1. Introduction
Cancer is a devastating disease distinguished by
increased cell division, ranking among the leading causes of
mortality worldwide. Its development and progression result from a
combination of genetic and epigenetic alterations (1). Simultaneous genomic and epigenetic
alterations lead to the dysregulation of numerous genes, including
the expression or suppression of tumor suppressor genes (2). These modifications are heritable and
have the potential for therapeutic intervention. DNA methylation
and other epigenetic processes have emerged as a rapidly expanding
field of investigation. They govern the critical stages of cell
development, differentiation and responses to environmental
factors, such as dietary variations, chemical exposure, radiation,
hormones and tobacco smoke. DNA methylation involves the addition
of a ‘methyl’ group at the fifth carbon position in cytosine
(3). Glucose-regulated protein 78,
Src, Toll like receptor 7, caveolin-1 and dopamine receptor D2 are
involved in JEV binding and entry into neurons, with these
receptors also known for their roles in carcinogenic activities
(4). Aberrant modifications in DNA
methylation patterns can either initiate cancer or serve as
indicators of malignant tumor growth (5). Hypermethylation involves an excessive
increase in the methylation of adenine and cytosine residues in
DNA. CpG region hypermethylation within tumor suppressor genes
causes transcriptional repression and the loss of their
tumor-suppressing functions, contributing to carcinogenesis
(6).
The hypermethylation of tumor suppressor genes
indicates their inactivation during cancer development. A notable
example is the frequently hypermethylated CpG island found within
the ras association domain family 1A promoter region. The
development of potent anticancer drugs often occurs through natural
compounds, structural alterations of existing natural compounds, or
the production of entirely novel molecules. The pursuit of improved
cytotoxic agents remains pivotal in the development of modern
anticancer drugs. The derivatives of several marine organisms,
plants and microorganisms are considered promising starting points
for enhancing their remedial efficacy through manipulation at the
molecular level, owing to the vast structural diversity and
bioactivity potential of natural chemicals (7). These bioactive anticancer compounds
can impede cancer cell proliferation, reverse DNA hypermethylation
in cancer cells, inhibit metastasis and halt the cell cycle.
Specific compounds, including naturally occurring polyphenols, can
function as hypermethylation agents, effectively counteracting the
epigenetic suppression of tumor suppressor genes. Notably, curcumin
has been discovered for its potential to target and regulate the
expression of numerous tumor suppressor genes (8). In cancers, such as esophageal,
prostate and cervical cancers, genes such as cyclin dependent
kinase inhibitor 2A (CDKN2A, also known as p16INK4a),
O-6-methylguanine-DNA methyltransferase (MGMT), retinoic acid
receptor (RAR)β, PC3 and suppressor of cytokine signaling 1
(SOCS1), which were previously hypermethylated, have been
demethylated through the action of natural compounds, such as
genistein and capsaicin (9). These
compounds demonstrate the potential of natural substances to
reverse epigenetic modifications, offering a promising avenue for
cancer therapy (9). Given the
limitations of current cancer treatments, there is an ultimate need
for novel therapeutic approaches. Natural compounds derived from
plants, with their diverse chemical structures and biological
activities, offer promising potential in cancer therapy. These
compounds have demonstrated the ability to reverse DNA
hypermethylation, inhibit cancer cell proliferation, and reactivate
silenced tumor suppressor genes. Additionally, some natural
substances may serve as biomarkers for early cancer diagnosis. The
present review focuses on the potential of natural compounds as
epigenetic modulators in cancer therapy, particularly their ability
to target and reverse DNA hypermethylation. By elucidating the
mechanisms underlying the anticancer effects of these molecules,
the present review aims to advance the development of more targeted
and effective cancer treatments (2).
2. Promoter hypermethylation
DNA methylation is the addition of a methyl group to
a DNA segment, which is a critical step in epigenetic regulation.
DNA methylation at specific sites typically inhibits the attachment
of proteins responsible for gene interpretation. Demethylation, a
process used to remove methyl groups, can activate genes (6). DNA methylation usually results in the
silencing of genes, whereas demethylation leads to gene activation.
In normal cells, DNA methylation is critical for proper cellular
function and development. However, cancerous cells exhibit abnormal
patterns in DNA methylation. Tumor-suppressor genes often
experience a surge in methylation levels, leading to their
silencing, whereas oncogenes, which promote tumor growth, exhibit
reduced methylation levels, resulting in their activation and
uncontrolled tumor growth. The dysregulation of DNA methylation is
pivotal in the development of diseases, such as cancer (10). DNA methyltransferases (DNMTs) are
crucial elements in DNA methylation that facilitate the
transmission of methyl groups to DNA sequences in a
sequence-specific manner. This enzymatic process occurs during DNA
replication and involves the recognition and binding of specific
DNA sequences by DNMTs. DNMTs add methyl groups, often from
S-adenosylmethionine, to cytosine residues, profoundly affecting
gene expression regulation and cellular function (11).
Hypermethylation promotes transcriptional repression
in the promoter region in CpG islands of tumor-suppressor genes,
thus silencing the gene, which is a hallmark of cancer. CpG island
hypermethylation is associated with numerous tumor types and
disrupts vital biological processes, including cell cycle control,
DNA repair, apoptosis and detoxification. Various hematological
disorders, including leukemia, are associated with
hypermethylation. Numerous genes, such as estrogen receptor,
syndecan 4, multidrug resistance 1, calcitonin, cyclin dependent
kinase inhibitor 2B, p21Cip1/Waf1 and others, exhibit
hypermethylation in these malignancies. The hypermethylation of
CASP8, TMS1 and DAPK genes was observed in childhood acute
lymphoblastic leukemia, exhibiting significant differences compared
to healthy controls (12). In lung
cancer, numerous genes display altered DNA methylation patterns,
with RARβ, Ras association domain family member 1A
(RASSF1A), cyclin dependent kinase inhibitor 2A and
APC being frequently hypermethylated (11). Dysregulated DNA methylation, either
hypomethylation or hypermethylation, disrupts the normal
functioning of genes and can lead to the development of various
disorders, including cancer. Hypermethylation, which is observed in
a number of types of cancer, inhibits tumor suppressor genes,
contributing to tumor formation and development. The promoter
hypermethylation of tumor suppressor genes, such as p14, p15, p16,
p21, p27, p57, p53, p73, RARβ2, fragile histidine triad
diadenosine triphosphatase, death associated protein kinase 1
(DAPK), STAT1 and RB1 has been detected in
paired biopsy and serum samples from cervical cancer patients
(13). Abnormal DNA methylation
can result in the development os various disorders, including
cancer. While there is a general decrease in global CpG
methylation, occasional events activate previously repressed
oncogenes. Conversely, CGI hypermethylation in gene promoters is a
significant marker of various types of cancer. One of the prompt
examples of this epigenetic silencing phenomenon was the
identification of hypermethylation in the promoter of
retinoblastoma tumor suppressor gene in individuals with
retinoblastoma (11).
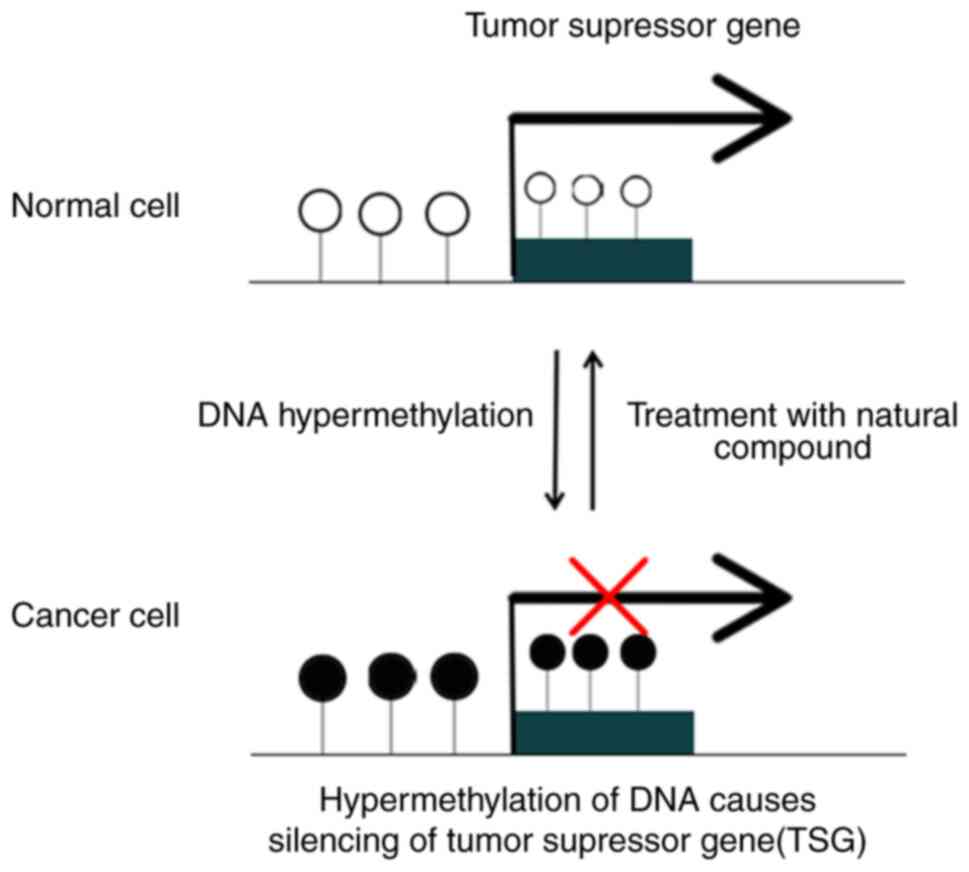
3. Reversal of DNA hypermethylation in
cancer cells
The process of the demethylation or reversal of
methylation removes methyl groups and allows the gene that has been
switched off to resume normal functioning. Research has provided
evidence that the dynamic process of reversible CpG island
methylation, mediated by DNA methyltransferases, exerts a
regulatory influence on the activity of important genes and
transcription factors that participate in the modulation of cell
progression (2) (Fig. 1).
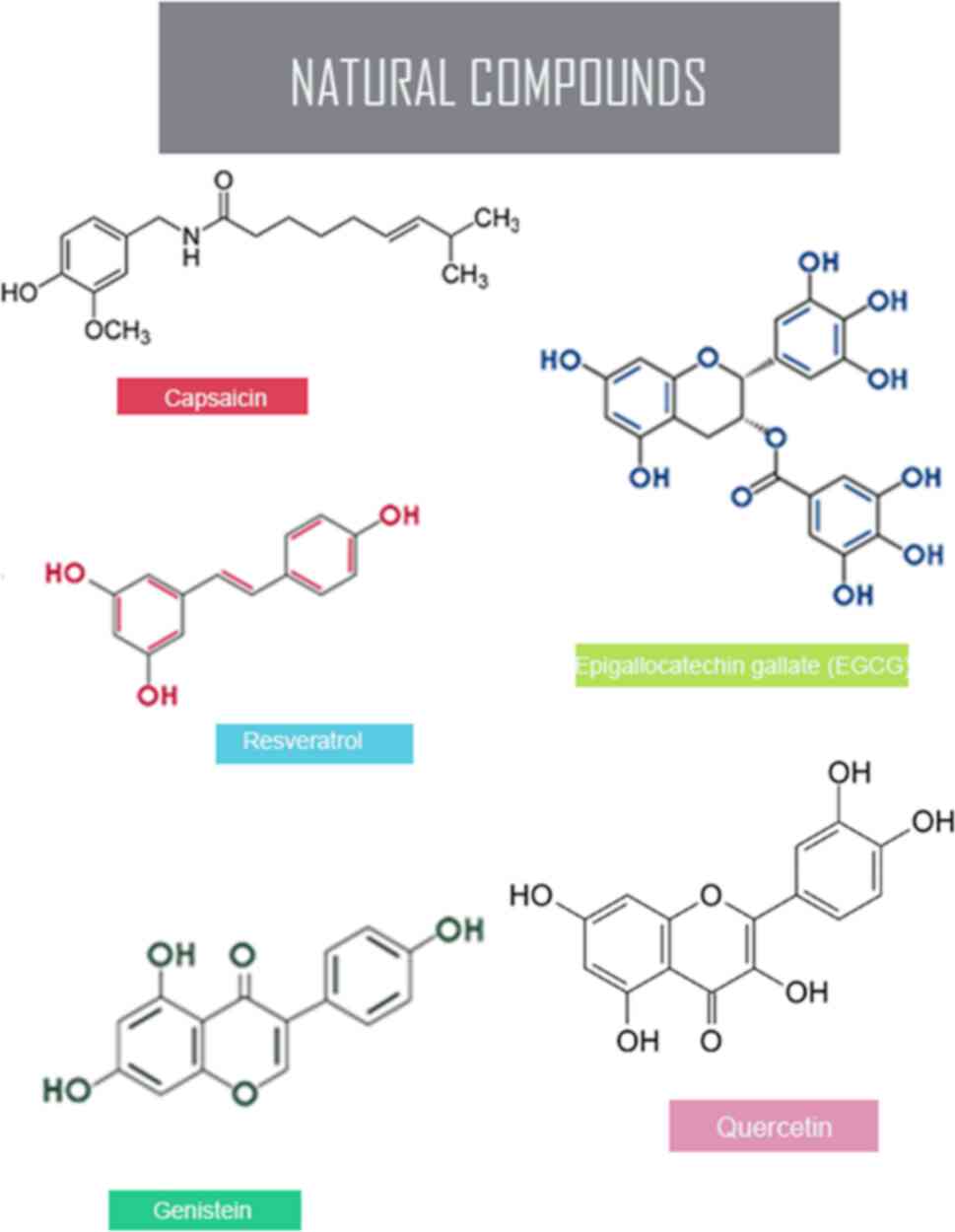
4. Reversal of hypermethylation by natural
compounds
In the pursuit of an effective cancer treatment
strategy, it is imperative to devise pharmaceutical agents capable
of targeting a spectrum of cellular processes encompassing gene
expression, signaling pathways, cellular proliferation,
intercellular associations and the extracellular matrix. An
essential consideration in the selection of these compounds is the
imperative need to ensure their safety profile, particularly with
regard to undesirable side effects such as cytotoxicity (9). Natural compounds have emerged as a
compelling alternative in this context due to their multifaceted
attributes. They can induce apoptosis, stimulate tumor suppressor
genes by resisting DNA methylation modifications, regulate the cell
cycle, activate cell survival proteins and modulate epigenetic
mechanisms. Notably, natural compounds have demonstrated the
capability to rectify aberrant epigenetic changes, such as DNA
hypermethylation, either by reversing hypermethylation or by
attenuating the activity of methyltransferases (2). The reversing ability of epigenetic
alteration has catalyzed the development of a class of therapeutic
agents specifically designed to target epigenetic processes. These
agents, known as epigenetic therapies, are engineered to obstruct
the activation of genes involved in the initiation of
hypermethylation in several cancers (14). The key epigenetic mechanism with
the pivotal role of natural compounds, such as genistein,
resveratrol, capsaicin, quercetin and epigallocatechin gallate
(EGCG) targets the hallmarks of cancer, DNA methylation and the
restraining of tumor suppressor genes to reverse the unusual
expression of these changes and restore the normal methylation
patterns (14) (Fig. 2 and Table I).
 | Table INatural compounds demonstrated to
have the ability to repair the overexpression of epigenetic
modifications, such as DNA methylation, by reversing
hypermethylation or by lowering the activity of methyl
transferase. |
Table I
Natural compounds demonstrated to
have the ability to repair the overexpression of epigenetic
modifications, such as DNA methylation, by reversing
hypermethylation or by lowering the activity of methyl
transferase.
| Compounds | Genes | Cell lines/cancer
type | (Refs.) |
|---|
| Genistein | p16INK4a,
MGMT and RARβ | KYSE 510, PC3and
LNCaP | (16) |
| | BTG3 | LNCaP and PC3
(prostrate cancer) | (17) |
| | GSTP1,
EPHB2, and BRCA1 | PC-3 and DU-145
(prostate cancer) | (18) |
| | APC,
ATM and PTEN | MDA-MB-231 and
MCF-7 (breast cancer) | (21) |
| | MGMT,
RAR, E-cadherin DAPK1 and p21 | HeLa cells
(cervical cancer) | (22) |
| Capsaicin | SOCS1 and
CADM1 | HeLa (cervical
cancer) | (27) |
| Epigallocatechin
gallate (EGCG) | p16INK4a,
RARβ, MGMT and hMLH1 | KYSE510 and PC3
(esophageal cancer) A431(epidermal cancer) | (30) |
| |
p16INK4a | HeLa (cervical
cancer) | (31) |
| | RAR, CDH1
and DAPK1 | KYSE510 (esophageal
cancer) | (29) |
| | RAR, p16,
hMLH1and MGMT | CAL-27(oral
squamous cell cancer) | (29) |
| | RECK | | (32) |
| Resveratrol | PTEN and
RARβ2 | MCF-7 (breast
cancer) | (36) |
| | BRCA-1 | MCF-7 (breast
cancer) | (37) |
| | RUNX3 | B16F10 (malignant
melanoma) | (39) |
| Quercetin | RASSF1A | HeLa (cervical
cancer) | (41,44) |
| | p16INK4a,
CDH1, and IGFBP7 | RKO cell lines
(colorectal cancer) | (42) |
| | MGMT,
GSTP1, APC, SOC51, CDH1 | HeLa (cervical
cancer) | (45) |
Genistein
Genistein is a significant nutraceutical compound
that is naturally present in soybean seeds and serves as a
precursor to phytoalexins found in legumes. Extensive research has
uncovered numerous potential health benefits associated with
dietary genistein (15). Moderate
exposure to genistein exerts inhibitory effects on several types of
cancer, such as cervical, colon, esophageal and prostate cancers.
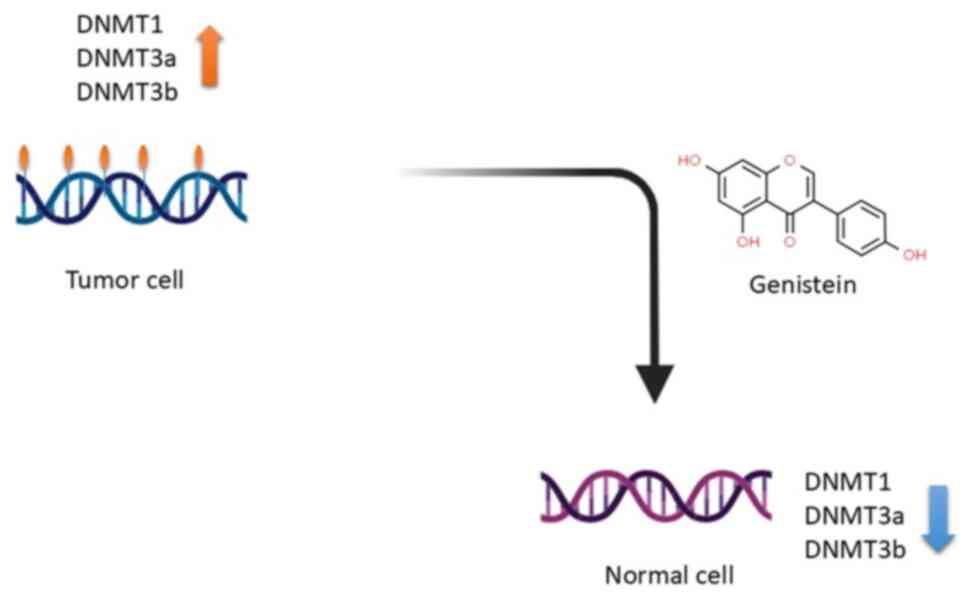
The process through which genistein achieves these effects involves
a reduction in the activity of DNA methyltransferase, consequently
leading to the reversal of DNA hypermethylation (2), as illustrated in Fig. 3. This reversal process leads to
gene reactivation that was initially suppressed due to methylation,
effectively counteracting epigenetic modifications, specifically
DNA hypermethylation (2). In the
esophageal cancer cell line, KYSE 510, genistein reactivates
previously suppressed genes, such as p16INK4a [which is a
cyclin-dependent kinase inhibitor that controls the cell cycle by
preventing retinoblastoma (Rb) protein phosphorylation, thus
blocking progression from the G1 to the S phase], MGMT (a
DNA repair enzyme that removes alkyl groups from the guanine O6
site, preventing replication errors and protecting the genome from
mutations) and RARβ (controls the expression of genes
involved in cell differentiation, apoptosis, and the suppression of
cell growth) (16). This
reactivation is attributed to the suppression of DNMT activity by
genistein, thereby causing the reversal of DNA hypermethylation
followed by the reactivation of methylation-silenced genes. Similar
effects were examined in prostate cancer PC3 and LNCaP cells, where
it altered DNA hypermethylation and activated the RARβ gene
(17).
Furthermore, in prostate cancer, specifically in the
LNCaP and PC3 cell lines, genistein treatment was proven to
transcriptionally reverse the expression of B-cell translocation
gene 3 (BTG3). These cell lines exhibited complete
methylation in the gene promoter region. Genistein treatment led to
an increase in BTG3 mRNA expression and the reactivation of the
BTG3 gene by reducing promoter methylation, primarily
through the inhibition of DNA methyltransferase activity, thus
modulating the hypermethylation pattern (17). The demethylating ability of
genistein has been demonstrated on the promoters of the glutathione
S-transferase Pi 1 (GSTP1), EPH receptor B2 (EPHB2)
and breast cancer susceptibility (BRCA)1 genes in PC-3 and
DU-145 prostate cancer cell lines. BRCA1 and BRCA2 are cellular
proteins involved in DNA repair. They are normally expressed in the
breast, ovaries, prostate and other tissues. Their germline
mutation is the cause of hereditary breast-ovarian cancer
syndromes. Prostate cancer cells proliferate when EPHB2 is
silenced. EPHB2 expression is decreased in prostate cancer tissues
and fully inactivated in the metastatic prostate cancer cell line,
DU145, suggesting a link to disease progression (18). While GSTP1 protects prostate
epithelial cells from DNA damage caused by carcinogens and
oxidative stress, its absence may leave prostate cells vulnerable
to genomic inserts. Using the Methyl Profiler DNA methylation
assay, the methylation levels of the prostate cancer cell lines
were previously quantified. It was demonstrated that the
methylation pattern of GSTP1, BRCA1 and EPHB2
promoters was decreased by genistein. Therefore, genistein may be a
preventive agent against prostate cancer (18).
Based on research studies on neuroblastoma,
extensive promoter hypermethylation has been observed in the
p53 and CHD5 genes. Genetic therapy regulates the
activity of CHD5 and p53 and reduces CHD5
hypermethylation levels. Genistein also functions as a DNMT
inhibitor, which considerably lowers DNMT3b expression. Genistein
functions in tandem with p53 to restrict neuroblastoma
growth, which is potentially mediated by the WNT pathway, by
lowering the methylation of the CHD5 promoter and increasing
CHD5 and p53 expression levels (19). In a study on the development of
breast tumors, the administration of genistein to SHR cells
inhibited cell growth in pre-cancerous cell populations. Notably,
this result has been linked with an upregulation in the activation
of crucial tumor suppressor genes, specifically p16 and p21, while
concurrently with the downregulation of tumor-promoting gene
expression, namely c-MYC and BMI1. These
genes, previously silenced due to aberrant epigenetic
modifications, are well-established contributors to cell cycle
arrest, exerting growth-inhibitory effects across various cancer
cell lines (20).
Genistein has been proven to reduce DNA methylation
in the tumor suppressor genes of the breast cancer MDA-MB-231 and
MCF-7 cell lines. The administration of genistein reactivated
methylation-silenced tumor suppressor genes, such as APC,
ATM and PTEN by direct contact with the DNMT1
enzymatic domain and suppressed DNMT1 activity in cancer cells
(21). In a previous study, it was
shown that genistein suppressed DNMT activity, induced
demethylation and activate methylation-silenced genes. The
treatment of KYSE 510 cells with genistein caused the reversal of
DNA hypermethylation and reactivation of the RARβ,
p16INK4a and MGMT genes (16). Studies on DNMTs in vitro and
in silico have been conducted to investigate the function of
DNMTs on human cervical cancer HeLa cells following treatment with
genistein. The effects of genistein treatment on tumor suppressor
genes, such as MGMT, RAR, E-cadherin DAPK1 and
p21 and the methylation pattern of their promoter regions
were observed. Genistein treatment reduced the enzymatic activity
of DNMTs. The promoter region methylated DNA was reversed, and gene
expression was restored following time-dependent exposure to
genistein (22). Prolonged therapy
with genistein increased expression in the MDA-MB-468 breast cancer
cell line and led to the demethylation of the promoter region.
Furthermore, the RARβ2 cistron in MCF-10A cells was
demethylated by genistein. The ability of genistein to demethylate
hypermethylated genes in MCF-10A, MDA-MB-468 and MCF-7 breast
cancer cells was further examined in these studies, although a
negligible effect was shown in the GSTP1 gene in MDA-MB-468
and MCF-7 breast cancer cells (21).
Genistein has been demonstrated to function as an
epigenetic modulator in a neuroblastoma model by inhibiting tumor
development and the methylation state of the CDH5 promoter region.
DNA methyltransferase activity was inhibited in vivo as a
result of the downregulation of DNMT3(19).
In general, natural compounds have been shown to be
able to reverse hypermethylation and the reactivation of tumor
suppressor genes (23).
Capsaicin
Red chili peppers contain a large amount of the
bioactive chemical capsaicin. In addition to its antifungal and
anti-inflammatory properties, capsaicin has anticancer properties.
The chemopreventive properties of capsaicin are extensive.
Capsaicin also affects the activation of genes involved in cancer
cell survival, the development of new blood vessels, metastasis and
the inhibition of cancer cell development by altering the activity
of enzymes involved in DNA methylation, thereby causing the
reversal of methylation. Capsaicin alters DNA expression by DNMTs.
By inhibiting their activity, capsaicin prevents DNA methylation,
thereby promoting a more standard epigenetic profile (24). Scientific research has shed
significant light on the medicinal potential of capsaicin in
treating the suppression of SOCS1 and cell adhesion molecule
1 (CADM1) in human papillomavirus cervical cancer cell line
expression caused by hypermethylation. Additional investigations
are warranted to explore the fundamental mechanisms and validate
the clinical significance of capsaicin as a specific intervention
for modifying DNA methylation and expression patterns of genes
associated with medical conditions (25). Capsaicin treatment affects the
Par-4 gene and exhibits the ability to lower the levels of
DNA methylation in the prostrate cancel cell line PC-3 gene.
The process of reversing hypermethylation holds significant
potential for reactivating tumor suppressor genes that were
initially suppressed. As a result, the development of cancer cells
can be effectively suppressed (26).
In a previous in silico study, tumor
suppressor in lung cancer 1 (TSLC-1), which has excessive
methylation, became less active and lost its function, leading to
the development of cervical tumors. TSLC1 encodes a cell adhesion
molecule essential for cell-cell interactions and regulating
proliferation. In cervical cancer, TSLC1 is often inactivated by
promoter hypermethylation or deletions, disrupting cell adhesion
and leading to increased proliferation, invasion, and metastasis.
This inactivation contributes to tumor progression and the
aggressive nature of the disease. Capsaicin was tested with TSLC-1
and exhibited a strong binding connection. Detailed analyses
revealed that capsaicin forms bonds with TSLC-1 in specific
ways. Considering this computer-based study, capsaicin also showed
promising results as a potential option to reactivate suppressed
TSLC-1 due to excessive methylation. This could have
implications for the treatment of cervical cancer (27). The DNA methylation of SOCS1
and CADM-1 was previously examined using
methylation-specific PCR on HeLa cells from cervical
adenocarcinoma. CADM1, a cell adhesion molecule vital for tissue
architecture, is downregulated in cervical adenocarcinoma due to
promoter hypermethylation. This disruption in cell adhesion
promotes tumor cell detachment, invasion and metastasis, while
SOCS1 encodes a protein that regulates cytokine signaling to
control cell growth and immune responses. In cervical
adenocarcinoma, its inactivation by promoter hypermethylation leads
to uncontrolled cytokine signaling, promoting tumor progression
through abnormal cell proliferation and immune evasion. The cells
were treated with capsaicin followed by 72 h of incubation and
treatment was repeated for a further 6 days. The findings of in
vitro analyses revealed that hypermethylation plays a
significant role in suppressing CADM1 and SOCS1 expression.
Capsaicin causes the reversal of hypermethylation in CADM1
and SOCS1 expression (25).
The effectiveness of capsaicin in restoring the expression of
CADM1 and SOCS1 was further supported by the evident
changes observed in methylation-specific and unmethylation-specific
patterns during methylation-specific PCR. These alterations
indicated a notable effect on the pattern of methylation associated
with the regulatory regions of CADM1 and SOCS1. The observed
changes in the patterns have proven that capsaicin treatment
affected the methylation status, triggering the restoration of
CADM1 and SOCS1 expression (25).
EGCG
EGCG is a potent bioactive compound belonging to the
family of catechins found in green tea. Its notable antioxidant,
anti-inflammatory and potential anticancer properties have
attracted considerable interest in recent times, particularly in
the context of chemoprevention and epigenetic modifications, such
as the methylation of DNA (2).
Epigenetic alterations such as hypermethylation are involved in the
growth of various types of cancers. Previous findings have shown
that EGCG can repair hypermethylation, providing a viable route for
cancer therapeutics based on epigenetics (28). The capacity of EGCG in
chemoprevention and its marked ability to mitigate
hypermethylation, demonstrate its prospects as an innovative
therapeutic agent (28). The
administration of EGCG to the KYSE 510 esophageal cancer cell line,
for durations spanning from 12 to 144 h, resulted in the
hypermethylation of p16INK4a, RARβ, MGMT and
hMLH1 to be reversed in a manner that was dependent on time
and concentration (29). It was
previously reported that in KYSE 150 esophageal cancer cells, PC-3
cells and HT-29 colon cells, EGCG demethylated the CpG regions and
reactivated methylation suppressed genes, such as RARβ,
MGMT, p16INK4a, glutathione S-transferase and human
mutL homologue 1(29).
Another study demonstrated that in epidermal
carcinoma A431 cells, the tumor suppressor gene p16INK4a was
reactivated after being silenced. EGCG administration reduced the
levels of methylation, DNMT activity and protein levels of DNMT1,
DNMT3a and DNMT3b in the A431 cancer cell line (30). It has also been demonstrated that
EGCG modifies epigenetic changes in HeLa cells. The interaction
between EGCG, DNMT3B, promoter methylation, and RAR, CDH1 and DAPK1
expression in EGCG-treated HeLa cells was previously examined using
molecular modeling. The treatment of HeLa cells with EGCG reduced
DNMT activity. Molecular modeling results established that EGCG
suppressed DNMT3B activity. The restoration of tumor suppressor
genes in HeLa cells was caused by significant alterations in the
pattern of methylation of these genes induced by EGCG (31). It was previously demonstrated that
EGCG treatment reactivated the hMLH1, p16INK4a,
RAR and MGMT genes in HT-29 and KYSE 150 cells
(29). It was also discovered that
EGCG administration reactivated the methylated silenced genes,
RAR, p16, hMLH1 and MGMT in the KYSE 510
esophageal cancer cell line. EGCG treatment resulted in the
activation of epigenetically silenced genes (29).
In a previous study, it was observed that in the
oral squamous cell carcinoma cell line, CAL-27, EGCG treatment
caused the partial hypermethylation of the RECK gene. RECK
is a tumor suppressor that controls matrix metalloproteinases
(MMPs), crucial for extracellular matrix degradation. In oral
squamous cell carcinoma, RECK is often downregulated due to
promoter hypermethylation or genetic changes, leading to an
increased MMP activity, which enhances tumor invasion and
metastasis (32). Reduced RECK
levels are associated with more aggressive tumors and a worse
prognosis. EGCG reverses the process of hypermethylation and
activates the RECK gene, restraining oral squamous cell
cancer invasiveness and translocation. EGCG was found to decrease
the DNA methylation of the RECK gene in the cell lines
(32). In another study,
quantitative PCR and methylation-specific PCR were employed to
assess the expression of the SOCS1 promoter methylation levels.
EGCG treatment induced an increase in SOCS1 expression and
the promoter demethylation of SOCS1 was also upregulated
(33). A previous study also
proved that EGCG affected DNA hypermethylation and head and neck
squamous cell carcinoma (HNSCC) development. EGCG treatment
decreased the broad DNA methylation levels in HNSCC cells (SCC-1
and FaDu). Following the administration of EGCG for a significant
amount of time, DNA hypermethylation was effectively reduced up to
70-80%. The transformation of 5-methylcytosine into
5-hydroxymethylcytosine in HNSCC cells provided evidence that DNA
hypermethylation was suppressed in these cells. Treatment with EGCG
markedly decreased DNMT activity in HNSCC cells, decreasing it to
80% in FaDu and 60% in SCC-1 cells (34). In another study, in the Apc(Min/+)
mouse model of colorectal carcinogenesis, EGCG treatment reduced
tumor formation and reversed RXRα gene silencing. EGCG decreased
CpG methylation in the RXRα promoter, restoring its expression and
highlighting the role of EGCG in epigenetically inhibiting
tumorigenesis (35).
Resveratrol
Resveratrol is a distinct polyphenolic compound
obtained from peanuts or grapes. The broad potential of resveratrol
includes the ability to fight various tumors, cell development,
cell death and angiogenesis It has gained the interest of
researchers involved in the exploration of chemopreventive
remedies. In a scientific review focusing on the non-invasive MCF-7
breast cancer cell line, it was demonstrated that treatment with
resveratrol induced the reversal of hypermethylation and the
re-activation of the tumor suppressor genes, PTEN and
RARβ2, and a concurrent reduction in the activity of DNMT1,
and an elevation in the levels of p21 (36). It has been reported that
resveratrol alters DNMT1 activity and the methyl binding domain
protein-2 with the promoter of the BRCA-1 gene to prevent
the tumor suppressor BRCA-1 from being epigenetically
silenced in MCF-7 breast cancer cells (37). Research has shown that resveratrol
demethylates RASSF1A in females with a higher risk of
developing breast cancer. In a previous study, individuals with an
elevated risk of developing breast cancer were treated with
resveratrol for 2 weeks. Mammary ductoscopy samples were used to
examine the methylation of cancer-associated genes, including
RASSF1A. An increase in trans-resveratrol levels reduced
RASSF1A methylation (10).
It has been reported that resveratrol causes the reversal of
promotor hypermethylation and the restoration of tumor suppressor
genes. Resveratrol therapy appears to reduce the activity of
DNMT1/3b and activate RASSF-1 in breast cancer cell lines (38). Resveratrol interacts with
hypermethylation in the RUNX3 promoter gene. Researchers
have used MSP to assess methylation following treatment, and the
results have revealed the marked methylation in the RUNX3
promoter of B16F10 cells of malignant melanoma, coupled with faint
unmethylated segments. Resveratrol doses for a >48 h gradually
attenuated RUNX3 promoter methylation and increased
unmethylated areas. Resveratrol exhibited a potent ability to
induce significant dose-dependent demethylation (39). In experimental models of mammary
carcinogenesis using ACI rats, trans-resveratrol has
demonstrated significant antineoplastic properties, primarily
through the downregulation of DNMT3 expression (40).
Quercetin
Quercetin is a potent natural antioxidant flavanol
that may be derived from a broad category of dietary sources, such
as apples, grains, onions, tea, red wine, citrus fruit, and a
number of other types of leaves (2). The anticancer effects of quercetin
are associated with altering mitogenic signaling, cancer cell
metastatic, angiogenesis, the cell cycle and apoptotic signaling
pathways, as well as by reversing epigenetic alterations, such as
DNA hypermethylation (41). It was
previously shown that quercetin modified the pattern of RASSF1A
methylation in the HeLa neoplastic cell line. The administration of
quercetin for 6 days resulted in the reverse hypermethylation of
the RASSF1A gene. The research outcomes revealed that the
administration of quercetin to cervical cells effectively altered
the hypermethylation level of RASSF1A, as documented by Lugli et
al (42). Another study was
performed to demonstrate the significance of quercetin at the
promoter methylation region of several genes that were silenced due
to DNA hypermethylation in colorectal cancer RKO cell lines. The
genes p16INK4a, CDH1 and IGFBP7 were examined.
These results suggest that quercetin can reverse the
methylated-promoter phenotype in the RKO colorectal cancer cell
line (43).
In vivo and in vitro studies have
shown that quercetin may function as a preventive measure against
colon cancer (2). The MTT assay
has been used in in vitro studies and has revealed the
capacity of quercetin to prevent the development of the colon
cancer cell line, RKO. The hypermethylation of p16INK4a was
successfully reversed after quercetin therapy for 120 h. The
expression of p16INK4a was restored in a concentration-dependent
manner. This demonstrates that quercetin exerts its chemopreventive
properties by demethylating the p16INK4a gene promoter
(2). Quercetin has the potential
to inhibit DNMTs in DU145 and PC3 prostate cancer (CaP) cell lines.
The hypermethylation of the androgen receptor (AR) gene promoter is
linked to CaP defiant to androgen-deprivation therapy with
antiandrogens. In AR-negative cell lines, quercetin and curcumin
restored AR mRNA and protein concentrations through global
hypomethylation, which led to the initiation of apoptosis by
mitochondrial depolarization (44). It has been proven that quercetin
can alter the methylation levels of RASSF1A in the HeLa cell line.
The hypermethylation of RASSF1A was reversed after 6 days of
treatment with quercetin. Quercetin was administered at different
doses for different periods of time to examine the reversal of
hypermethylation in the RASSF1A gene. The results
demonstrated that the RASSF1A gene in HeLa cells was
demethylated after 6 days of therapy (14).
It has been proven that quercetin strongly affects
the activity of DNMTs. In a previous study, nuclear extracts were
observed to suppress the activity of the DNMTs by 32 and 49%,
respectively, when quercetin was added to the mixture in contrast
to that of the untreated extract, and it was discovered that the
broad methylation levels of the HeLa cell line were decreased.
Almost 50% of the methylation was reduced by quercetin treatment
within 24 h, and further administration for 24 and 48 h decreased
the methylation of DNA to 15 and 36% of the control, respectively.
The methylation levels of the TSGs to DAPK1 (7%),
RASSF1 (9%), VHL (10%), PTEN (11%),
RARB (19%), MGMT (22%), GSTP1 (24%),
APC (31%), SOC51 (58%) and CDH1 (60%) were all
attenuated by quercetin treatment (45). Another study was conducted to
examine the impact of quercetin on cellular immunity against cancer
via the IL15 gene. The cell lines A549 and HeLa were grown in
vitro and exposed to quercetin at various doses. RT-qPCR was
performed to examine the transcription activity of IL15 and DNA
methyltransferase. Treatment with quercetin in HeLa and A549 cells
reduced the expression Quercetin reduced the production of IL15 by
increasing the methylation of the IL15 promoter, thus inhibiting
the expansion of malignant cells (46). In xenograft models of MCF-7 (breast
cancer) and CT-26 (colon cancer), quercetin administration at
various doses (50, 100, and 200 mg/kg) resulted in a notable
reduction in tumor volume. This decrease was observed as early as
the 18th day in CT-26 tumors and the 20th day in MCF-7 tumors
post-treatment, demonstrating quercetin antitumor potential through
epigenetic regulation mechanisms (47).
5. Conclusion and future perspectives
It can be concluded that bioactive compounds of
natural origin can serve as inhibitors of DNMT activity and
mitigate promoter hypermethylation. In addition, the demonstrated
the capability to facilitate the re-expression of genes previously
subjected to methylation-induced silencing, thereby orchestrating
the reversal of epigenetic alterations implicated in oncogenesis.
Compounds such as genistein, quercetin, resveratrol, capsaicin and
EGCG can reverse the hypermethylation of various genes that cause
cancer. Further research into natural compounds as therapeutic
epigenetic agents is essential to fully explore their potential in
cancer treatment. While some compounds have yielded promising
results, further research is required in order to fully elucidate
their mechanisms of action and effectiveness. By expanding studies
to include a wider range of natural substances, novel, potent
epigenetic agents can be identified that may offer novel approaches
to cancer therapy. Research on natural compounds could lead to the
development of more targeted, less toxic treatments, ultimately
improving patient outcomes and advancing the field of oncology.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
F conducted a significant portion of the literature
search and drafted substantial parts of the manuscript. KSS
contributed to editing specific sections of the manuscript. RM
edited and proofread the manuscript. AKJ contributed to the initial
idea and scope of the review, provided critical review and feedback
on the manuscript, and approved the final version of the
manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Aggarwal R, Jha M, Shrivastava A and Jha
AK: Natural compounds: Role in reversal of epigenetic changes.
Biochemistry (Mosc). 80:972–989. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Suter MA and Aagaard-Tillery KM:
Environmental influences on epigenetic profiles. Semin Reprod Med.
27:380–390. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Yadav P, Chakraborty P, Jha NK, Dewanjee
S, Jha AK, Panda SP, Mishra PC, Dey A and Jha SK: Molecular
mechanism and role of Japanese encephalitis virus infection in
central nervous System-mediated diseases. Viruses.
14(2686)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ehrlich M: DNA hypermethylation in
disease: Mechanisms and clinical relevance. Epigenetics.
14:1141–1163. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gordaliza M: Natural products as leads to
anticancer drugs. Clin Transl Oncol. 9:767–776. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kumar U, Sharma U and Rathi G: Reversal of
hypermethylation and reactivation of glutathione S-transferase pi 1
gene by curcumin in breast cancer cell line. Tumour Biol.
39(1010428317692258)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li JW and Vederas JC: Drug discovery and
natural products: End of an era or an endless frontier? Science.
325:161–165. 2009.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Dammann RH, Richter AM, Jiménez AP, Woods
M, Küster M and Witharana C: Impact of natural compounds on DNA
methylation levels of the tumor suppressor gene RASSF1A in cancer.
Int J Mol Sci. 18(2160)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Das PM and Singal R: DNA methylation and
cancer. J Clin Oncol. 22:4632–4642. 2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nikbakht M, Jha AK, Malekzadeh K, Askari
M, Mohammadi S, Marwaha RK, Kaul D and Kaur J: Aberrant promoter
hypermethylation of selected apoptotic genes in childhood acute
lymphoblastic leukemia among North Indian population. Exp Oncol.
39:57–64. 2017.PubMed/NCBI
|
|
13
|
Jha AK, Sharma V, Nikbakht M, Jain V,
Sehgal A, Capalash N and Kaur J: A comparative analysis of
methylation status of tumor suppressor genes in paired biopsy and
serum samples from cervical cancer patients among north Indian
population. Russ J Genet. 52:226–230. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Saloni Sharma A, Goel H, Pal S, Rai P,
Rawat K, Syeda S, Shrivastava A and Kumar Jha A: Demethylation of
RASSF1A gene by quercetin and eugenol in heLa cancer cell line. Int
J Health Sci Res. 9:29–34. 2019.
|
|
15
|
Dixon RA and Ferreira D: Genistein.
Phytochemistry. 60:205–211. 2002.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fang MZ, Chen D, Sun Y, Jin Z, Christman
JK and Yang CS: Reversal of hypermethylation and reactivation of
p16INK4a, RARbeta, and MGMT genes by genistein and other
isoflavones from soy. Clin Cancer Res. 11:7033–7041.
2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Majid S, Dar AA, Shahryari V, Hirata H,
Ahmad A, Saini S, Tanaka Y, Dahiya AV and Dahiya R: Genistein
reverses hypermethylation and induces active histone modifications
in tumor suppressor gene B-Cell translocation gene 3 in prostate
cancer. Cancer. 116:66–76. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Adjakly M, Bosviel R, Rabiau N, Boiteux
JP, Bignon YJ, Guy L and Bernard-Gallon D: DNA methylation and soy
phytoestrogens: Quantitative study in DU-145 and PC-3 human
prostate cancer cell lines. Epigenomics. 3:795–803. 2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li H, Xu W, Huang Y, Huang X, Xu L and Lv
Z: Genistein demethylates the promoter of CHD5 and inhibits
neuroblastoma growth in vivo. Int J Mol Med. 30:1081–1086.
2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
He F and Chen JQ: Consumption of soybean,
soy foods, soy isoflavones and breast cancer incidence: Differences
between Chinese women and women in Western countries and possible
mechanisms. Food Sci Hum Wellness. 2:146–161. 2013.
|
|
21
|
Xie Q, Bai Q, Zou LY, Zhang QY, Zhou Y,
Chang H, Yi L, Zhu JD and Mi MT: Genistein inhibits DNA methylation
and increases expression of tumor suppressor genes in human breast
cancer cells. Genes Chromosomes Cancer. 53:422–431. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sundaram MK, Ansari MZ, Al Mutery A,
Ashraf M, Nasab R, Rai S, Rais N and Hussain A: Genistein induces
alterations of epigenetic modulatory signatures in human cervical
cancer cells. Anticancer Agents Med Chem. 18:412–421.
2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Saha D, Vaishnav N, Ahsan Z, Rani N,
Mathur R and Jha AK: Reversal of hypermethylation and reactivation
of tumor suppressor genes due to natural compounds in breast cancer
cells. Int J Biol Innovations. 2:63–75. 2020.
|
|
24
|
Chapa-Oliver AM and Mejía-Teniente L:
Capsaicin: From plants to a cancer-suppressing agent. Molecules.
21(931)2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sharan M, Jha M, Chandel R, Syeda S,
Mathur R, Jha NK, Jha AK, Goel H, Shrivastava A, Chauhan S, et al:
Demethylation of CADM1 and SOCS1 using capsaicin in cervical cancer
cell line. Naunyn Schmiedebergs Arch Pharmacol. 396:649–657.
2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sanchez AM, Sanchez MG, Malagarie-Cazenave
S, Olea N and Diaz-Laviada I: Induction of apoptosis in prostate
tumor PC-3 cells and inhibition of xenograft prostate tumor growth
by the vanilloid capsaicin. Apoptosis. 11:89–99. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sharan M, Ravinuthala RP, Mathur R, Khare
N and Jha AK: In silico study of the effect of capsaicin against
hypermethylation of TSLC1 to cure cervical cancer. GIS Sci J.
8:1159–1176. 2021.
|
|
28
|
Li Y and Tollefsbol TO: Impact on DNA
methylation in cancer prevention and therapy by bioactive dietary
components. Curr Med Chem. 17:2141–2151. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H,
Welsh W and Yang CS: Tea polyphenol (-)-epigallocatechin-3-gallate
inhibits DNA methyltransferase and reactivates methylation-silenced
genes in cancer cell lines. Cancer Res. 63:7563–7570.
2003.PubMed/NCBI
|
|
30
|
Nandakumar V, Vaid M and Katiyar SK:
(-)-Epigallocatechin-3-gallate reactivates silenced tumor
suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA
methylation and increasing histones acetylation in human skin
cancer cells. Carcinogenesis. 32:537–544. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Khan MA, Hussain A, Sundaram MK, Alalami
U, Gunasekera D, Ramesh L, Hamza A and Quraishi U:
(-)-Epigallocatechin-3-gallate reverses the expression of various
tumor-suppressor genes by inhibiting DNA methyltransferases and
histone deacetylases in human cervical cancer cells. Oncol Rep.
33:1976–1984. 2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Long NK, Kato K, Yamashita T, Makita H,
Toida M, Hatakeyama D, Hara A, Mori H and Shibata T:
Hypermethylation of the RECK gene predicts poor prognosis in oral
squamous cell carcinomas. Oral Oncol. 44:1052–1058. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Alizadeh M, Nafari A, Safarzadeh A,
Veiskarami S, Almasian M and Asghar Kiani A: The impact of EGCG and
RG108 on SOCS1 promoter DNA methylation and expression in U937
leukemia Cells. Rep Biochem Mol Biol. 10:455–461. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Agarwal A, Kansal V, Farooqi H, Prasad R
and Singh VK: Epigallocatechin gallate (EGCG), an active phenolic
compound of green tea, inhibits tumor growth of head and neck
cancer cells by targeting DNA hypermethylation. Biomedicines.
11(789)2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Morris J, Moseley VR, Cabang AB, Coleman
K, Wei W, Garrett-Mayer E and Wargovich MJ: Reduction in promotor
methylation utilizing EGCG (epigallocatechin-3-gallate) restores
RXRα expression in human colon cancer cells. Oncotarget.
7:35313–35326. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Stefanska B, Salamé P, Bednarek A and
Fabianowska-Majewska K: Comparative effects of retinoic acid,
vitamin D and resveratrol alone and in combination with adenosine
analogues on methylation and expression of phosphatase and tensin
homologue tumour suppressor gene in breast cancer cells. Br J Nutr.
107:781–790. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Thakur VS, Deb G, Babcook MA and Gupta S:
Plant phytochemicals as epigenetic modulators: Role in cancer
chemoprevention. AAPS J. 16:151–163. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Khan MI, Rath S, Adhami VM and Mukhtar H:
Targeting epigenome with dietary nutrients in cancer: Current
advances and future challenges. Pharmacol Res. 129:375–387.
2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kang S, Wang Z, Li B, Gao X, He W, Cao S
and Chen H: Anti-tumour effects of resveratrol on malignant
melanoma is associated with promoter demethylation of RUNX3 gene.
Pharmazie. 74:163–167. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Qin W, Zhang K, Clarke K, Weiland T and
Sauter ER: Methylation and miRNA effects of resveratrol on mammary
tumors vs. normal tissue. Nutr Cancer. 66:270–277. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gibellini L, Pinti M, Nasi M, Montagna JP,
De Biasi S, Roat E, Bertoncelli L, Cooper EL and Cossarizza A:
Quercetin and cancer chemoprevention. Evid Based Complement
Alternat Med. 2011(591356)2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lugli E, Ferraresi R, Roat E, Troiano L,
Pinti M, Nasi M, Nemes E, Bertoncelli L, Gibellini L, Salomoni P,
et al: Quercetin inhibits lymphocyte activation and proliferation
without inducing apoptosis in peripheral mononuclear cells. Leuk
Res. 33:140–150. 2009.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Samuel T, Fadlalla K, Yehualaeshet T and
Turner T: Abstract 2005: Modulation of tumor suppressor gene
DNA-methylation by quercetin and dietary indoles. Cancer Res.
71:2005. 2011.
|
|
44
|
Sharma V, Kumar L, Mohanty SK, Maikhuri
JP, Rajender S and Gupta G: Sensitization of androgen refractory
prostate cancer cells to anti-androgens through re-expression of
epigenetically repressed androgen receptor-Synergistic action of
quercetin and curcumin. Mol Cell Endocrinol. 431:12–23.
2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kedhari Sundaram M, Hussain A, Haque S,
Raina R and Afroze N: Quercetin modifies 5' CpG promoter
methylation and reactivates various tumor suppressor genes by
modulating epigenetic marks in human cervical cancer cells. J Cell
Biochem. 120:18357–18369. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang M, Lu A, Wang H and Yang J:
Quercetin downregulates the expression of IL15 in cancer cells
through DNA methylation. Eur Rev Med Pharmacol Sci. 27:2580–2590.
2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Volate SR, Muga SJ, Issa AY, Nitcheva D,
Smith T and Wargovich MJ: Epigenetic modulation of the retinoid X
receptor alpha by green tea in the azoxymethane-Apc Min/+ mouse
model of intestinal cancer. Mol Carcinog. 48:920–933.
2009.PubMed/NCBI View Article : Google Scholar
|