Introduction
Worldwide, cancer is the primary cause of mortality
and a key obstacle to extending life expectancy. According to
estimates from the World Health Organization (WHO) in 2019, cancer
was the first or second major cause of mortality prior to the age
of 70 years in 112 of 183 countries (1). Worldwide, an estimated 19.3 million
new cancer cases and almost 10.0 million cancer-related deaths
occurred in 2020. The global cancer burden is expected to be 28.4
million cases by the year 2040, a 47% increase from 2020, with a
larger increase in transitioning (64 to 95%) vs. transitioned (32
to 56%) countries. Some types of cancer are very common, such as
lung, cervical, liver and pancreatic cancer (2). According to GLOBOCAN, lung cancer was
the second most frequently diagnosed type of cancer and the leading
cause of cancer-related mortality in 2020, accounting for almost
one in every ten (11.4%) diagnosed cases of cancer and one in every
five (18.0%) related deaths, with an anticipated 2.2 million new
cancer cases and 1.8 million deaths (2). Cervical cancer is the fourth most
commonly diagnosed disease and the fourth most common cause of
cancer-related death among females, with 604,000 new cases and
342,000 related deaths globally (2). In 2020, primary liver cancer was the
sixth most commonly diagnosed type of cancer and the third major
cause of cancer-related mortality worldwide, with ~906,000 new
cases and 830,000 fatalities. Pancreatic cancer is the seventh most
common cause of cancer-related mortality, accounting for almost as
many deaths (466,000) as diagnoses (496,000), owing to its poor
prognosis (2). The pathological
type and clinical stage of cancers are not the only factors used to
identify the overall survival (OS) and recurrence-free survival of
patients with cancer. The expression and pathways regulated by
tumor genes also play a crucial role in the survival of patients
with cancer (3). Identifying
cancer-specific genes or biomarkers involved in cancer development
and progression helps to understand cancer pathophysiology and
identify therapeutic targets (4).
These biomarkers can be used to predict the risk, occurrence of
cancer and patient outcomes (5).
Previous studies have suggested the significance of abnormally
expressed genes in early diagnosis, prognosis, disease monitoring
and response to therapy in liver, lung, pancreatic and cervical
cancers (6-9).
Therefore, research on survival-associated biomarkers may be
beneficial for the treatment of these types of cancer, which may
ultimately improve the OS rate of patients with cancer.
Currently, a large amount of functional genomic data
have been generated owing to the development of high-throughput
sequencing technology, which makes it possible to identify
survival-associated cancer biomarkers by analyzing differentially
expressed genes (DEGs) (10). The
common issue with biomarker identification using high-throughput
sequencing technologies lies in their selection and validation
processes owing to the generation of large amounts of data
(11). Other issues include a
limited sample size, variations in sampling and experimental
processes and high costs (11). To
date, a number of gene expression profiling studies have been
carried out in liver, lung, pancreatic and cervical cancers to
identify potential biomarkers related to diagnosis, prognosis,
survival, development and response to therapy (12-15).
For instance, Sun et al (16) investigated the role of exosomal
copine III in colorectal cancer diagnosis and prognosis. Similarly,
based on DEGs, a prognostic survival model was constructed for
pancreatic cancer (9). To date,
these biomarkers have not been used in clinical settings. In recent
years, bioinformatics has been widely used for the functional
analysis of genomic and proteomic data of tumors for cancer
management. Advancements in bioinformation technology, the
establishment of multiple public databases, and the use of
analytical methodologies have resulted in the development of
sophisticated tools for analyzing and identifying DEGs (17).
The aim of the present study was to identify common
biomarkers associated with the survival of patients with liver,
lung, cervical and pancreatic cancers. The present study identified
seven common biomarkers that affect survival in four types of
cancers. These biomarkers can be further used for the therapeutic
targeting of cancers.
Materials and methods
Study design
In the present study, an in silico analysis
using RNA-Seq data from The Cancer Genome Atlas (TCGA) was
performed (9) to identify
potential biomarkers and/or therapeutic targets for four types of
cancer: Lung, cervical, liver and pancreatic cancer. TCGA is a
database containing a large number of molecularly characterized
datasets of >20,000 tumors and matched normal samples, along
with matching clinical information, such as drug exposure and
survival rates (18). These genes
were filtered out to identify common and unique genes for the
different cancer types used in the present study. Genes that had a
significant impact on the survival of patients with cancer were
screened and then validated using the cBioPortal database
(https://www.cbioportal.org/). The
expression of genes common to at least two types of cancers was
also evaluated in serum samples from patients with oral cancer. The
identification of these common genes will help to identify the risk
of cancer progression or therapeutic intervention in general.
Cancer-associated unique biomarkers were also identified that can
be used for the prognosis, diagnosis, or therapeutic management of
the cancers studied in the present study.
Data resources
All available RNA-sequencing data from TCGA were
retrieved using TCGAbiolinks (19). Briefly, gene count data from the
available pre-processed data types were selected for four different
types of cancer: i) Lung squamous cell carcinoma (TCGA-LUSC: 504
patients; age range, 41-84 years; disease stage, I, II, III and IV;
normal samples, 51); ii) cervical squamous cell carcinoma and
endocervical adenocarcinoma (TCGA-CESC: 307 patients; age range,
20-88 years; disease stage, I, II, III and IV; normal samples, 3);
iii) liver hepatocellular carcinoma (TCGA-LIHC: 377 patients; age
range, 16-90 years; disease stage, I, II and III; normal samples:
50); and iv) pancreatic adenocarcinoma (TCGA-PAAD: 185 patients;
age range, 40-88 years; disease stage, I, II, III and IV; normal
samples, 4).
Ethics approval
A total of 10 patients with oral cancer referred by
the Parul Sevashram Hospital (Vadodara, India) were enrolled in the
present study in 2023 according to the following inclusion and
exclusion criteria. Inclusion criteria: Patients with biopsy proven
oral cancer and who provided consent for the study were included.
Exclusion criteria: i) Patients with other types of cancer; ii)
patients who had undergone prior chemotherapy treatment and had
known additional malignancies that progressed or required active
treatment over the past 2 years; iii) patients having salivary
gland disease; iv) those who did not provide consent. The blood
samples (3 ml) were collected before commencing therapy and serum
(~1 ml) was extracted. In addition, 5 healthy volunteers were
included as the control group. All samples were collected in
between February, 2023 to October, 2023. The present study was
approved by the Ethics Committee of Parul University
(PUIECHR/PIMSR/00/081734/5307). All methods were performed
according to the relevant guidelines and regulations provided by
the ethics committee of Parul University.
Differential expression analysis
To identify DEGs, the ‘DESeq2’ package was employed.
To detect statistically significant genes, the package leverages
the negative binomial distribution algorithm using the ‘DESeq’
function (20). Genes with counts
of <100 were filtered out. Genes with an absolute log2 fold
change (LFC) >1 and <1 were considered upregulated and
downregulated, respectively. Genes with a false discovery rate
(FDR)-adjusted P-value of <0.001 were considered statistically
significant.
Interaction network construction
The protein-protein interaction (PPI) network of
DEGs was constructed using the STRING database (http://string-db.org) (21). Furthermore, the list of genes was
uploaded to Cytoscape software to visualize the PPI network and
analyze the structural properties of the constructed network. The
high confidence (value=0.7) was used to analyze the degree of
connectivity in the networks in Cytoscape software (version 3.9.1).
The top genes were screened based on their high degree values.
Heatmaps construction and survival
analysis
The selected hub genes were used to construct a
heatmap for the common genes present in various types of cancer
using the ComplexHeatmap package (22). OS analysis of TCGA data was
performed using the gene expression profiling interactive analysis
(GEPIA) online tool (23). To
date, GEPIA has >400 citations (24). GEPIA performs a survival analysis
based on gene expression levels. GEPIA uses the log-rank test,
often termed the Mantel-Cox test, for hypothesis evaluation. The
Cox proportional hazard ratio and 95% confidence interval were also
included in the survival plot. The log-rank test has optimum power
under the assumption of proportional hazard rates. However, this
assumption is often violated, particularly when two survival curves
cross each other. In figs. 3-6, late crossing of survival curve can
be seen. The authors were not able to restrict the time range in
GEPIA to remove the late-stage crossover. Therefore, this may be
the limitation of the present study. The cut-off criteria for log2
fold change (Log2FC) and log-rank p were set to ≥1 and <0.05,
respectively.
Gene Ontology (GO) and pathway
analysis
Functional enrichment analyses were performed using
the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt), which
includes GO and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analyses (25). This
analysis was performed to determine the involvement of the DEGs in
various pathways and their functions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The lung is the primary site of oral metastatic
tumors, with no proven treatment (26). A recent study also supported the
risk of developing lung metastasis in oral cancer (27). It has also been shown that liver
cancer can metastasize to oral cancer (28-30).
Therefore, the present study included patients with oral cancer to
identify more effective and targeted treatments that can enhance
the survival of patients with cancers that are capable of
metastasizing to oral tissues. RNA extraction from the serum
samples of 10 patients with oral cancer was performed using QIAzol
reagent (cat. no. 79306, Qiagen Inc.), as previously described
(31). Complementary DNA (cDNA)
was synthesized from 500 ng RNA using a G-Biosciences cDNA
synthesis kit (cat. no. 786-5020). The resulting cDNA was used for
qPCR (Rotor-Gene Q, Qiagen, Inc.). qPCR was performed in triplicate
with 2X SYBR-Green qPCR Master Mix from G-Biosciences (cat. no.
786-5062) under the following conditions: 95˚C for 3 min, followed
by 40 cycles at 95˚C for 15 sec and 60˚C for 60 sec. The relative
expression levels of the target gene mRNAs were calculated using
the comparative Cq method (relative expression=2-IICq),
using β-actin as an internal control (32). The primer sequences used were as
follows: Human NDC80 kinetochore complex component (NDC80)
forward, 5'-CCTCTCCATGCAGGAGTTAAGA-3' and reverse,
GGTCTCGGGTCCTTGATTTTCT; human minichromosome maintenance complex
component (MCM)3 forward, TGGCCTCCATTGATGCTACC and reverse,
GGACGACTTTGGGACGAACT; human disks large-associated protein 5
(DLGAP5) forward, AAGTGGGTCGTTATAGACCTGA and reverse,
TGCTCGAACATCACTCTCGTTAT; human DNA topoisomerase II alpha
(TOP2A) forward, CATTGAAGACGCTTCGTTATGG and reverse,
CAGAAGAGAGGGCCAGTTGTG; human β-actin forward, GGACTTCGAGCAAGAGATGG
and reverse, AGCACTGTGTTGGCGTACAG.
Molecular docking
Small compounds or inhibitors were selected from the
connectivity map (CMap) database (https://clue.io/about). It is an online database
containing information on the relationship between small-molecule
compounds and various genes (33).
Briefly, the names of DEGs were uploaded into the CMap database
using its query module. The connectivity score defined the
correlation between DEGs and small-molecule compounds. Negative
scores indicate the therapeutic potential of a drug molecule.
Therefore, the maximum negative scores were selected to predict the
inhibitors of specific gene types. The structure of cytochalasin B
(NDC80 inhibitor) was obtained from PubChem with its
compound identifier (compound identification no. 5311281). No
inhibitor was found for cell division cycle 45 (CDC45),
MCM and origin recognition complex subunit 1 (ORC1).
Molecular docking was performed between the drug and NDC80
(Protein Data Bank ID: 2IGP) using PyRx, a virtual screening tool
(34).
Molecular dynamics (MD)
simulations
A simulation was performed between cytochalasin B
(the drug with the highest binding affinity) and NDC80 using
the NAMD3 system by applying a CHARMM force-field (35). The system build-in commands of
NAMD3 were used to calculate the root mean square deviation (RMSD),
root mean square fluctuation (RMSF) and radius of gyration (Rg),
which were plotted using the Bio3D v2.3-0 package (http://thegrantlab.org/bio). The simulation was
conducted for 50 nsec. The binding free energy (ΔG) of the
drug-protein complex was calculated using the NAMD energy plugin
(35).
c-BioPortal analysis
To observe the effects of the expression of common
and unique genes on cancer survival, RNA-seq data were downloaded
from cBioPortal (TCGA, PanCancer Atlas) (https://www.cbioportal.org/datasets) (36). Samples that held data for the
following parameters were selected: RNA-seq data of cancers,
American Joint Committee on Cancer (AJCC) pathological staging and
survival data. AJCC staging is based on the evaluation of the T
(tumor), N (nodes) and M (metastasis) components of the primary
cancer and the assignment of a stage grouping. Samples were divided
into high- and low-risk groups based on the cut-off value of gene
expression, which was determined using the median ± 3 standard
error of the mean (SEM).
Statistical analysis
Common and unique DEGs were validated using
available data from the cBioPortal database (36). Gene expression values were
classified as low or high based on the cut-off values. The cut-off
value was obtained using the following formula: Median ± 3SEM. The
anticipated rate of distant relapse was calculated using the
Breslow-type estimator of the survival function (OriginLab version
2019). The receiver operating characteristic (ROC) curves and all
statistical computations were performed using OriginPro version
2019. OriginPro offers advanced statistical analysis tools and apps
[Origin: Data Analysis and Graphing Software (originlab.com)]. These ROC curves are based on the
classification model at all classification thresholds and include
two parameters: True positive rate (sensitivity) and false positive
rate (1-specificity). The results of RT-qPCR were analyzed using
the Students t-test (unpaired). A value of P<0.05 was considered
to indicate a statistically significant difference.
Results
Identification of common biomarkers in
four different solid cancers
To explore potential biomarkers of the four
different tumors, mRNA sequencing (RNA-seq) analysis was performed
to identify the DEGs between the tumor and normal tissues. An
integrative analysis of the RNA-seq database of TCGA-LUSC (lung),
TCGA-CESC (cervical), TCGA-LIHC (liver) and TCGA-PAAD (pancreatic)
cancers was performed using the TCGAbiolinks package. After
processing the data, a total of 16,365, 24,777, 24,893 and 25,253
genes were differentially expressed in lung, cervical, liver and
pancreatic cancer, respectively, when compared to normal tissues.
These DEGs were further filtered using the criteria of log fold
change ≥1 and FDR <0.001. A total of 5,809 (35.49%)
(upregulated, 3,156; downregulated, 2,653) genes in lung cancer,
3,043 (12.28%) (upregulated, 1,942; downregulated, 1,101) genes in
cervical cancer, 6,003 (24.11%) (upregulated, 3,937; downregulated,
2,066) genes in liver cancer and 41 (0.16%) (upregulated, 32;
downregulated, 9) genes in pancreatic cancer exhibited altered
transcript levels when compared with normal tissues. The DEGs were
then compared to identify common genes present in all four types of
cancer. A total of 490 common genes (393 upregulated and 97
downregulated) were found in lung, cervical and liver cancers
(Table SI). However, no genes
were found to be common among the four cancer types. Common genes
were used to establish a network to identify closely related genes.
These networks were analyzed and 229 genes were extracted based on
a high confidence score (Fig. 1).
The top 70 genes were selected for further analysis based on their
degree and closeness values (Table
SII). Heatmap analysis revealed the upregulation of the top 70
genes in lung (Fig. 2A), cervical
(Fig. 2B) and liver cancer
(Fig. 2C).
Survival curves of the top 70 common
genes
The survival plot demonstrated that only six genes
(8.57%) in lung cancer, nine genes (12.85%) in cervical cancer, and
41 genes (58.57%) in liver cancer significantly affected the
survival of patients with cancer (Table I). These results suggested that,
irrespective of the significant upregulation of the remaining DEGs
in the cancers examined, they play no significant role in the
survival of these patients with cancer.
 | Table ICommon hub genes significantly
affecting the survival of patients with lung, cervical and liver
cancer |
Table I
Common hub genes significantly
affecting the survival of patients with lung, cervical and liver
cancer
| Lung | Cervical | Liver |
|---|
| DLGAP5, NDC80,
POLE2, CASC5, CHEK1, WDHD1 | MCM2, ORC1, CDC45,
MCM3, TOP2A, PCNA, RFC4, RRM2, EXO1 | CDK1, CCNA2, CDC20,
BUB1B, CCNB1, BUB1, KIF11, PLK1, KIF20A, DLGAP5, TOP2A, DBF4, MCM3,
RAD51, MCM10, ASPM, HMMR, MCM2, MCM6, SPAG5, NDC80, ORC1, AURKA,
TPX2, BIRC5, KIF4A, CLSPN, CDC45, CDC6, CDC7, MCM5, MCM4, NCAPG,
FOXM1, CDCA3, TKK, CENPA, NEK2, CENPF, KIF14, CCNA2 |
RT-qPCR analysis of common genes
present in at least two types of tumors
The present study then attempted to identify DEGs
that were present in at least two types of tumors and had a
significant impact (P<0.05) on OS. Based on these criteria, only
two genes (DLGAP5 and NDC80) in lung and liver
cancers, and five genes (MCM2, MCM3, ORC1,
CDC45 and TOP2A) in cervical and liver cancers were
identified. It was also observed that the upregulation of these
genes decreased the survival of patients with liver cancer
(Figs. 3A and 4A). However, the opposite results were
obtained for lung (Fig. 3B) and
cervical cancers (Fig. 4B). High
levels of these DEGs are closely associated with longer OS in lung
and cervical cancers, which may be attributed to their different
intracellular locations (37).
Subsequently, the significant impact of these seven genes on
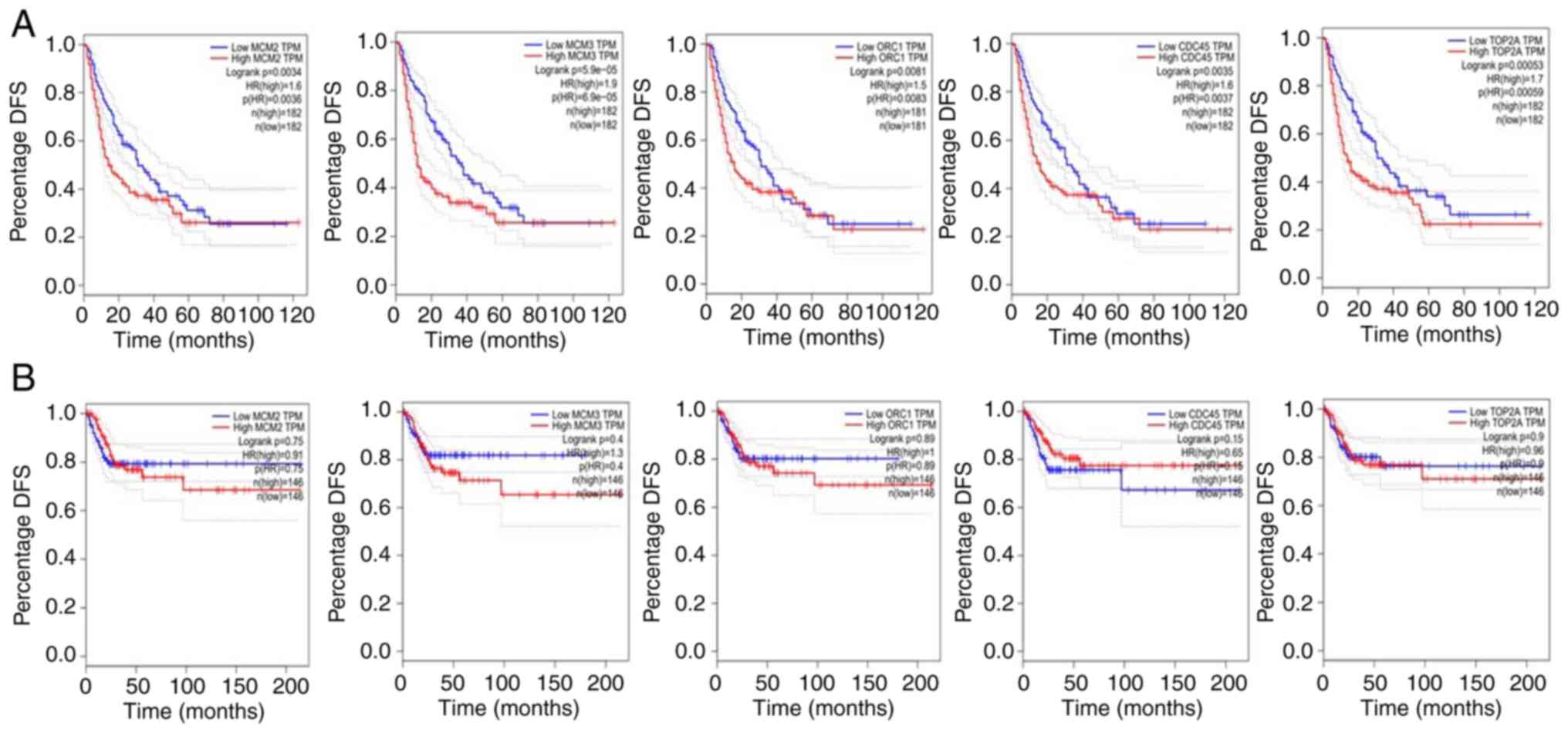
disease-free survival (DFS) was only observed in liver cancer
(Figs. 5A and 6A). None of the genes significantly
affected the DFS of lung (Fig. 5B)
and cervical cancers (Fig. 6B),
suggesting that these genes significantly affect the DFS of only
patients with liver cancer. The log-rank test has optimum power
under the assumption of proportional hazard rates. However, this
assumption is often violated, particularly when two survival curves
cross each other. In figs. 3-6, late crossing of survival curve can
be seen. The authors were not able to restrict the time range in
GEPIA to remove the late-stage crossover. Therefore, this may be
the limitation of the present study. Notably, it has been proposed
that when each sample size is ≥100, all the tests
(Kolmogorov-Smirnov statistic, Cramér-von Mises test, Maximum of
the WKM tests, and Renyi test) demonstrate powers >98%
regardless of the censoring rate (38). In all the figures (Fig. 3, Fig.
4, Fig. 5 and Fig. 6), the sample size is >150 and
thus it should not affect the statistical analysis.
To increase the broad spectrum of the study, after
analyzing the data on four cancers, the authors wish to evaluate
the expression of common genes in other types of cancer that are
relevant to the studied cancers (lung, liver, cervical and
pancreatic cancer). Therefore, oral cancer samples were added to
increase the broad spectrum of the study and the expression levels
of all genes in serum samples from patients with oral cancer were
assessed. As shown in Fig. 7A-C,
in the oral cancer samples, the expression of DLGAP5
(1.53±0.22; no significant difference; Fig. 7A), MCM3 (3.56±0.42;
P<0.05; Fig. 7B) and
NDC80 (5.01±0.35; P<0.05; Fig. 7C) was markedly higher compared with
the control (healthy donor) samples. However, the expression of
TOP2A (Fig. 7D) was not
markedly altered in the samples from patients with oral cancer
(0.96±0.12), when compared with the healthy samples. The expression
of other genes was not measurable in the serum samples of both the
patients and normal samples.
GO functional and pathway enrichment
analysis
GO functional and KEGG pathway enrichment analyses
were performed to identify the potential target genes. The enriched
GO functions for the target genes are presented in Table SIII, including the microtubule
cytoskeleton organization, mitotic cell cycle, DNA metabolic
process, microtubule-based process, cell cycle, regulation of
mitotic cell cycle, cell cycle process, negative regulation of cell
cycle, negative regulation of mitotic cell cycle, regulation of
cell cycle in the biological process category; cytoskeletal part,
microtubule cytoskeleton, chromosome, chromosomal part, microtubule
organizing center, nuclear chromosome, nuclear chromosome part,
chromosomal region, condensed chromosome, condensed nuclear
chromosome in the cellular component category; and adenosine
tri-phosphate binding, microtubule binding, drug binding, tubulin
binding, purine nucleotide binding, adenyl nucleotide binding,
ribonucleotide binding, purine ribonucleotide binding, adenyl
ribonucleotide binding, purine ribonucleoside triphosphate binding
in the molecular function category. The enriched KEGG pathways for
the target genes included DNA replication, progesterone-mediated
oocyte maturation, cell cycle, oocyte meiosis, cellular senescence,
human T-cell leukemia virus 1 infection and the p53 signaling
pathway (Tables SIII and SIV).
Validation of selected common genes in
the cBioPortal database
The seven common genes identified by TCGA data
analysis were validated using c-BioPortal data for the specific
cancer type. Patients were divided into high- and low-risk groups
based on the cut-off value (median ± SEM) of gene expression and on
the basis of their impact on overall survival. First, the data for
two common genes (NDC80 and DLGAP5) in lung and liver
cancer were validated. It was observed that out of the 133 patients
with lung cancer, NDC80 was increased in only 40 patients
(low-risk) (30.07%) and decreased in only 32 patients (24.06%)
(high-risk). For lung cancer, the data of DLGAP5 were not
available in the c-BioPortal database. Out of the 371 patients with
liver cancer, a high expression of NDC80 was observed in 131
patients (high-risk) (35.30%) and a low expression was observed in
143 patients (38.54%) (low-risk). Similarly, a high expression of
DLGAP5 was observed in 134 patients (high-risk) (36.11%) and
a low expression was observed in 136 patients (low-risk) (36.65%).
The effects of NDC80 and DLGAP5 gene expression on
the survival of these patients are presented in Table II. The results suggested that only
NDC80 had a significant effect on both the OS and DFS of
patients with liver cancer (Table
II).
 | Table IISurvival analysis of the common DEGs
identified in lung and liver cancer from the cBioPortal
database. |
Table II
Survival analysis of the common DEGs
identified in lung and liver cancer from the cBioPortal
database.
| SN | Gene symbol | Cancer type | Risk of disease
(no. of patients) | Survival/deceased
status (no. of patients) | P-value (OS) | Disease free
survival/recurred status (no. of patients) | P-value (DFS) |
|---|
| 1 | NDC80 | Lung | High risk (32) | OS (23) | 0.09 | DFS (21) | 0.100348 |
| | |
(CO=0.070±0.001) | | Deceased (9) | | Recurred (11) | |
| | | | Low risk (40) | OS (35) | | DFS (33) | |
| | | | | Recurred (5) | | Recurred (7) | |
| 2 | | Liver | High risk
(131) | OS (91) | 0.036a | DFS (48) |
0.002974a |
| | |
(CO=11.26±0.78) | | Deceased (40) | | Recurred (83) | |
| | | | Low risk (143) | OS (115) | | DFS (78) | |
| | | | | Deceased (28) | | Recurred (65) | |
| 3 | DLGAP5 | Liver | High risk
(134) | OS (89) |
0.004378a | DFS (49) | 0.675183 |
| | |
(CO=9.01±0.79). | | Deceased (45) | | Recurred (40) | |
| | | | Low risk (136) | OS (111) | | DFS (71) | |
| | | | | Deceased (25) | | Recurred (65) | |
Subsequently, the data of five common genes
(MCM2, ORC1, CDC45, MCM3 and
TOP2A) identified by TCGA data analysis of cervical and
liver cancer were validated using the c-BioPortal database. Of
these five genes, only MCM3, CDC45 and ORC1
were found to exert significant effects on both the OS and DFS of
patients with liver cancer (Table
III). None of these genes significantly affected the survival
rate of patients with cervical cancer.
 | Table IIISurvival analysis of the common DEGs
identified in cervical and liver cancer from cBioPortal
database. |
Table III
Survival analysis of the common DEGs
identified in cervical and liver cancer from cBioPortal
database.
| SN | Gene symbol | Cancer type | Expression | Details | P-value | Details | P-value |
|---|
| 1 | MCM2 | Cervical | High risk
(103) | OS (88) | 0.48812 | DFS (84) | 0.996133 |
| | | CO=137.56±3.94 | | Deceased (15) | | Recurred (19) | |
| | | | Low risk (114) | OS (101) | | DFS (93) | |
| | | | | Deceased (13) | | Recurred (21) | |
| 2 | | Liver | High risk
(132) | OS (89) |
0.015686a | DFS (49) | 0.506711 |
| | | CO=44.62±3.54 | | Deceased (43) | | Recurred (39) | |
| | | | Low risk (133) | OS (107) | | DFS (68) | |
| | | | | Deceased (26) | | Recurred (65) | |
| 3 | MCM3 | Cervical | High risk
(106) | OS (94) | 0.953041 | DFS (89) | 0.3708 |
| | | CO=99.87±1.98 | | Deceased (12) | | Recurred (17) | |
| | | | Low risk (121) | OS (107) | | DFS (96) | |
| | | | | Deceased (14) | | Recurred (25) | |
| 4 | | Liver | High risk
(134) | OS (94) |
0.033606a | DFS (52) |
0.011974a |
| | | CO=68.32±3.07 | | Deceased (40) | | Recurred (82) | |
| | | | Low risk (129) | OS (105) | | DFS (70) | |
| | | | | Deceased (26) | | Recurred (59) | |
| 5 | ORC1 | Cervical | High risk
(109) | OS (93) | 0.564918 | DFS (85) | 0.211026 |
| | | CO=11.22±0.25 | | Deceased (16) | | Recurred (24) | |
| | | | Low risk (116) | OS (102) | | DFS-98 | |
| | | | | Deceased (14) | | Recurred-18 | |
| 6 | | Liver | High risk
(121) | OS (85) |
0.01449a | DFS (45) |
0.018018a |
| | | CO=5.50±0.47 | | Deceased (36) | | Recurred (76) | |
| | | | Low risk (141) | OS (117) | | DFS (73) | |
| | | | | Deceased (24) | | Recurred (68) | |
| 7 | CDC45 | Cervical | High risk
(101) | OS (89) | 0.931737 | DFS (79) | 0.272893 |
| | | CO=18.92±0.54 | | Deceased (12) | | Recurred (22) | |
| | | | Low risk (113) | OS (100) | | DFS (95) | |
| | | | | Deceased (13) | | Recurred (79) | |
| 8 | | Liver | High risk
(137) | OS (94) |
0.005112a | DFS (52) |
0.007663a |
| | | CO=8.11±0.64 | | Deceased (43) | | Recurred (85) | |
| | | | Low risk (141) | OS (117) | | DFS (76) | |
| | | | | Deceased (24) | | Recurred (65) | |
| 9 | TOP2A | Cervical | High risk
(104) | OS (91) | 0.784725 | DFS (87) | 0.708667 |
| | | CO=130.7±3.68 | | Deceased (13) | | Recurred (17) | |
| | | | Low risk (115) | OS (102) | | DFS (94) | |
| | | | | Deceased (13) | | Recurred (21) | |
| 10 | | Liver | High (141) | OS (99) | 0.098857 | DFS (54) |
0.015001a |
| | | CO=68.27±6.49 | | Deceased (42) | | Recurred (87) | |
| | | | Low (120) | OS-95 | | DFS (64) | |
| | | | | Deceased (25) | | Recurred (56) | |
In addition, ROC analysis was performed for these
four genes (MCM3, NDC80, CDC45 and
ORC1), which were found to exert a significant effect on
both the OS and DFS of patients with liver cancer. The results
suggested that out of these genes, only CDC45 (Fig. S1A) and ORC1 (Fig. S1B) had comparatively high area
under the curve (AUC) values (0.57) compared with NDC80
(Fig. S1C) and MCM3 (AUC
<0.43; Fig. S1D).
Molecular models and molecular docking
of common DEGs with inhibitors
AlphaFold was used [AlphaFold Protein Structure
Database (ebi.ac.uk)] to produce the tertiary
structure of DEGs expressed in tumor samples through in
silico projection processing of their molecular structures.
Using the CMap online portal, we identified 48 inhibitors that
inhibit common genes. However, the structures of 13 inhibitors were
not available in the PubChem database. No inhibitors of
MCM2, MCM3, CDC45 and ORC1 were found.
The binding affinities of the 35 inhibitors to the remaining common
genes (NDC80, DLGAP5 and TOP2A) are presented
in Table SV. The inhibitor
(cytochalasin B) with the highest binding affinity, was selected
for MD simulations.
MD simulations
The RMSD of the backbone atoms was used to analyze
the stability of the complex, as shown in Fig. 8A. The average RMSD of the complex
(NDC80 and cytochalasin B) was 1.32±0.25 (8A). Furthermore,
the RMSF for the NDC80-cytochalasin B complex were
calculated. In the RMSF plot, residues of NDC80 and the
cytochalasin B complex were found to have fewer fluctuations
(average, 0.81±0.12 SEM) (Fig.
8B). Protein compactness was assessed by plotting the radius of
gyration (Rg) (Fig. 8C). The Rg
plot revealed the stability and compactness of the docked complex.
The average value of the radius of gyration was 28.78±0.20 A˚ and
it remained stable after 20 nsec, indicating the stability of the
3D protein structure during MD simulation. The results also
revealed that the electrostatic (-888±90.72) and van der Waals
energies (-347±16.96) of the docked complex were negative, which
ultimately resulted in a negative binding energy (-122±6.25
kcal/mol). These results suggest that cytochalasin B can be used as
a therapeutic drug in cancers where a high expression of
NDC80 significantly affects the survival of patients.
Identification of unique biomarkers
based on DEGs in liver, lung, cervical and pancreatic cancers
TCGA data analysis demonstrated that 58.40%
(3393/5809), 41.51% (1263/3042), 67.24% (4037/6003) and 21.95%
(9/41) DEGs were only present in the lungs, cervical, liver and
pancreatic tumors, respectively, and were absent in other tumors.
Furthermore, these genes were filtered based on their impact on the
survival of patients with cancer. It was observed that only three,
eight and 23 genes significantly affected the survival of patients
with lung, cervical and liver tumors, respectively (Table IV). The impact of these genes on
the survival of patients with these cancers differed (Table SVI). These genes were then
validated using c-BioPortal data of the specific cancer type. In
lung cancer, none of the three genes had a significant effect on OS
or DFS. In cervical cancer, out of the eight genes, the data for
only six genes were available in the c-BioPortal database [Fanconi
anemia complementation group M (FANCM), ubiquitin-specific
peptidase 18 (USP18), colony stimulating factor 2
(CSF2), DnaJ heat shock protein family (Hsp40) member C9
(DNAJC9), fatty acid synthase (FASN) and Runt-related
transcription factor 1 (RUNX1]. Notably, all patients with
cervical cancer exhibited a high expression of CSF2 on the basis of
cut-off value. Of the remaining five genes, only FASN was
found to exert a significant impact on both the OS and DFS of
patients with cervical cancer (Table
V). Similarly, the expression of cytochrome P450 family 2
subfamily C member 9 (CYP2C9) and acyl-CoA dehydrogenase
short chain (ACADS) in patients with liver cancer had a
significant effect on both the OS and DFS.
 | Table IVUnique genes significantly impacting
the survival of lung, cervical and liver cancer. |
Table IV
Unique genes significantly impacting
the survival of lung, cervical and liver cancer.
| Lung | Cervical | Liver |
|---|
| ALDOC, FN1,
SNPRG | CSF2, SCD, DNAJC9,
FANCM, USP18, FASN, RUNX1, NASP | RPL38, RPS21,
CYP2C9, RPL8, CYP2C8, NDUFS3, MT-CO1, MT-CYB, ABAT, ACADS, AKR1D1,
AGXT2, ACAA1, SHARPIN, RPSA, RBCK1, RIPK2, RPS5, TRAF5, APOC3,
ECHS1, SARDH, HSD17B8 |
 | Table VSurvival analysis of the unique DEGs
identified in lung, cervical and liver cancer from the cBioPortal
database. |
Table V
Survival analysis of the unique DEGs
identified in lung, cervical and liver cancer from the cBioPortal
database.
| SN | Cancer type | Total no. of
patients | Gene symbol | Risk status (no. of
patients) | Survival/deceased
status (no. of patients) | P-value | Disease free
survival/recurrence status (no. of patients) | P-value |
|---|
| 1 | Lung | 133 | ALDOC | High risk (37) | OS-(29) | 0.860901 | DFS (27) | 0.839395 |
| | | | | | Deceased (8) | | Recurred (10) | |
| | | | | Low risk (40) | OS-(32) | | DFS (30) | |
| | | | | | Deceased (8) | | Recurred (10) | |
| 2 | | | SNRPG | High risk (43) | OS (34) | 0.617942 | DFS (32) | 0.858053 |
| | | | | | Deceased (9) | | Recurred (11) | |
| | | | | Low risk (35) | OS (26) | | DFS (32) | |
| | | | | | Deceased (9) | | Recurred (12) | |
| 3 | | | FN1 | High risk (9) | OS (6) | 0.701063 | DFS (6) | 0.856395 |
| | | | | | Deceased (3) | | Recurred (3) | |
| | | | | Low risk (19) | OS (14) | | DFS (12) | |
| | | | | | Deceased (5) | | Recurred (7) | |
| 4 | Cervical | 304 | FANCM | High risk
(108) | OS (98) | 0.003825 | DFS (91) | 0.923519 |
| | | | | | Deceased (10) | | Recurred (17) | |
| | | | | Low risk (111) | OS (98) | | DFS (93) | |
| | | | | | Deceased (30) | | Recurred (18) | |
| 5 | | | USP18 | High risk
(104) | OS (89) | 0.521714 | DFS (82) | 0.414601 |
| | | | | | Deceased (15) | | Recurred (22) | |
| | | | | Low risk (113) | OS (100) | | DFS (94) | |
| | | | | | Deceased (13) | | Recurred (19) | |
| 6 | | | DNAJC9 | High risk
(109) | OS (93) | 0.582174 | DFS (83) | 0.171057 |
| | | | | | Deceased (16) | | Recurred (26) | |
| | | | | Low risk (115) | OS (101) | | DFS (96) | |
| | | | | | Deceased (14) | | Recurred (19) | |
| 7 | | | FASN | High risk
(103) | OS (85) |
0.016737a | DFS (77) |
0.007345a |
| | | | | | Deceased (18) | | Recurred (26) | |
| | | | | Low risk (115) | OS (107) | | DFS (102) | |
| | | | | | Deceased (8) | | Recurred (13) | |
| 8 | | | RUNX1 | High risk
(113) | OS (97) | 0.179014 | DFS (86) | 0.029777 |
| | | | | | Deceased (16) | | Recurred (27) | |
| | | | | Low risk (117) | OS (107) | | DFS (102) | |
| | | | | | Deceased (10) | | Recurred (15) | |
| 9 | Liver | 371 | RPL38 | High risk
(127) | OS (92) | 0.635329 | DFS (53) | 0.519766 |
| | | | | | Deceased (35) | | Recurred (74) | |
| | | | | Low risk (100) | OS (73) | | DFS (46) | |
| | | | | | Deceased (24) | | Recurred (54) | |
| 10 | | | RPS21 | High risk
(119) | OS (84) | 0.226981 | DFS (47) | 0.247757 |
| | | | | | Deceased (35) | | Recurred (72) | |
| | | | | Low risk -(95) | OS (74) | | DFS (45) | |
| | | | | | Deceased (21) | | Recurred (50) | |
| 11 | | | RPL8 | High risk
(117) | OS (90) | 0.989298 | DFS (54) | 0.864929 |
| | | | | | Deceased (27) | | Recurred (63) | |
| | | | | Low risk (100) | OS (77) | | DFS (45) | |
| | | | | | Deceased (23) | | Recurred (55) | |
| 12 | | | CYP2C9 | High risk 116) | OS (78) |
0.013493a | DFS (41) |
0.025993a |
| | | | | | Deceased (38) | | Recurred (75) | |
| | | | | Low risk (149) | OS (120) | | DFS (73) | |
| | | | | | Deceased (29) | | Recurred (76) | |
| 13 | | | CYP2C8 | High risk
(130) | OS (92) | 0.133773 | DFS (51) | 0.104747 |
| | | | | | Deceased (38) | | Recurred (79) | |
| | | | | Low risk (145) | OS (114) | | DFS (71) | |
| | | | | | Deceased (31) | | Recurred (74) | |
| 14 | | | NDUFS3 | High risk
(136) | OS (101) | 0.672898 | DFS (60) | 0.90272 |
| | | | | | Deceased (35) | | Recurred (76) | |
| | | | | Low risk (136) | OS (104) | | DFS (59) | |
| | | | | | Deceased (32) | | Recurred (77) | |
| 15 | | | ABAT | High risk
(125) | OS (85) | 0.038715 | DFS (47) | 0.050122 |
| | | | | | Deceased (40) | | Recurred (78) | |
| | | | | Low risk (152) | OS (120) | | DFS (75) | |
| | | | | | Deceased (32) | | Recurred (77) | |
| 16 | | | ACADS | High risk
(127) | OS (85) |
0.014895a | DFS (49) |
0.007165a |
| | | | | | Deceased (42) | | Recurred (78) | |
| | | | | Low risk (136) | OS (109) | | DFS (75) | |
| | | | | | Deceased (27) | | Recurred (61) | |
| 17 | | | AKR1D1 | High risk (95) | OS (66) | 0.125758 | DFS (36) | 0.071368 |
| | | | | | Deceased (29) | | Recurred (59) | |
| | | | | Low risk (131) | OS (102) | | DFS (65) | |
| | | | | | Deceased (28) | | Recurred (65) | |
| 18 | | | AGXT2 | High risk
(131) | OS (95) | 0.187204 | DFS (55) | 0.188999 |
| | | | | | Deceased (36) | | Recurred (76) | |
| | | | | Low risk (136) | OS (108) | | DFS (68) | |
| | | | | | Deceased (28) | | Recurred (68) | |
| 19 | | | ACAA1 | High risk
(139) | OS (99) | 0.121543 | DFS (53) | 0.024599 |
| | | | | | Deceased (40) | | Recurred (86) | |
| | | | | Low risk-(144) | OS (114) | | DFS (77) | |
| | | | | | Deceased (30) | | Recurred (76) | |
| 20 | | | SHARPIN | High risk
-(132) | OS (103) | 0.777398 | DFS (61) | 0.786172 |
| | | | | | Deceased (28) | | Recurred (70) | |
| | | | | Low risk
-(132) | OS (98) | | DFS (57) | |
| | | | | | Deceased (29) | | Recurred (70) | |
| 21 | | | RPSA | High risk
(122) | OS (86) | 0.165274 | DFS (49) | 0.218615 |
| | | | | | Deceased (37) | | Recurred (74) | |
| | | | | Low risk (113) | OS (88) | | DFS (54) | |
| | | | | | Deceased-(25) | | Recurred (59) | |
| 22 | | | RBCK1 | High risk
(128) | OS (94) | 0.801019 | DFS (65) | 0.354948 |
| | | | | | Deceased (34) | | Recurred (63) | |
| | | | | Low risk (131) | OS (98) | | DFS (59) | |
| | | | | | Deceased (33) | | Recurred (72) | |
| 23 | | | RIPK2 | High risk
(127) | OS (91) | 0.265504 | DFS (53) | 0.08948 |
| | | | | | Deceased (36) | | Recurred (74) | |
| | | | | Low risk (130) | OS (101) | | DFS (68) | |
| | | | | | Deceased (29) | | Recurred (62) | |
| 24 | | | RPS5 | High risk
(114) | OS (78) | 0.136647 | DFS (45) | 0.348421 |
| | | | | | Deceased (36) | | Recurred (69) | |
| | | | | Low risk (114) | OS (88) | | DFS (52) | |
| | | | | | Deceased (26) | | Recurred (62) | |
| 25 | | | TRAF5 | High risk
(139) | OS (100) | 0.36903 | DFS (55) | 0.161267 |
| | | | | | Deceased (39) | | Recurred (84) | |
| | | | | Low risk (129) | OS (99) | | DFS (62) | |
| | | | | | Deceased (30) | | Recurred (67) | |
| 26 | | | APOC3 | High risk
(121) | OS (87) | 0.215545 | DFS (49) | 0.07654 |
| | | | | | Deceased (34) | | Recurred (72) | |
| | | | | Low risk (144) | OS (113) | | DFS (74) | |
| | | | | | Deceased (31) | | Recurred (70) | |
| 27 | | | ECHS1 | High risk
(128) | OS (97) | 0.630766 | DFS (54) | 0.20164 |
| | | | | | Deceased (31) | | Recurred (74) | |
| | | | | Low risk (138) | OS (108) | | DFS (69) | |
| | | | | | Deceased (30) | | Recurred (69) | |
| 28 | | | SARDH | High risk
(133) | OS (92) | 0.071347 | DFS (52) | 0.046716 |
| | | | | | Deceased (41) | | Recurred (81) | |
| | | | | Low risk (128) | OS (111) | | DFS (72) | |
| | | | | | Deceased (30) | | Recurred (69) | |
| 29 | | | HSD17B8 | High risk
(127) | OS (91) | 0.436891 | DFS (48) | 0.06633 |
| | | | | | Deceased (36) | | Recurred (79) | |
| | | | | Low risk (107) | OS (107) | | DFS (69) | |
| | | | | | Deceased (34) | | Recurred (72) | |
Recurrence analysis
Recurrence analysis was performed only for
FASN in cervical cancer, and for CYP2C9 and
ACADS in patients with liver cancer. For the recurrence
analysis, subjects with no follow-up data and recurrence status
were excluded. The AUC value of FASN was 0.57, and the AUC
value of CYP2C9 and ACADS was 0.56 and 0.59,
respectively. Due to the low number of patients with stage II, III
and IV diseases, both the patients with cervical and liver cancer
were divided into two groups. Group I included all patients with
stage I cancer (low risk), and group II included all other
remaining patients with stage II, III or IV cancer (high risk). ROC
analysis was conducted using OriginPro statistical software, and
the AUC value was calculated. A total of 66 patients with cervical
cancer had AJCC stage I tumors, while 52 patients were classified
as either stage II, III, or stage IV. Combining the gene expression
of FASN with tumor stage in cervical cancer slightly
decreased the AUC value (0.56). These results suggest that tumor
stage does not play a significant role in the recurrence of
cervical cancer. A total of 265 patients with liver cancer had
stage I tumors, while 69 patients had stage II, III, or IV tumors.
Combining CYP2C9 and ACADS expression with TNM stage
enhanced the AUC value to 0.71 in liver cancer, suggesting that the
expression of these genes together with tumor stage can be used as
prognostic markers for liver cancer recurrence (39).
Discussion
Surgery, radiotherapy and chemotherapy are the
standard treatments for the types of cancer examined in the present
study. However, in recent years, treatments targeting specific
genes or proteins have also been used for cancer treatment,
especially in cases of metastatic disease. For example, monoclonal
antibodies targeting specific receptors, such as bevacizumab
(avastin), are used in non-small cell lung carcinoma and cervical
cancer (40), whereas nivolumab,
pembrolizumab, ramucirumab, nivolumab/ipilimumab,
atezolizumab/bevacizumab, and tremelimumab/durvalumab are used for
hepatocellular carcinoma (41).
None of the monoclonal antibodies have been approved for pancreatic
cancer. However, no significant difference in the OS of patients
has been found in recent years compared to previous years. For
example, it was shown that the OS of avastin-treated patients with
lung cancer was 18.5 months in 2014(42) and 16.3 months in 2021(43). Hence, the management of recurrence
and OS remains a challenge in the field of cancer therapeutics.
Therefore, it is critical to identify survival-related biomarkers
that can be subsequently used to separate patients into high- or
low-risk groups to enhance treatment efficacy.
The present study focused on genes affecting the OS
and DFS of patients with four types of cancer. A dataset from TCGA
for these types of cancer was used to identify the DEGs. Liver and
pancreatic cancers had the largest and smallest number of DEGs,
respectively. The present study identified the top 70 DEGs that
were common in the three types of cancer, excluding pancreatic
cancer. Of these 70 common genes, only a few genes in each cancer
type had a significant impact on OS. For instance, only two genes
(DLGAP4 and NDC80) in liver and lung cancer and five genes
(MCM2, MCM3, ORC1, CDC45 and
TOP2A) in liver and cervical cancer were common, which
significantly affected the OS of patients with cancer. These common
genes have also been reported previously. For example, DLGAP4 and
NDC80 act as effective prognostic markers for liver and lung
cancers, and are closely related to tumor progression and
metastasis (44-48).
DLGAP5 is a microtubule-associated protein that plays an
oncogenic role in tumorigenesis, including lung cancer and
hepatocellular carcinoma (49).
For instance, it has been shown that the high expression of DLGAL5
is associated with a poor response to immunotherapy and promotes
proliferation via cell cycle-related pathways in lung cancer, such
as p53 and DNA replication (46).
Similarly, the OS of patients with hepatocellular carcinoma
exhibiting a high expression of DLGAP5 has been found to be low
(45). It has also been shown that
PRC1 and DLGAP5 are co-expressed in proliferative
T-cells that actively participate in immune escape by liver cancer
cells and thereby are independent risk factors for poor survival
(50). These studies suggest that
DLGAP5 plays a key role in suppressing the immune response to
immunotherapy, and thus, inhibitors targeting DLGAP5 along with
immunotherapy may enhance the immune response to immunotherapy.
NDC80 (also known as Hec1), a fundamental component of the
outer kinetochore and mitotic regulator, is of particular
relevance, as it has a demonstrable link with cancer progression
(47). For example, a high
NDC80 expression has been found to be associated with the
poor survival of patients with hepatocellular carcinoma and liver
cancer cell lines (51). The
silencing of NDC80 has been shown to significantly reduce
hepatic cancer cell proliferation, colony formation, increased
apoptosis, cell cycle arrest at the S-phase and hepatitis B
virus-related hepatocellular carcinoma (47,52).
Similarly, the increased expression of NDC80 has been shown
to induce therapeutic radioresistance by promoting autophagy in
lung cancer (48). These studies
suggest the significance of NDC80 silencing or inhibition in
reducing cancer proliferation and therapeutic resistance. The
results of the present study also suggest that the expression of
DLGAP5 and NDC80 are significantly increased in both
liver and lung cancer. However, the results of GEPIA2 analysis
demonstrated that the high expression of these two genes enhanced
the survival of patients with lung cancer, while reducing the OS of
patients with liver cancer patients. The opposite role of these
genes in lung and liver cancer may be due to various factors, such
as intracellular location of genes and associated mechanisms, which
might affect drug sensitivity and thus, survival in these cancer
patients. In the present study, it was observed that cytochalasin B
was a potent NDC80 inhibitor that can be used to enhance the
survival of patients with cancer, where a high expression
contributes to poor survival.
The expression of MCM2, MCM3,
ORC1, CDC45 and TOP2A has also been shown to
be significantly increased in liver and cervical cancers (37,53-60).
These genes are involved in DNA replication and cell cycle
(58,60-63).
It was found that all five genes were common in liver and cervical
cancer, and the expression of these genes significantly affected
the survival of both cancers. The results of the GEPIA2 analysis
demonstrated the opposite effect on survival in patients with liver
and cervical cancer. For example, the OS of patients with cervical
cancer exhibiting high transcript per million of these genes was
high, whereas the opposite results were observed for patients with
liver cancer. Additionally, all seven common genes only affected
DFS in the case of liver cancer. Several anticancer drugs, such as
pembrolizumab, entrectinib and larotrectinib have been used in
clinical settings to treat tumors with common molecular features
(64). Furthermore,
pharmacogenomics-guided therapeutic decisions help enhance the
precision of cancer therapy and improve the outcomes of patients in
clinical practice. However, the present study suggests that
although the common genes may be used for the diagnosis or
prognosis of patients with cancer, they should be used to identify
the risk of cancer or for therapeutic intervention only after
verifying their role in cancer survival. This is due to the reason
that although the genes could be differentially expressed at the
significant level, they may not significantly affect survival.
Moreover, the effect of specific genes on survival may vary
depending on cancer type. After validating TCGA data with the data
from the c-BioPortal database, it was suggested that only two
genes, CDC45 and ORC1, can be used for the prognosis
of patients with liver cancer progression due to their effects on
OS and DFS, and a comparatively high AUC value.
The present study also identified three, eight and
23 unique DEGs in lung, cervical and liver tumors, respectively,
which had a significant effect on survival. These genes are unique
and are only present in specific types of cancer. To determine
whether these DEGs could be used for the survival prediction of
these cancers, the data were validated using the cBioPortal
database. FASN has been found to be associated with several
hallmarks of cancer and to promote cell proliferation through
membrane biosynthesis. A high gene expression has been observed in
advanced stages of cervical cancer, and it has been shown that the
use of a FASN inhibitor arrests the cell cycle and autophagy
in cervical cancer (65). The
present study also observed a high expression of FASN and
its association with both OS and DFS. However, due to its low ROC
value, this gene cannot be used for the diagnosis or prognosis of
patients with cervical cancer. The results support the protein
atlas data, where the expression of FASN was not considered
a prognostic biomarker in cervical cancer. CYP2C9 is an enzyme that
is involved in drug metabolism. Hypoxia-inducible CYP2C9 enhances
drug resistance in liver cancer stem cells (66). ACADS encodes key metabolic
enzymes that are associated with the metabolic reactions involved
in liver cancer proliferation and metastasis and is highly likely
that ACADS could be a novel therapeutic target in liver
cancer (67). According to the
protein atlas, both CYP2C9 and ACADS can be used as
prognostic markers for liver cancer. The present study also
observed that CYP2C9 and ACADS in liver cancer
significantly affected the OS and DFS of patients. Previous studies
have demonstrated that tumor stage and a combination of biomarker
panels can better predict the prognosis of recurrence or survival
(68,69). Therefore, the present study
analyzed the effects of these genes together with the AJCC
pathological staging on cancer recurrence. It was suggested that
the expression of CYP2C9 and ACADS, together with
tumor stage, may be used as prognostic markers for liver cancer
recurrence due to their comparatively higher ROC values (vs. other
genes). However, furthers investigation of the site identification
of DEGs are required, which may lead to the discovery of specific
targeted drugs and more precise targeted therapy.
In conclusion, the present study identified two
common genes in liver and lung cancers, and five common genes in
liver and cervical cancers. However, only two common genes,
CDC45 and ORC1, can be used for the prognosis of
liver cancer owing to their effects on OS, DFS and comparatively
higher AUC values. Similarly, three, eight and 23 genes were unique
to lung, cervical and liver tumors, respectively. However, when
these data were validated using the cBioPortal dataset, it was
identified that only the CYP2C9 and ACADS genes,
along with tumor stage, could be used for the prognosis or
diagnosis of recurrence in liver cancer when compared with lung and
cervical cancer. However, the present study did not measure the
expression of DEGs at translational levels in either of the cancers
examined. Thus, this is a limitation of the present study. In
future studies, the authors aim to investigate the expression of
common genes at both the transcriptional and translational labels
in all the types of cancer examined herein.
It is suggested that the identification of
therapeutic targets in future studies should focus on DEGs and
their impact on survival, cancer type and stage. The reason for
this is that the genes can be differentially expressed at
significant levels, but they may not significantly affect survival
or recurrence.
Supplementary Material
AUC curve of (A) CDC45, (B)
ORC1, (C) NDC80, and (D) MCM3 in samples from
patients with liver cancer. AUC, area under the curve;
CDC45, cell division cycle 45; ORC1, origin
recognition complex subunit 1; NDC80, human NDC80
kinetochore complex component; MCM3, human minichromosome
maintenance complex component 3.
AUC curve of (A) FASN alone and
with tumor stages in cervical cancer (B) CYP2C9, ACADS
alone and with tumor stages in liver cancer. AUC, area under the
curve; CYP2C9, cytochrome P450 family 2 subfamily C
member 9; ACADS, acyl-CoA dehydrogenase short
chain.
Significant differentially expressed
genes in lung, liver, cervical and pancreatic cancer.
Top 70 common hub genes that are
expressed in lung, cervical and liver cancer.
Go function and pathway enrichment
analysis of seven common genes in lung, cervical and liver
cancer.
KEGG pathways analysis of seven common
genes in lung, cervical, and liver cancer.
Binding affinity of inhibitors with
the common genes present in at least two types of cancer.
Effects of the high expression of
unique DEGs on the survival of patients with lung, cervical and
liver cancer.
Acknowledgements
Not applicable.
Funding
Funding: Intramural funding was received from Parul University
for the present study (grant no. CR4D/IMSL/084).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
PS, NK and DB performed all the analyses. PS, MP
and SKP performed the experimental work on the patient samples. RG
was responsible for the conceptualization of the study and for the
drafting of the manuscript. PS and RG confirm the authenticity of
all the raw data. All the authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Parul University (PUIECHR/PIMSR/00/081734/5307). All
methods were performed according to the relevant guidelines and
regulations provided by the Ethics Committee of Parul University.
Informed consent was obtained from all the participants to
participate in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization (WHO): Global
health estimates 2020: Deaths by cause, age, sex, by country and by
region, 2000-2019. WHO, Geneva, 2019.
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang Z, Guo M, Ai X, Cheng J, Huang Z, Li
X and Chen Y: Identification of potential diagnostic and prognostic
biomarkers for colorectal cancer based on GEO and TCGA databases.
Front Genet. 11(602922)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sathishkumar K, Chaturvedi M, Das P,
Stephen S and Mathur P: Cancer incidence estimates for 2022 &
projection for 2025: Result from national cancer registry
programme, India. Indian J Med Res. 156:598–607. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang PW, Chen L, Huang T, Zhang N, Kong
XY and Cai YD: Classifying ten types of major cancers based on
reverse phase protein array profiles. PLoS One.
10(e0123147)2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gao S, Gang J, Yu M, Xin G and Tan H:
Computational analysis for identification of early diagnostic
biomarkers and prognostic biomarkers of liver cancer based on GEO
and TCGA databases and studies on pathways and biological functions
affecting the survival time of liver cancer. BMC Cancer.
21(791)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu P, Li H, Liao C, Tang Y, Li M, Wang Z,
Wu Q and Zhou Y: Identification of key genes and biological
pathways in Chinese lung cancer population using bioinformatics
analysis. PeerJ. 10(e12731)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yang HJ, Xue JM, Li J, Wan LH and Zhu YX:
Identification of key genes and pathways of diagnosis and prognosis
in cervical cancer by bioinformatics analysis. Mol Genet Genomic
Med. 8(e1200)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xie LY, Huang HY, Fang T, Liang JY, Hao
YL, Zhang XJ, Xie YX, Wang C, Tan YH and Zeng L: A prognostic
survival model of pancreatic adenocarcinoma based on
metabolism-related gene expression. Front Genet.
13(804190)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yin LK, Yuan HY, Liu JJ, Xu XL, Wang W,
Bai XY and Wang P: Identification of survival-associated biomarkers
based on three datasets by bioinformatics analysis in gastric
cancer. World J Clin Cases. 11:4763–4787. 2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Patel KD, Vora HH and Patel PS:
Transcriptional biomarkers in oral cancer: An integrative analysis
and the cancer genome atlas validation. Asian Pac J Cancer Prev.
22:371–380. 2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Veyel D, Wenger K, Broermann A,
Bretschneider T, Luippold AH, Krawczyk B, Rist W and Simon E:
Biomarker discovery for chronic liver diseases by multi-omics-a
preclinical case study. Sci Rep. 10(1314)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang F, Su Q and Li C: Identidication of
novel biomarkers in non-small cell lung cancer using machine
learning. Sci Rep. 12(16693)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
O'Neill RS and Stoita A: Biomarkers in the
diagnosis of pancreatic cancer: Are we closer to finding the golden
ticket? World J Gastroenterol. 27:4045–4087. 2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kori M and Arga KY: Potential biomarkers
and therapeutic targets in cervical cancer: Insights from the
meta-analysis of transcriptomics data within network biomedicine
perspective. PLoS One. 13(e0200717)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sun B, Li Y, Zhou Y, Ng TK, Zhao C, Gan Q,
Gu X and Xiang J: Circulating exosomal CPNE3 as a diagnostic and
prognostic biomarker for colorectal cancer. J Cell Physiol.
234:1416–1425. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Han J, Chen M, Wang Y, Gong B, Zhuang T,
Liang L and Qiao H: Identification of biomarkers based on
differentially expressed genes in papillary thyroid carcinoma. Sci
Rep. 8(9912)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Neary B, Zhou J and Qiu P: Identifying
gene expression patterns associated with drug-specific survival in
cancer patients. Sci Rep. 11(5004)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res.
44(e71)2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261.
2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560.
2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liao Y, Wang J, Jaehnig EJ, Shi Z and
Zhang B: WebGestalt 2019: Gene set analysis toolkit with revamped
UIs and APIs. Nucleic Acids Res. 47:W199–W205. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Picot E, Jouan R, Bach E, Murcier G and
Borgnat F: Oral metastasis of pulmonary adenocarcinoma: Diagnosis
and treatment. J Oral Med Oral Surg. 25:9–12. 2019.
|
|
27
|
Yu D, Guo R and Zhu L: The risk and
prognostic factors for lung metastases in oral squamous cell
carcinoma: A population-based analysis of the SEER database. J
Stomatol Oral Maxillofac Surg. 125(101713)2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hong JH, Lee K, Kim J and Ahn KM:
Prognosis of hepatocellular carcinoma metastasizing to the oral
cavity. Maxillofac Plast Reconstr Surg. 43(9)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Misra SR, Shankar YU, Rastogi V and
Maragathavalli G: Metastatic hepatocellular carcinoma in the
maxilla and mandible, an extremely rare presentation. Contemp Clin
Dent. 6:S117–S121. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pires RS, Sagarra R, Corrêa ME, Pereira
CM, Vargas PA and Lopes MA: Oral metastasis of a hepatocellular
carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod.
97:359–368. 2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yin L, Gupta R, Vaught L, Grosche A,
Okunieff P and Vidyasagar S: An amino acid-based oral rehydration
solution (AA-ORS) enhanced intestinal epithelial proliferation in
mice exposed to radiation. Sci Rep. 6(37220)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Youens-Clark K, Faga B, Yap IV, Stein L
and Ware D: CMap 1.01: A comparative mapping application for the
internet. Bioinformatics. 25:3040–3042. 2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Dallakyan S and Olson AJ: Small-molecule
library screening by docking with PyRx. Methods Mol Biol.
1263:243–250. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Phillips JC, Hardy DJ, Maia JDC, Stone JE,
Ribeiro JV, Bernardi RC, Buch R, Fiorin G, Hénin J, Jiang W, et al:
Scalable molecular dynamics on CPU and GPU architectures with NAMD.
J Chem Phys. 153(044130)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sun Y, Cheng Z and Liu S: MCM2 in human
cancer: Functions, mechanisms, and clinical significance. Mol Med.
28(128)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10(e0116774)2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
von Eyben FE, Madsen EL, Fritsche H, Suciu
G, Liu F and Amato R: A new prognostic model for testicular germ
cell tumours. APMIS. 111:100–105. 2003.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Aghbash PS, Hemmat N, Fathi H and Baghi
HB: Monoclonal antibodies in cervical malignancy-related HPV. Front
Oncol. 12(904790)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Psilopatis I, Damaskos C, Garmpi A,
Sarantis P, Koustas E, Antoniou EA, Dimitroulis D, Kouraklis G,
Karamouzis MV, Vrettou K, et al: FDA-approved monoclonal antibodies
for unresectable hepatocellular carcinoma: What do we know so far?
Int J Mol Sci. 24(2685)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhou CC, Bai CX, Guan ZZ, Jiang GL, Shi
YK, Wang MZ, Wu YL, Zhang YP and Zhu YZ: Safety and efficacy of
first-line bevacizumab combination therapy in Chinese population
with advanced non-squamous NSCLC: Data of subgroup analyses from
MO19390 (SAiL) study. Clin Transl Oncol. 16:463–468.
2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wan R, Dong X, Chen Q, Yu Y, Yang S, Zhang
X, Zhang G, Pan Y, Sun S, Zhou C, et al: Efficacy and safety of
MIL60 compared with bevacizumab in advanced or recurrent
non-squamous non-small cell lung cancer: A phase 3 randomized,
double-blind study. EClinicalMedicine. 42(101187)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Dong C, Huang S, Sun L, Yao J, Yan J and
Yin X: DLGAP4 acts as an effective prognostic predictor for
hepatocellular carcinoma and is closely related to tumour
progression. Sci Rep. 12(19775)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liao W, Liu W, Yuan Q, Liu X, Ou Y, He S,
Yuan S, Qin L, Chen Q, Nong K, et al: Silencing of DLGAP5 by siRNA
significantly inhibits the proliferation and invasion of
hepatocellular carcinoma cells. PLoS One. 8(e80789)2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Tang X, Zhou H and Liu Y: High expression
of DLGAP5 Indicates poor prognosis and immunotherapy in lung
adenocarcinoma and promotes proliferation through regulation of the
cell cycle. Dis Markers. 2023(9292536)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ju LL, Chen L, Li JH, Wang YF, Lu RJ, Bian
ZL and Shao JG: Effect of NDC80 in human hepatocellular carcinoma.
World J Gastroenterol. 23:3675–3683. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chen X, He Q, Zeng S and Xu Z:
Upregulation of nuclear division cycle 80 contributes to
therapeutic resistance via the promotion of autophagy-related
protein-7-dependent autophagy in lung cancer. Front Pharmacol.
13(985601)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zhang H, Liu Y, Tang S, Qin X, Li L, Zhou
J, Zhang J and Liu B: Knockdown of DLGAP5 suppresses cell
proliferation, induces G(2)/M phase arrest and apoptosis in ovarian
cancer. Exp Ther Med. 22(1245)2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yang CL, He JT, Li NN, Song R, Ni HH,
Huang JT, Liu GQ, Wang JD, Li YK, Zhan GH, et al: Clinical value of
PRC1 and DLGAP5 and immunosuppressive T cells overexpressing them
in HCC based on transcriptome data. ResarchSquare. 2023. doi:
10.21203/rs.3.rs-2616803/v1.
|
|
51
|
Chen X, Li W, Xiao L and Liu L: Nuclear
division cycle 80 complex is associated with malignancy and
predicts poor survival of hepatocellular carcinoma. Int J Clin Exp
Pathol. 12:1233–1247. 2019.PubMed/NCBI
|
|
52
|
Liu B, Yao Z, Hu K, Huang H, Xu S, Wang Q,
Yang Y and Ren J: ShRNA-mediated silencing of the Ndc80 gene
suppress cell proliferation and affected hepatitis B virus-related
hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
40:297–303. 2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Cao T, Yi SJ, Wang LX, Zhao JX, Xiao J,
Xie N, Zeng Z, Han Q, Tang HO, Li YK, et al: Identification of the
DNA replication regulator MCM complex expression and prognostic
significance in hepatic carcinoma. Biomed Res Int.
2020(3574261)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang XK, Wang QQ, Huang JL, Zhang LB, Zhou
X, Liu JQ, Chen ZJ, Liao XW, Huang R, Yang CK, et al: Novel
candidate biomarkers of origin recognition complex 1, 5 and 6 for
survival surveillance in patients with hepatocellular carcinoma. J
Cancer. 11:1869–1882. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yang C, Xie S, Wu Y, Ru G, He X, Pan HY,
Wang S and Tong X: Prognostic implications of cell division cycle
protein 45 expression in hepatocellular carcinoma. PeerJ.
9(e10824)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Cai H, Zhu X, Qian F, Shao B, Zhou Y,
Zhang Y and Chen Z: High expression of TOP2A gene predicted poor
prognosis of hepatocellular carcinoma after radical hepatectomy.
Transl Cancer Res. 9:983–992. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Del Moral-Hernandez O, Hernandez-Sotelo D,
Alarcon-Romero LDC, Mendoza-Catalan MA, Flores-Alfaro E,
Castro-Coronel Y, Ortiz-Ortiz J, Leyva-Vázquez MA, Ortuño-Pineda C,
Castro-Mora W and Illades-Aguiar B: TOP2A/MCM2, p16(INK4a), and
cyclin E1 expression in liquid-based cytology: A biomarkers panel
for progression risk of cervical premalignant lesions. BMC Cancer.
21(39)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
He Z, Wang X, Yang Z, Jiang Y, Li L, Wang
X, Song Z, Wang X, Wan J, Jiang S, et al: Expression and prognosis
of CDC45 in cervical cancer based on the GEO database. PeerJ.
9(e12114)2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Chen X, Xiong D, Ye L, Wang K, Huang L,
Mei S, Wu J, Chen S, Lai X, Zheng L and Wang M: Up-regulated lncRNA
XIST contributes to progression of cervical cancer via regulating
miR-140-5p and ORC1. Cancer Cell Int. 19(45)2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Ma H, Liu Z, Li H, Guo X, Guo S, Qu P and
Wang Y: Bioinformatics analysis reveals MCM3 as an important
prognostic marker in cervical cancer. Comput Math Methods Med.
2021(8494260)2021.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yuan J, Lan H, Huang D, Guo X, Liu C, Liu
S, Zhang P, Cheng Y and Xiao S: Multi-Omics analysis of MCM2 as a
promising biomarker in pan-cancer. Front Cell Dev Biol.
10(852135)2022.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wu L, Chen H and Yang C: Origin
recognition complex subunit 1(ORC1) is a potential biomarker and
therapeutic target in cancer. BMC Med Genomics.
16(243)2023.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wang T, Lu J, Wang R, Cao W and Xu J:
TOP2A promotes proliferation and metastasis of hepatocellular
carcinoma regulated by miR-144-3p. J Cancer. 13:589–601.
2022.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Kyritsis KA, Akrivou MG, Giassafaki LP,
Grigoriadis NG and Vizirianakis IS: Analysis of TCGA data of
differentially expressed EMT-related genes and miRNAs across
various malignancies to identify potential biomarkers. World
Academy Of Sciences Journal. 3(6)2021.
|
|
65
|
Nascimento J, Mariot C, Vianna DRB,
Kliemann LM, Chaves PS, Loda M, Buffon A, Beck RCR and Pilger DA:
Fatty acid synthase as a potential new therapeutic target for
cervical cancer. An Acad Bras Cienc. 94(e20210670)2022.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Myung SJ, Yoon JH and Yu SJ: STAT3 &
Cytochrome P450 2C9: A novel signaling pathway in liver cancer stem
cells. Biomed Pharmacother. 66:612–616. 2012.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Chen D, Feng X, Lv Z, Xu X, Lu Y, Wu W, Wu
H, Liu H, Cao L, Ye S, et al: ACADS acts as a potential methylation
biomarker associated with the proliferation and metastasis of
hepatocellular carcinomas. Aging (Albany NY). 11:8825–8844.
2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Liersch T, Langer C, Ghadimi BM, Kulle B,
Aust DE, Baretton GB, Schwabe W, Häusler P, Becker H and Jakob C:
Lymph node status and TS gene expression are prognostic markers in
stage II/III rectal cancer after neoadjuvant fluorouracil-based
chemoradiotherapy. J Clin Oncol. 24:4062–4068. 2006.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Chen TH, Chiu JY and Shih KH: A 23-gene
prognostic classifier for prediction of recurrence and survival for
Asian breast cancer patients. Biosci Rep.
40(BSR20202794)2020.PubMed/NCBI View Article : Google Scholar
|