244–1260.e16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Rosato V, Bosetti C, Levi F, Polesel J,
Zucchetto A, Negri E and La Vecchia C: Risk factors for young-onset
colorectal cancer. Cancer Causes Control. 24:335–341. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Murata M: Inflammation and cancer. Environ
Health Prev Med. 23:502018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ohnishi S, Ma N, Thanan R, Pinlaor S,
Hammam O, Murata M and Kawanishi S: DNA damage in
inflammation-related carcinogenesis and cancer stem cells. Oxid Med
Cell Longev. 2013:3870142013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wang RH: From reflux esophagitis to
Barrett's esophagus and esophageal adenocarcinoma. World J
Gastroenterol. 21:5210–5219. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ioannou GN: Epidemiology and
risk-stratification of NAFLD-associated HCC. J Hepatol.
75:1476–1484. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Powell EE, Wong VW and Rinella M:
Non-alcoholic fatty liver disease. Lancet. 397:2212–2224. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Pouwels S, Sakran N, Graham Y, Leal A,
Pintar T, Yang W, Kassir R, Singhal R, Mahawar K and Ramnarain D:
Non-alcoholic fatty liver disease (NAFLD): A review of
pathophysiology, clinical management and effects of weight loss.
BMC Endocr Disord. 22:632022. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Duell EJ, Lucenteforte E, Olson SH, Bracci
PM, Li D, Risch HA, Silverman DT, Ji BT, Gallinger S, Holly EA, et
al: Pancreatitis and pancreatic cancer risk: A pooled analysis in
the international pancreatic cancer case-control consortium
(PanC4). Ann Oncol. 23:2964–2970. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Sipponen P and Maaroos HI: Chronic
gastritis. Scand J Gastroenterol. 50:657–667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Tempera PJ, Michael M, Tageldin O and
Hasak S: Gastric cancer due to chronic H. pylori infection:
What we know and where we are going. Diseases. 10:572022.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Kim J and Lee HK: Potential role of the
gut microbiome in colorectal cancer progression. Front Immunol.
12:8076482022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Xu B, Zhou X, Li X, Liu C and Yang C:
Diabetes mellitus carries a risk of esophageal cancer: A
meta-analysis. Medicine (Baltimore). 96:e79442017. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Li M, Park JY, Sheikh M, Kayamba V, Rumgay
H, Jenab M, Narh CT, Abedi-Ardekani B, Morgan E, de Martel C, et
al: Population-based investigation of common and deviating patterns
of gastric cancer and oesophageal cancer incidence across
populations and time. Gut. 72:846–854. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Chari ST, Leibson CL, Rabe KG, Ransom J,
de Andrade M and Petersen GM: Probability of pancreatic cancer
following diabetes: A population-based study. Gastroenterology.
129:504–511. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lim JH, Shin CM, Han KD, Lee SW, Jin EH,
Choi YJ, Yoon H, Park YS, Kim N and Lee DH: Association between the
persistence of obesity and the risk of gastric cancer: A nationwide
population-based study. Cancer Res Treat. 54:199–207. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Guo J, Liu C, Pan J and Yang J:
Relationship between diabetes and risk of gastric cancer: A
systematic review and meta-analysis of cohort studies. Diabetes Res
Clin Pract. 187:1098662022. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Soltani G, Poursheikhani A, Yassi M,
Hayatbakhsh A, Kerachian M and Kerachian MA: Obesity, diabetes and
the risk of colorectal adenoma and cancer. BMC Endocr Disord.
19:1132019. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Plaz Torres MC, Jaffe A, Perry R,
Marabotto E, Strazzabosco M and Giannini EG: Diabetes medications
and risk of HCC. Hepatology. 76:1880–1897. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Guo Z, Zhang J, Fan L, Liu J, Yu H, Li X
and Sun G: Long noncoding RNA (lncRNA) small nucleolar RNA host
gene 16 (SNHG16) predicts poor prognosis and sorafenib resistance
in hepatocellular carcinoma. Med Sci Monit. 25:2079–2086. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhang QJ, Li DZ, Lin BY, Geng L, Yang Z
and Zheng SS: SNHG16 promotes hepatocellular carcinoma development
via activating ECM receptor interaction pathway. Hepatobiliary
Pancreat Dis Int. 21:41–49. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhang C, Huang Q and He F: Correlation of
small nucleolar RNA host gene 16 with acute respiratory distress
syndrome occurrence and prognosis in sepsis patients. J Clin Lab
Anal. 36:e245162022. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Sun J, Xin K, Leng C and Ge J:
Down-regulation of SNHG16 alleviates the acute lung injury in
sepsis rats through miR-128-3p/HMGB3 axis. BMC Pulm Med.
21:1912021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Xia L, Zhu G, Huang H, He Y and Liu X:
LncRNA small nucleolar RNA host gene 16 (SNHG16) silencing protects
lipopolysaccharide (LPS)-induced cell injury in human lung
fibroblasts WI-38 through acting as miR-141-3p sponge. Biosci
Biotechnol Biochem. 85:1077–1087. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhou Z, Zhu Y, Gao G and Zhang Y: Long
noncoding RNA SNHG16 targets miR-146a-5p/CCL5 to regulate
LPS-induced WI-38 cell apoptosis and inflammation in acute
pneumonia. Life Sci. 228:189–197. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Xie C, Zhu B, Gu J and Sun M: The
correlation of lncRNA SNHG16 with inflammatory cytokines, adhesion
molecules, disease severity, and prognosis in acute ischemic stroke
patients. J Clin Lab Anal. 36:e244392022. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Zhao Y, Wang H, Tang Y, Wang J, Wu X, He
Z, He Y and Tang Z: SNHG16/miR-205/HDAC5 is involved in the
progression of renal fibrosis. J Biochem Mol Toxicol.
38:e236172024. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Luo J: KRAS mutation in pancreatic cancer.
Semin Oncol. 48:10–18. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Mei ZB, Duan CY, Li CB, Cui L and Ogino S:
Prognostic role of tumor PIK3CA mutation in colorectal cancer: A
systematic review and meta-analysis. Ann Oncol. 27:1836–1848. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Cox AD, Fesik SW, Kimmelman AC, Luo J and
Der CJ: Drugging the undruggable RAS: Mission possible? Nat Rev
Drug Discov. 13:828–851. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Busuttil RA, Zapparoli GV, Haupt S,
Fennell C, Wong SQ, Pang JM, Takeno EA, Mitchell C, Di Costanzo N,
Fox S, et al: Role of p53 in the progression of gastric cancer.
Oncotarget. 5:12016–12026. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Li Y, Wu J, Li E, Xiao Z, Lei J, Zhou F,
Yin X, Hu D, Mao Y, Wu L and Wenjun L: TP53 mutation detected in
circulating exosomal DNA is associated with prognosis of patients
with hepatocellular carcinoma. Cancer Biol Ther. 23:439–445. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wang W, Shao F, Yang X, Wang J, Zhu R,
Yang Y, Zhao G, Guo D, Sun Y, Wang J, et al: METTL3 promotes tumour
development by decreasing APC expression mediated by APC mRNA
N(6)-methyladenosine-dependent YTHDF binding. Nat Commun.
12:38032021. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zeineldin M and Neufeld KL: Understanding
phenotypic variation in rodent models with germline Apc mutations.
Cancer Res. 73:2389–2399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Cui R, Kamatani Y, Takahashi A, Usami M,
Hosono N, Kawaguchi T, Tsunoda T, Kamatani N, Kubo M, Nakamura Y
and Matsuda K: Functional variants in ADH1B and ALDH2 coupled with
alcohol and smoking synergistically enhance esophageal cancer risk.
Gastroenterology. 137:1768–1775. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Ilango S, Paital B, Jayachandran P, Padma
PR and Nirmaladevi R: Epigenetic alterations in cancer. Front
Biosci (Landmark Ed). 25:1058–1109. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Zhang K, Chen J, Song H and Chen LB:
SNHG16/miR-140-5p axis promotes esophagus cancer cell
proliferation, migration and EMT formation through regulating ZEB1.
Oncotarget. 9:1028–1040. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zhang L, Liang H, Zhang J, Yang Y, Ling X
and Jiang H: Long non-coding RNA SNHG16 facilitates esophageal
cancer cell proliferation and self-renewal through the
microRNA-802/PTCH1 axis. Curr Med Chem. 29:6084–6099. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Ren L, Fang X, Shrestha SM, Ji Q, Ye H,
Liang Y, Liu Y, Feng Y, Dong J and Shi R: LncRNA SNHG16 promotes
development of oesophageal squamous cell carcinoma by interacting
with EIF4A3 and modulating RhoU mRNA stability. Cell Mol Biol Lett.
27:892022. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Xie X, Xu X, Sun C and Yu Z: Long
intergenic noncoding RNA SNHG16 interacts with miR-195 to promote
proliferation, invasion and tumorigenesis in hepatocellular
carcinoma. Exp Cell Res. 383:1115012019. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Li S, Qi Y, Huang Y, Guo Y, Huang T and
Jia L: Exosome-derived SNHG16 sponging miR-4500 activates HUVEC
angiogenesis by targeting GALNT1 via PI3K/Akt/mTOR pathway in
hepatocellular carcinoma. J Physiol Biochem. 77:667–682. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Li W, Xu W, Song JS, Wu T and Wang WX:
LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19
axis in hepatocellular carcinoma. Neoplasma. 66:397–404. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Li S, Peng F, Ning Y, Jiang P, Peng J,
Ding X, Zhang J, Jiang T and Xiang S: SNHG16 as the miRNA let-7b-5p
sponge facilitates the G2/M and epithelial-mesenchymal transition
by regulating CDC25B and HMGA2 expression in hepatocellular
carcinoma. J Cell Biochem. 121:2543–2558. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Jing Z, Ye X, Ma X, Hu X, Yang W, Shi J,
Chen G and Gong L: SNGH16 regulates cell autophagy to promote
sorafenib resistance through suppressing miR-23b-3p via sponging
EGR1 in hepatocellular carcinoma. Cancer Med. 9:4324–4338. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Xu Y, Luan G, Li Z, Liu Z, Qin G and Chu
Y: Tumour-derived exosomal lncRNA SNHG16 induces telocytes to
promote metastasis of hepatocellular carcinoma via the
miR-942-3p/MMP9 axis. Cell Oncol (Dordr). 46:251–264.
2023.PubMed/NCBI
|
|
161
|
Zhang J and Lou W: A key mRNA-miRNA-lncRNA
competing endogenous RNA triple sub-network linked to diagnosis and
prognosis of hepatocellular carcinoma. Front Oncol. 10:3402020.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Chen S, Zhao Z, Wang X, Zhang Q, Lyu L and
Tang B: The predictive competing endogenous RNA regulatory networks
and potential prognostic and immunological roles of cyclin A2 in
pan-cancer analysis. Front Mol Biosci. 9:8095092022. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Shao X, Zhu J, Shi Y, Fang H, Chen J,
Zhang Y, Wang J, Jian H, Lan S, Jiang F, et al: Upregulated UBE4B
expression correlates with poor prognosis and tumor immune
infiltration in hepatocellular carcinoma. Aging (Albany NY).
14:9632–9646. 2022.PubMed/NCBI
|
|
164
|
Lou W, Wang W, Chen J, Wang S and Huang Y:
ncRNAs-mediated high expression of SEMA3F correlates with poor
prognosis and tumor immune infiltration of hepatocellular
carcinoma. Mol Ther Nucleic Acids. 24:845–855. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Xu F, Zha G, Wu Y, Cai W and Ao J:
Overexpressing lncRNA SNHG16 inhibited HCC proliferation and
chemoresistance by functionally sponging hsa-miR-93. Onco Targets
Ther. 11:8855–8863. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Xu H, Miao X, Li X, Chen H, Zhang B and
Zhou W: LncRNA SNHG16 contributes to tumor progression via the
miR-302b-3p/SLC2A4 axis in pancreatic adenocarcinoma. Cancer Cell
Int. 21:512021. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Yu Y, Zou YF, Hong RQ, Chen WJ, Chen L,
Chen WQ, Wang HP and Yu Y: Long non-coding RNA SNHG16 decreased
SMAD4 to induce gemcitabine resistance in pancreatic cancer via
EZH2-mediated epigenetic modification. Kaohsiung J Med Sci.
38:981–991. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Yu Y, Dong JT, He B, Zou YF, Li XS, Xi CH
and Yu Y: LncRNA SNHG16 induces the SREBP2 to promote lipogenesis
and enhance the progression of pancreatic cancer. Future Oncol.
15:3831–3844. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Ding Y, Gao S, Zheng J and Chen X:
Blocking lncRNA-SNHG16 sensitizes gastric cancer cells to 5-Fu
through targeting the miR-506-3p-PTBP1-mediated glucose metabolism.
Cancer Metab. 10:202022. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Lian D, Amin B, Du D and Yan W: Enhanced
expression of the long non-coding RNA SNHG16 contributes to gastric
cancer progression and metastasis. Cancer Biomark. 21:151–160.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Zhao JJ, Liu JJ, Zhang YY, Xia Y, Du H,
Yan ZQ, Zhou CH, Xia WS, Zellmer L, Liao DJ, et al: SNHG16 lncRNAs
are overexpressed and may be oncogenic in human gastric cancer by
regulating cell cycle progression. Neoplasma. 69:49–58. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Zhou C, Zhao J, Liu J, Wei S, Xia Y, Xia
W, Bi Y, Yan Z and Huang H: LncRNA SNHG16 promotes
epithelial-mesenchymal transition via down-regulation of DKK3 in
gastric cancer. Cancer Biomark. 26:393–401. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Yang Z, Pu M, Dong X, Yang H, Chang W, Liu
T and Zhang X: CTCF-activated SNHG16 facilitates gastrointestinal
stromal tumor by targeting miR-128-3p/CASC3 axis. Exp Cell Res.
417:1131312022. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Wang X, Kan J, Han J, Zhang W, Bai L and
Wu H: LncRNA SNHG16 functions as an oncogene by sponging MiR-135a
and promotes JAK2/STAT3 signal pathway in gastric cancer. J Cancer.
10:1013–1022. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Zhou W, Zhang S, Li HB, Cai Z, Tang S,
Chen LX, Lang JY, Chen Z and Chen XL: Development of prognostic
indicator based on autophagy-related lncRNA analysis in colon
adenocarcinoma. Biomed Res Int. 2020:98079182020. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Chen ZY, Wang XY, Yang YM, Wu MH, Yang L,
Jiang DT, Cai H and Peng Y: LncRNA SNHG16 promotes colorectal
cancer cell proliferation, migration, and epithelial-mesenchymal
transition through miR-124-3p/MCP-1. Gene Ther. 29:193–205. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Tan P, Xu M, Nie J, Qin J, Liu X, Sun H,
Wang S and Pan Y: LncRNA SNHG16 promotes colorectal cancer
proliferation by regulating ABCB1 expression through sponging
miR-214-3p. J Biomed Res. 36:231–241. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
He X, Ma J, Zhang M, Cui J and Yang H:
Long non-coding RNA SNHG16 activates USP22 expression to promote
colorectal cancer progression by sponging miR-132-3p. Onco Targets
Ther. 13:4283–4294. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Ke D and Wang Q, Ke S, Zou L and Wang Q:
Long-non coding RNA SNHG16 supports colon cancer cell growth by
modulating miR-302a-3p/AKT axis. Pathol Oncol Res. 26:1605–1613.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Huang E, Ma T, Zhou J, Ma N, Yang W, Liu
C, Hou Z, Chen S, Zong Z, Zeng B, et al: A novel
senescence-associated LncRNA signature predicts the prognosis and
tumor microenvironment of patients with colorectal cancer: A
bioinformatics analysis. J Gastrointest Oncol. 13:1842–1863. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Duan L, Xia Y, Li C, Lan N and Hou X:
Identification of autophagy-related LncRNA to predict the prognosis
of colorectal cancer. Front Genet. 13:9069002022. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Christensen LL, True K, Hamilton MP,
Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen
JB, Pedersen JS, et al: SNHG16 is regulated by the Wnt pathway in
colorectal cancer and affects genes involved in lipid metabolism.
Mol Oncol. 10:1266–1282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Xiang Z, Huang G, Wu H, He Q, Yang C, Dou
R, Liu Q, Song J, Fang Y, Wang S and Xiong B: SNHG16
upregulation-induced positive feedback loop with YAP1/TEAD1 complex
in colorectal cancer cell lines facilitates liver metastasis of
colorectal cancer by modulating CTCs epithelial-mesenchymal
transition. Int J Biol Sci. 18:5291–5308. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Wu T, Lei MS, Gao XZ, Xiong TG, Yang K,
Gong Q, Tang R, Tian YP and Fu XH: lncRNA SNHG16 mediates cell
proliferation and apoptosis in cholangiocarcinoma by directly
targeting miR-146a-5p/GATA6 axis. Biochem Genet. 59:1311–1325.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Dasari A, Shen C, Halperin D, Zhao B, Zhou
S, Xu Y, Shih T and Yao JC: Trends in the incidence, prevalence,
and survival outcomes in patients with neuroendocrine tumors in the
United States. JAMA Oncol. 3:1335–1342. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
He W, Li Q, Lu Y, Ju D, Gu Y, Zhao K and
Dong C: Cancer treatment evolution from traditional methods to stem
cells and gene therapy. Curr Gene Ther. 22:368–385. 2022.
View Article : Google Scholar : PubMed/NCBI
|