Introduction
History: Overview of natural products
(NPs)
NPs are primary and secondary metabolites produced
and used by living organisms for defending mechanisms or adapting
actions. These molecules have been naturally selected and modified
for millions of years to acquire specificity and cover a wide range
of biological mechanisms, depending on the originating species, the
environment, and the specific biological action involved in the
corresponding organism (1).
Considering these beneficial features, NPs have been used as
therapeutic agents for thousands of years from the beginning of
rational medicine and to date, continue to be the most important
source of novel candidate therapeutic agents in the pharmaceutical
industry (2,3).
NPs, derived mostly from herb plants, have been used
as the major source of therapeutics for traditional medicine
throughout history and continue to be the basis for a number of
pharmaceuticals currently used (4). The pharmaceutical properties of herb
plants were described on Assyrian clay tablets dating back to 2000
B.C. and were also reported in ancient Greek culture by Hippocrates
(400 B.C.) and Galinos (160 A.C). Currently, NPs are the principal
source of compounds for modern medicine, and the extent of the
mining of such compounds has increased during the 20th century
(5). The advantage of the use of
NPs for the development of novel drugs and other chemicals derives
from their innate affinity for biological receptors and their
interaction in a number of biological functions. NPs have provided
the most effective antiparasitic, anti-infection and antidiabetic
compounds known to date (5-7).
Over the past decades, huge libraries of
fractionated NPs have been screened with impressive hit rates in
several diseases and pathogenic conditions. Of note, a number of
cases are known where the crude biological extract is more
pharmacologically effective against the purified most active
chemical compound from this extract (8). In several instances, other compounds
present in the extract with no pharmacological activity function
synergistically with the primary compound (9). To date, the effective compounds are
screened using advanced cell-based assay techniques, the candidate
targets in the cell are probed, and possible synergies are
identified (10,11).
Use of NPs in biology, pharmacology
and medicine
Traditional medicines and NPs provide valuable
insight towards the discovery of novel medicinal agents. Crude
biological extracts may help to enlarge the drug discovery paradigm
from ‘identifying novel entity drugs’ to ‘combining existing
agents’ and may even direct the combinations between such
NP-derived agents (12,13). Recent structural comparisons
between NPs and modern drugs or candidates identified 35% of NPs
that are structurally similar or identical in structure with modern
therapeutic agents (14). Although
modern pharmaceutical drugs were born from botanical medicine and
are mainly NPs, synthetic approaches to drug development have more
recently turn out to be standard. Based on recent studies on human
drugs introduced between the early 1980s until 2014, 62% of the new
small-molecule drugs were either NPs, derived from NPs (often
semi-synthetically), and NPs derived from pharmacophores
(considered as NPs analogs) (4,15).
However, the synthetic combinatorial chemistry and high-throughput
screening (HTS) of potential modern drug targets disconnected the
link between NPs and medicines. In the early 1990s, the
pharmaceutical industry turned towards the HTS of chemical
libraries against potential pharmacological targets, while the
screening of NPs was diminished. There was a feeling that NPs were
an ‘obsolete science’ and not a scientifically valid paradigm for
the modern discovery of novel drugs or chemicals. More recently,
modern pharmaceutical research has acknowledged this oversight and
stimulated new interest in the potential of NPs as novel
pharmaceutical agents (15). In a
great effort to recover and recombine the abandoned fragmented
information of the pharmaceutical properties of NPs, the
Hippo(crates) NP database, currently containing multidimensional
knowledge based on the collected and analyzed and cross-correlated
information for each NPs, was developed.
Use of chemoinformatics and new
goals
Chemoinformatics provide computer methods for the
organization, analysis and visualization of chemical information,
and is used extensively in drug discovery and development. It is a
rapidly evolving field, particularly due to the advent of
high-throughput experimental techniques, the widespread
availability of public databases, and the development of machine
learning algorithms (16,17). Successful results of
chemoinformatics approaches, such as Quantitative
Structure-Activity Relationship (QSAR) or Quantitative
Structure-Property Relationship (QSPR) and drug design, depend
critically on the quality of data and the representation of
chemical structure information through chemical descriptors and
high-dimensional vectors termed fingerprints. Several fingerprint
methods and similarity coefficients are used in similarity-based
virtual screening applications in order to identify database
compounds with probably similar bioactivity to a query compound
(18).
Pharmacophore is another concept integral to
computer-aided drug design. It is the ensemble of steric and
electronic features necessary to ensure the optimal supramolecular
interactions with a specific biological target structure and to
trigger (or to block) its biological response (19). Pharmacophore models can be derived
from experimentally determined protein-ligand complexes
(receptor-based pharmacophores), or from known active compounds
(ligand-based pharmacophores) (20,21).
Recently, polypharmacology, the ability of a single
agent to interact with multiple receptors and modulate several
processes, has drawn attention (22). Apart from studying drug
side-effects, polypharmacology facilitates the repurposing of ‘old’
drugs to treat both common and rare diseases, hopefully reducing
costs and accelerating drug development (23,24).
Ligand-based computational methods of predicting small molecule's
unknown targets involve similarity searches in databases containing
information on the activity of compounds and their protein targets
(25).
Hippo(crates), an updated atlas of
NPs
Different databases have been shared in recent
years, providing information required to develop the exploration
and exploitation in NPs. As expected, each database has been
specialized in a different field and presents the NPs from a
different point of view, including DrugBank, Natural Product
Activity and Species Source (NPASS), NPCARE and Open National
Cancer Institute (NCI) (Table I)
(26-30,32).
The Hippo(crates) database aims to facilitate the combination and
correlation of different fields of NP knowledge in a unified
platform, providing knowledge of each NP in an updated atlas of
NPs. In the same direction, several tools and algorithms have been
incorporated in the Hippo(crates) Database Graphical User Interface
(HDGUI) in order to export collective results and provide
beneficial knowledge of individual NPs or a group or category.
 | Table IThe six source databases and studies
that were used for the synthesis of the Hippo(crates) database. |
Table I
The six source databases and studies
that were used for the synthesis of the Hippo(crates) database.
| A/A | Source/(Refs.) | Sample | Common
identifiers | Dataset |
|---|
| 1 | Selleckchem | 16550 | - CAS Number | Natural
products |
| | | | - Canonical
SMILE | Synthetic
drugs |
| 2 | Open NCI | 15000 | - CAS Number | Natural
products |
| | | | - InChIKey | Synthetic
drugs |
| 3 | DrugBank | 700 | - Canonical
SMILE | Natural
products |
| | | | - InChIKey | |
| 4 | NPCARE | 9100 | - Canonical
SMILE | Natural
products |
| 5 | NPASS | 30000 | - Canonical
SMILE | Natural
products |
| | | | - CID | |
| 6 | Newman and Cragg
(15) | 1376 | - Name | Natural
products |
| | | | | Synthetic
drugs |
The Hippo(crates) database aims to assist the
pharmaceutical research for novel potential candidate
pharmacological agents and pharmacological targets. The user can
perform searches using the HDGUI with a combination of several
preset parameters, features, properties and keywords related to the
NPs and chemical compounds. The HDGUI applies various filtering,
processing and annotation techniques towards identifying and
visualizing the most probable dominant NPs and chemicals based on
the user preset parameters. The HDGUI identifies all the candidate
NPs using the up-to-date curated Hippo(crates) database and
provides each chemical compound information guided by explanatory
information from the annotation and data mining analyses, as well
as direct links to several online databases, such as PubChem
(https://pubchem.ncbi.nlm.nih.gov/),
Protein Data Bank (https://www.rcsb.org/) and a chemical 3D viewer.
(https://molview.org/).
Data: From collection to clustering
Data collection and filtering
The NP derivatives and synthetic compounds with
available experimentally-determined quantitative activity,
chemical, physicochemical properties and relative information were
extracted from the Selleckchem available catalog (https://www.selleckchem.com/) (31), the Open NCI database (https://cactus.nci.nih.gov/download/nci/) (26,32),
the DrugBank database (https://go.drugbank.com/) (27,28,33),
the NPCARE database (http://silver.sejong.ac.kr/npcancer/) (30) and the NPASS database (http://bidd.group/NPASS/) (29) by using combinations of keywords
related to the term ‘natural products’. Moreover, the final dataset
with the results of the NP chemical study by Newman and Cragg
(15) has been included in the
present study. A detailed content comparison among the six
extracted chemical datasets is provided in Table I. All the retrieved information has
been analyzed towards identifying common chemical characteristic
identifiers in the extracted chemical datasets, including ‘Name’,
‘Canonical SMILES’, ‘CAS registry number’, ‘International Chemical
Identifier (InChIKey)’ and PubChem id ‘CID’ (34,35).
The Hippo(crates) database has been synthetized from the described
chemical datasets, and each chemical entry has been checked to be
unique in the demo version of the database by using the common
chemical characteristic identifiers information (Fig. 1). All additional information from
the common chemical entries between the six extracted chemical
datasets has been included in the unique entries of the
Hippo(crates) database for each duplicate chemical as described in
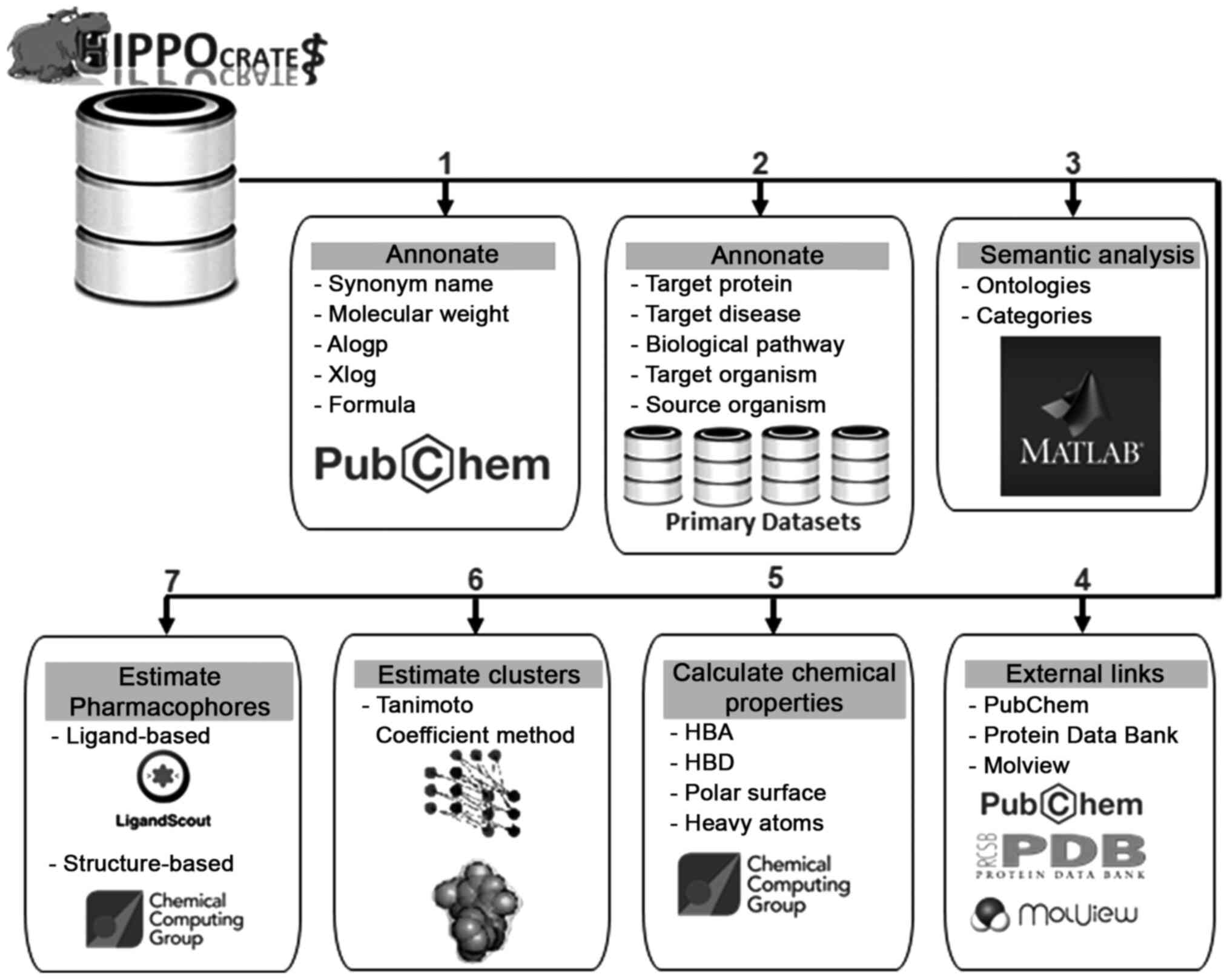
the data annotation and processing step. The main pipeline of the
described procedure is presented in Figs. 1 and 2.
Data annotation and processing
Hippo(crates) database entries have been annotated
with information from several fields contained in the PubChem
Database by using one or a combination of the four primary
identifiers (Name, CAS Number, InChIKey and CID) describing each
chemical compound (Fig. 1). More
specifically, from the PubChem database, the extracted information
contains the name, synonym_name, canonical_SMILES, InChiKey,
Formula, Molecular_Weight, alogp and Xlog (Fig. 2). The second level of annotation
has been performed with the use of the four common identifiers on
the primary datasets of the chemical compounds. According to the
type of knowledge contained in the primary datasets, information
has been extracted and annotated for each entry, including
target_protein, target_disease biological_pathway, target_organism
and source_organism (Fig. 2).
Subsequently, data mining and semantic analyses have been performed
using the Matlab Bioinformatics toolbox towards identifying the
major representative ‘ontologies’ per specific field with the
entries being characterized on the basis of the recognized
ontologies in categories (Fig. 2
and Table II) (36,37).
Through the data annotation, the major goal was to find the hidden
correlations between the chemical compounds and their connection
with other international chemical and structural databases.
Chemical compounds have been linked to the PubMed database using
the ‘CID’ and with the Protein Data Bank using the ‘InChIKey’,
respectively (Fig. 2). Last but
not least, specific chemical properties have been calculated using
the program MOE, including ‘hba’, ‘hbd’, polarsurface, ‘rings’ and
‘heavyatoms’ (Fig. 2) (38,39).
 | Table IIList of the 32 major categories
present in the Hippo(crates) database. |
Table II
List of the 32 major categories
present in the Hippo(crates) database.
| A/A | Category | A/A | Category |
|---|
| 1 | Natural
Product | 17 | Epigenetics |
| 2 | Anticancer | 18 | FDA Approved |
| 3 | Antidiabetic | 19 | GPCR related |
| 4 | Antiinfection | 20 | Immunology
inflammation |
| 5 | Antibacterial | 21 | Inhibitors |
| 6 |
Antihypertensive | 22 | Ion channels
related |
| 7 | Antiviral | 23 |
Kinase_inhibitor |
| 8 | Antiparasitic | 24 | MAPK inhibitor |
| 9 | Antifungal | 25 | Metabolism
compound |
| 10 | Antiulcer | 26 | Neuronal
signaling |
| 11 | Apoptosis | 27 | PI3K |
| 12 | Autophagy | 28 | Protease
inhibitor |
| 13 | Bioactive
compound | 29 | Pdb_related |
| 14 | Clinical | 30 | Stem cell
signaling |
| 15 | Calcium
metabolism | 31 | Target
selective |
| 16 | Drug
repurposing | 32 | Tyrosine kinase
inhibitor |
Data clustering and pharmacophore
design
Compound fingerprints were calculated using the
CACTVS Chemoinformatics Toolkit (40). The Sphere Exclusion algorithm
(41,42) complemented with Tanimoto
coefficient (43) was applied to
select diverse subsets from the Hippo(crates) database. This
procedure was repeated several times using different parameter sets
in order to identify the optimal thresholds and separate the
dataset in internal clusters. This was followed by pharmacophore
determination for the NPs and chemical compounds that were
annotated into the Hippo(crates) database using the analyzed
information from the previous processing step. In the present
study, both ligand-based and structure-based pharmacophore model
design was performed (Fig. 2). The
Tanimoto coefficient for clustered compounds was employed to
generate ligand-based pharmacophores using LigandScout (44). The structure-based pharmacophores
were constructed using the chemical compounds that were
co-crystalized with proteins in experimentally determined complexes
from the Protein Data Bank (PDB). A specialized analysis has been
setup towards estimating the corresponding protein cavities for
each chemical compound and then using their characteristic features
for designing the corresponding pharmacophore models using MOE
(45,46).
Structure of the Hippo(crates) database
HDGUI) webserver
The Hippo(crates) database of NPs and chemical
compounds is publicly available online at http://www.openscreen.aua.gr/login.php. The HDGUI runs
on a Secure HTTP Apache web server hosted at the HDGUI web server,
using the LINUX Operating System, Apache Technology, PHP,
JavaScript, R, and parallel computing architecture on the computing
facility of the School of Applied Biology and Biotechnology at the
Agricultural University of Athens (AUA). HDGUI has been designed in
a way to enable the user to retrieve NPs and chemical compounds
through various developed toolboxes. Additionally, specialized
toolbars that have been added to the interface enable the user to
make 2D and 3D chemical similarity searches (molecular similarity
score of ≥0.9) in the PubChem database using the extracted chemical
SMILES from the Hippo(crates) database (47,48).
The Hippo(crates) database
The Hippo(crates) database is an integrated resource
for NPs, chemical compounds derived from NPs and chemical compounds
considered as NPs analogs, and other chemical compounds. The
Hippo(crates) database currently holds 45,300 entries, which are
divided into 32 major categories, as presented in Table II. Moreover, from the annotation
and semantic analyses, 45,293 chemical SMILES, 45,183 CAS numbers,
45,292 PubChem IDs, 4,758 target ontologies, 27,802 disease
ontologies, 22,830 source organisms and 6,070 connected protein 3D
structures from the PDB have been correlated (Fig. 3). The information within the
database is structured in 32 different fields, and the knowledge is
organized in a specific manner in order to serve the webserver
application immediately and in a timely manner.
Functionality of the Hippo(crates)
database
Chemical clusters and
pharmacophores
The Hippo(crates) database provides a well-organized
atlas of interconnected NPs and other chemical compounds using both
advanced bioinformatics and chemoinformatics techniques. The
contents of the database were analyzed using specialized techniques
such as the Tanimoto coefficient for the analysis of chemical
compounds (43). Based on the
results, 3,046 different clusters were identified containing 2 to
1,600 chemical compounds (Fig. 4).
A second level of chemical analysis was performed towards
generating the representative pharmacophore models for each entry
(Fig. 2). Specifically, 2,100
ligand-based and 673 structure-based pharmacophore models were
constructed. The results of the chemoinformatic analysis are
interlinked in the Hippo(crates) database and are accessible
through a customized toolbox.
Usability and applications
HDGUI webserver
The HDGUI webserver aids the chemical and medical
experts, pharmacists and other users in searching and identifying
NPs, NP-derived and other synthetic chemicals with identified
chemical properties through a characteristic set of keywords and
ontologies. This is achieved through filtering web tools and the
summarized knowledge under ‘key’ terms is presented in smart lists.
Users are able to perform complex filtering operations using
chemical properties, fingerprints, disease, target proteins,
biological pathways, source organisms and several other specific
keywords under specific domain ontologies. In addition, the HDGUI
webserver enables users who may not be familiar with chemical
molecular structures (Fig. 5) to
be able to discover, filter and classify, and easily present NPs
and other chemical compounds contained in the Hippo(crates)
database. Furthermore, users are able to select and list
efficiently chemical compounds that are associated with ‘key’
terms, domain ontologies, or characteristic fingerprints
(SMILES).
The HDGUI filtering options are separated into seven
major webtools (Fig. 5). The
default simple search provides the ability to the user to discover
chemical compounds based on a specific international identifier
such as chemical name, SMILES, CAS number and InCHIkey or
characteristic chemical properties, or a combination of those
terms. The second web tool named ‘Species Search’ provides the user
with the ability to extract chemical compounds that are extracted
from a specific source organism such as plants, herbs, fungi,
flatworms and other living organisms. The species search webtool is
well-organized with a directory and smart list in an effective
manner from which the user can identify the main genera and the
species contained in the Hippo(crates) database. The third and the
fourth web tools named ‘Target Search’ and ‘Disease or Target
Organism’ provide the user with lists of key terms identified,
classified and summarized from a semantic analysis study. By
clicking a specific term from the lists, the user is able to
discover a cluster of chemical compounds that are correlated with
this specific term. The fifth webtool name ‘Smart Search’ enables
the user to perform a filtering search in the contents of the
Hippo(crates) database based on a number of specific keywords of
preference. The sixth webtool named ‘Blast Chem’ is an integrated
service from where the user selects similar 2D and 3D chemical
compounds with a 95% similarity cutoff from the PubChem database
using the SMILES identifier. Finally, the last webtool named
‘Pharmacophore Search’ provides the user with the option to
discover the possible ligand-based and structure-based
pharmacophore models by using a SMILES entry of a chemical
compound. Furthermore, in this webtool, the user can download the
dataset of the cluster of the chemical compounds from which the
pharmacophoric model has been generated (Fig. 6).
The HDGUI output is an HTML file that describes the
chemical compounds profile through a smart array which contains the
specific fields, including ‘name’, ‘category’, ‘species’, ‘target’,
‘disease’, ‘viewer’, ‘pubchem’, ‘PDB’, ‘cas_number’, ‘SMILES’,
‘molecular_weight’, ‘formula’, ‘alogp’, ‘hba’, ‘hbd’,
‘polar_surface’, ‘rotatable_bound’, ‘heavy_atoms’, ‘rings’, ‘info’
and ‘synonyms’ (Fig. 7). The user
may make a second level filtering pass on the generated results
through a keyword search on the smart array in order to separate
entries of interest. The HDGUI output from the ‘pharmacophore
search’ differs significantly from the other web tools. It tables
the output results in a downloadable smart array with the major
fields, such as ‘grid’, ‘SMILES’, ‘name’, ‘pubchem’, ‘pdb’,
‘ligand-based’, ‘structural-based’ and ‘tanimoto80’ (Fig. 6). Further characterization of the
NPs as drugs in clinical research phases is under way.
An example
The Hippo(crates) interface has been used towards
extracting beneficial knowledge and corresponding natural products
through various example searches (Species, Target, Disease, Smart,
Blast Chem or Pharmacophore) located at http://openscreen.aua.gr/examples.php. For the genus
‘Ganoderma’, a well-known basidiomycete used in Chinese traditional
medicine with pharmacological effects for several ailments. By
typing the keyword Ganoderma in the ‘Smart Search’ tab, and
clicking ‘Search’, the user creates a query for the database. The
search of the Hippo(crates) database results, in a list of 281
entries with several natural products related to Ganoderma.
Moreover, by inspecting the results and specially the ‘Info’ list,
where information correlated to the query genus is provided, it can
clearly be seen that Ganoderma related natural products have
therapeutic potential as hepatoprotective, anticancer,
anti-inflammatory, immunomodulatory, antioxidant and antiviral
agents. Several recent systematic studies have established and
confirmed these findings based on the literature (49,50).
Conclusion
Recent advances in genetics, clinical genomics and
personalized medicine have led to the need of discovering effective
therapeutic agents for several pathological conditions (51-54).
The discovery and correlation of NPs has shed light on the
seemingly unrelated correlation between human diseases and certain
molecules, leading to novel biologically active drugs. Natural
products are considered as a rich source of therapeutic agents
endowed with various significant pharmacological properties. The
Hippo(crates) database today lists 45,300 NPs and other chemical
compounds categorized into 32 major categories. Moreover, several
NPs have been associated with various organisms of origin. A total
of 22,500 different organisms were associated with various chemical
compounds, and more than 32,500 ontologies identified and presented
in the ‘target’ and ‘diseases’ fields of the database. Currently,
several NP databases facilitate the research on the field by
classifying the chemical compounds based on 2D and 3D molecular
similarity. However, to date, to the best of our knowledge, there
was a lack of a NP database that comprises information regarding
chemical compounds and pharmacophore models. The present study,
with the Hippo(crates) database, aimed to fill this essential gap
by providing >2,500 representative pharmacophore models.
Finally, the HDGUI provides significant assistance for the
discovery of NPs as it facilitates access to information relating
substances with their known implication in biological activities,
originating source, disease association, targeting biomolecule
(protein, nucleic acid and carbohydrate), co-crystallized 3D
structures, pharmacophoric models, and other beneficial
knowledge.
Acknowledgements
Not applicable.
Funding
The present study was funded by the project ‘(OPENSCREEN-GR) An
Open-Access Research Infrastructure of Chemical Biology and
Target-Based Screening Technologies for Human and Animal Health,
Agriculture and the Environment’ (grant no. MIS 5002691), which is
implemented under the Action ‘Reinforcement of the Research and
Innovation Infrastructure’, funded by the Operational Program
‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014-2020)
and co-financed by Greece and the European Union (European Regional
Development Fund). The project OPENSCREEN-GR aims to integrate
high-capacity screening platforms throughout Greece which jointly
use a rationally selected compound collection and offer to
researchers from academic institutions, SME's and industrial
organizations open access to its shared resources.
Availability of data and materials
The Hippo(crates) database is publicly available
online at: http://www.openscreen.aua.gr/login.php.
Authors' contributions
LP and EE participated in the construction of the
database. LP, AA, EC, KB, DV, TT and EE were involved in the
validation and visualization of the database. TT and EE
participated in methodology and TT in the Tanimoto analysis. AA,
EC, KB and DV searched the literature and performed data collection
and curation. LP and EE wrote the original draft of the manuscript
and were involved in further writing, reviewing and editing along
with AA and TT. EE was involved in the conceptualization and design
of the study, as well as in funding acquisition. LP and EE have
confirmed the authenticity of all the raw data and all authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hong J: Role of natural product diversity
in chemical biology. Curr Opin Chem Biol. 15:350–354.
2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bernardini S, Tiezzi A, Laghezza Masci V
and Ovidi E: Natural products for human health: An historical
overview of the drug discovery approaches. Nat Prod Res.
32:1926–1950. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Beutler JA: Natural Products as a
Foundation for Drug Discovery. Curr Protoc Pharmacol.
46:9.11.1–9.11.21. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Newman DJ and Cragg GM: Natural products
as sources of new drugs over the last 25 years. J Nat Prod.
70:461–477. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Thomford NE, Senthebane DA, Rowe A, Munro
D, Seele P, Maroyi A and Dzobo K: Natural Products for Drug
Discovery in the 21st Century: Innovations for Novel Drug
Discovery. Int J Mol Sci. 19(E1578)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kayser O, Kiderlen AF and Croft SL:
Natural products as antiparasitic drugs. Parasitol Res. 90 (Suppl
2):S55–S62. 2003.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Salam AM and Quave CL: Opportunities for
plant natural products in infection control. Curr Opin Microbiol.
45:189–194. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Sut S, Dall'Acqua S, Zengin G, Senkardes
I, Bulut G, Cvetanović A, Stupar A, Mandić A, Picot-Allain C, Dogan
A, et al: Influence of different extraction techniques on the
chemical profile and biological properties of Anthemis
cotula L.: Multifunctional aspects for potential pharmaceutical
applications. J Pharm Biomed Anal. 173:75–85. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Salis C, Papageorgiou L, Papakonstantinou
E, Hagidimitriou M and Vlachakis D: Olive Oil Polyphenols in
Neurodegenerative Pathologies. Adv Exp Med Biol. 1195:77–91.
2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Treml J, Gazdová M, Šmejkal K, Šudomová M,
Kubatka P and Hassan STS: Natural Products-Derived Chemicals:
Breaking Barriers to Novel Anti-HSV Drug Development. Viruses.
12(E154)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Koumandou VL, Papageorgiou L, Tsaniras SC,
Papathanassopoulou A, Hagidimitriou M, Cosmidis N and Vlachakis D:
Microbiome Hijacking Towards an Integrative Pest Management
Pipeline. Adv Exp Med Biol. 1195:21–32. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kong DX, Li XJ and Zhang HY: Where is the
hope for drug discovery? Let history tell the future. Drug Discov
Today. 14:115–119. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wagner H and Ulrich-Merzenich G: Synergy
research: Approaching a new generation of phytopharmaceuticals.
Phytomedicine. 16:97–110. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Calixto JB: The role of natural products
in modern drug discovery. An Acad Bras Cienc. 91 (Suppl
3)(e20190105)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Newman DJ and Cragg GM: Natural Products
as Sources of New Drugs from 1981 to 2014. J Nat Prod. 79:629–661.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Agrafiotis DK, Bandyopadhyay D, Wegner JK
and Vlijmen H: Recent advances in chemoinformatics. J Chem Inf
Model. 47:1279–1293. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lo YC, Rensi SE, Torng W and Altman RB:
Machine learning in chemoinformatics and drug discovery. Drug
Discov Today. 23:1538–1546. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Muegge I and Mukherjee P: An overview of
molecular fingerprint similarity search in virtual screening.
Expert Opin Drug Discov. 11:137–148. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fernandes JPS: The Importance of Medicinal
Chemistry Knowledge in the Clinical Pharmacist's Education. The
Importance of Medicinal Chemistry Knowledge in the Clinical
Pharmacist's Education. Am J Pharm Educ. 82(6083)2018.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Mitsis T, Papageorgiou L, Efthimiadou A,
Bacopoulou F, Vlachakis D, Chrousos GP and Eliopoulos E: A
comprehensive structural and functional analysis of the ligand
binding domain of the nuclear receptor superfamily reveals highly
conserved signaling motifs and two distinct canonical forms through
evolution. World Acad Sci J. 1:264–274. 2019.
|
|
21
|
Papageorgiou L, Shalzi L, Pierouli K,
Papakonstantinou E, Manias S, Dragoumani K, Nicolaides N,
Giannakakis A, Bacopoulou F, Chrousos G, et al: An updated
evolutionary study of the nuclear receptor protein family. World
Acad Sci J. 3(51)2021.
|
|
22
|
Chaudhari R, Tan Z, Huang B and Zhang S:
Computational polypharmacology: A new paradigm for drug discovery.
Expert Opin Drug Discov. 12:279–291. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Pushpakom S, Iorio F, Eyers PA, Escott KJ,
Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, et
al: Drug repurposing: Progress, challenges and recommendations. Nat
Rev Drug Discov. 18:41–58. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vlachakis D, Papageorgiou L, Papadaki A,
Georga M, Kossida S and Eliopoulos E: An updated evolutionary study
of the Notch family reveals a new ancient origin and novel
invariable motifs as potential pharmacological targets. PeerJ.
8(e10334)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Awale M, Visini R, Probst D, Arús-Pous J
and Reymond JL: Chemical Space: Big Data Challenge for Molecular
Diversity. Chimia (Aarau). 71:661–666. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Voigt JH, Bienfait B, Wang S and Nicklaus
MC: Comparison of the NCI open database with seven large chemical
structural databases. J Chem Inf Comput Sci. 41:702–712.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: A
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36 (Suppl 1):D901–D906. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wishart DS, Feunang YD, Guo AC, Lo EJ,
Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, et al:
DrugBank 5.0: A major update to the DrugBank database for 2018.
Nucleic Acids Res. 46 (D1):D1074–D1082. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zeng X, Zhang P, He W, Qin C, Chen S, Tao
L, Wang Y, Tan Y, Gao D, Wang B, et al: NPASS: Natural product
activity and species source database for natural product research,
discovery and tool development. Nucleic Acids Res. 46
(D1):D1217–D1222. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Choi H, Cho SY, Pak HJ, Kim Y, Choi JY,
Lee YJ, Gong BH, Kang YS, Han T, Choi G, et al: NPCARE: Database of
natural products and fractional extracts for cancer regulation. J
Cheminform. 9(2)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cheng S, Zhu C, Chu C, Huang T, Kong X and
Zhu LC: Prediction of bioactive compound pathways using chemical
interaction and structural information. Comb Chem High Throughput
Screen. 19:161–169. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ihlenfeldt WD, Voigt JH, Bienfait B,
Oellien F and Nicklaus MC: Enhanced CACTVS browser of the Open NCI
Database. J Chem Inf Comput Sci. 42:46–57. 2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Knox C, Law V, Jewison T, Liu P, Ly S,
Frolkis A, Pon A, Banco K, Mak C, Neveu V, et al: DrugBank 3.0: A
comprehensive resource for ‘omics’ research on drugs. Nucleic Acids
Res. 39 (Database):D1035–D1041. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hähnke VD, Kim S and Bolton EE: PubChem
chemical structure standardization. J Cheminform.
10(36)2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Heller SR, McNaught A, Pletnev I, Stein S
and Tchekhovskoi D: InChI, the IUPAC International Chemical
Identifier. J Cheminform. 7(23)2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Reyes-Aldasoro CC: The proportion of
cancer-related entries in PubMed has increased considerably; is
cancer truly ‘The Emperor of All Maladies’? PLoS One.
12(e0173671)2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Dunn MJ, Jorde LB, Little PFR and
Subramaniam S (eds): Encyclopedia of Genetics, Genomics, Proteomics
and Bioinformatics, 8 Volume Set. John Wiley & Sons Ltd.,
Hoboken NJ, 2005.
|
|
38
|
Vilar S, Cozza G and Moro S: Medicinal
chemistry and the molecular operating environment (MOE):
Application of QSAR and molecular docking to drug discovery. Curr
Top Med Chem. 8:1555–1572. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Mhlanga P, Wan Hassan WA, Hamerton I and
Howlin BJ: Using combined computational techniques to predict the
glass transition temperatures of aromatic polybenzoxazines. PLoS
One. 8(e53367)2013.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ihlenfeldt WD, Takahashi Y, Abe H and
Sasaki S: Computation and management of chemical properties in
CACTVS: An extensible networked approach toward modularity and
compatibility. J Chem Inf Comput Sci. 34:109–116. 1994.
|
|
41
|
Huggins DJ, Venkitaraman AR and Spring DR:
Rational methods for the selection of diverse screening compounds.
ACS Chem Biol. 6:208–217. 2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hudson BD, Hyde RM, Rahr E, Wood J and
Osman J: Parameter Based Methods for Compound Selection from
Chemical Databases. Quant Struct-Act Rel. 15:285–289. 1996.
|
|
43
|
Bajusz D, Rácz A and Héberger K: Why is
Tanimoto index an appropriate choice for fingerprint-based
similarity calculations? J Cheminform. 7(20)2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wolber G and Langer T: LigandScout: 3-D
pharmacophores derived from protein-bound ligands and their use as
virtual screening filters. J Chem Inf Model. 45:160–169.
2005.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Shaker B, Yu MS, Lee J, Lee Y, Jung C and
Na D: User guide for the discovery of potential drugs via protein
structure prediction and ligand docking simulation. J Microbiol.
58:235–244. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhou Y, Tang S, Chen T and Niu MM:
Structure-Based Pharmacophore Modeling, Virtual Screening,
Molecular Docking and Biological Evaluation for Identification of
Potential Poly. Structure-Based Pharmacophore Modeling, Virtual
Screening, Molecular Docking and Biological Evaluation for
Identification of Potential Poly (ADP-Ribose) Polymerase-1 (PARP-1)
Inhibitors. Molecules. 24(E4258)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Bolton EE, Chen J, Kim S, Han L, He S, Shi
W, Simonyan V, Sun Y, Thiessen PA, Wang J, et al: PubChem3D: A new
resource for scientists. J Cheminform. 3(32)2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kim S, Bolton EE and Bryant SH: Similar
compounds versus similar conformers: Complementarity between
PubChem 2-D and 3-D neighboring sets. J Cheminform.
8(62)2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wachtel-Galor S, Yuen J, Buswell JA and
Benzie IFF: Ganoderma lucidum (Lingzhi or Reishi): A Medicinal
Mushroom. In: Herbal Medicine: Biomolecular and Clinical Aspects.
Benzie IFF and Wachtel-Galor S (eds). 2nd edition. CRC Press/Taylor
& Francis, Boca Raton, FL, 2011.
|
|
50
|
Xu J, Chen F, Wang G, Liu B, Song H and Ma
T: The Versatile Functions of G. Lucidum Polysaccharides and G.
Lucidum Triterpenes in Cancer Radiotherapy and Chemotherapy. Cancer
Manag Res. 13:6507–6516. 2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Papageorgiou L, Zervou MI, Vlachakis D,
Matalliotakis M, Matalliotakis I, Spandidos DA, Goulielmos GN and
Eliopoulos E: Demetra Application: An integrated genotype analysis
web server for clinical genomics in endometriosis. Int J Mol Med.
47(115)2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Spreafico R, Soriaga LB, Grosse J, Virgin
HW and Telenti A: Advances in Genomics for Drug Development. Genes
(Basel). 11(E942)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Cardon LR and Harris T: Precision
medicine, genomics and drug discovery. Hum Mol Genet. 25
(R2):R166–R172. 2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sonehara K and Okada Y: Genomics-driven
drug discovery based on disease-susceptibility genes. Inflamm
Regen. 41(8)2021.PubMed/NCBI View Article : Google Scholar
|