Introduction
The incidence of colorectal cancer (CRC), which
affects the colon, large intestine and rectum, is increasing
globally, particularly in developing countries (1). In addition, the incidence and
mortality rates due to CRC are expected to increase in the coming
decades (2). As per global cancer
statistics (2020), the incidence of CRC (in males and females) was
10%, the third highest after breast and lung cancers. From a
mortality perspective, CRC is the second leading cause of
cancer-related death (9.4%) (3).
According to the 2018 Cancer Incidence Report, the most common
cancers among Saudi nationals were breast cancer (17.9%) and CRC
(12.2%). In CRC, the occurrence of colon cancer (males, 60.2%;
females, 64.9%) is higher than that of rectal cancer (4). A report by the World Health
Organization, stated that the incidence and mortality rates
associated with CRC in Saudi Arabia were estimated to be 14.6 and
15.2%, respectively (5).
The first line of defense in the standard treatment
plan for the CRC is surgery, along with chemotherapy and
radiotherapy as adjuvant and neoadjuvant settings to reduce the
tumor mass and stabilize the tumor (6). However, 25% of CRC cases are not
manifested until the advanced stages of the disease. In such
instances, surgical treatment alone may not result in a positive
prognosis and may lead to death (7). Notably, targeted therapy in the form
of immunotherapy enhances the overall survival rate of patients
with CRC. Several pathways and checkpoints have been identified,
and targeted agents have been developed against specific pathways
or proteins to control disease progression. The Food and Drug
Administration has approved agents to target specific proteins,
such as vascular endothelial growth factor receptor, epidermal
growth factor receptor (EGFR) and programmed cell death protein
1(8).
It has been well established that genetic and
environmental factors increase the risk of developing CRC. However,
patients with Crohn's disease and ulcerative colitis are at a
greater risk of developing CRC, which increases with age (9). Several studies have demonstrated that
chronic inflammation, family history, lifestyle factors (such as a
sedentary lifestyle, regular alcohol consumption and smoking) and
diet (such as the frequent consumption of red meat and processed
meats) are major risk factors for the development of CRC (10). CRC is genetically diverse,
involving various pathways and mechanisms in its development. CRC
cells consist of several mutations and different gene expression
profiles (11). Several genomic
aberrations, such as genetic mutations in the BRAF, NRAS and KRAS
genes, are associated with resistance to treatment with EGFR
antibody. This phenomenon has raised the issue of selecting
suitable agents in the treatment of metastatic CRC (12). Several CRC studies have reported
genetic alterations in the genes of various pathways, including
p53, TGF-β, WNT-β-catenin, EGFR, MAPK and phosphatidylinositol 3
kinase (PI3K)/Akt (13-18).
Previous research has demonstrated the association
between genetic mutation profiles and the prognosis of patients
with CRC (19). In CRC, the
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
alpha (PIK3CA), KRAS and BRAF genes may be of
prognostic significance (20,21).
The PI3KCA gene encodes for p110α, a catalytic subunit of
PI3K. PI3K functions as a phosphorylating agent for downstream
signaling molecules, which play a key role in the pathogenesis of
several types of cancer, including CRC, by regulating cell growth
and proliferation via the Akt-mTOR signaling pathway (21). PIK3CA mutations are observed
in various human cancers, including breast, head and neck squamous
cell carcinoma, colorectal, endometrial, brain, ovarian, lung,
thyroid and cervical cancers, with varying frequencies (22-24).
However, additional studies are required to explore the somatic
mutations of the genes involved in the development of CRC,
including PIK3CA, and their association with the
pathogenesis of CRC and the prognosis of affected patients. An
in-depth understanding of the gene mutation profiles in these
pathways, the association of these pathways with CRC development
and the identification of patient suitability for the different
treatment options are all required in this era of personalized
medicine. Hence, in the present study, the mutational spectrum in
the PIK3CA gene (exons 9 and 20) in the PI3K pathway was
investigated to assess the frequency of somatic mutations and their
association with clinicopathological parameters in a cohort of
patients with CRC.
Patients and methods
Patient samples
A total of 71 formalin-fixed paraffin-embedded
(FFPE) tissue samples collected at the King Fahd Hospital of the
University (KFHU; Al Khobar, Saudi Arabia) were used in the present
study. All the samples were obtained from patients who had received
a confirmed diagnosis of CRC (by histological analysis). The
inclusion criteria included the following: i) A confirmed diagnosis
of CRC with >70% tumor cells in the FFPE block; ii) the absence
of a family history of cancer; and iii) an age >18 years.
Samples with <70% tumor content in the FFPE block were excluded
from the study. Tumor cell percentage was calculated using
hematoxylin and eosin staining (this staining was performed at the
hospital laboratory as a routine procedure on paraffin-embedded
tissues). The present study received Institutional review board
approval from Imam Abdulrahman Bin Faisal University, Dammam, Saudi
Arabia (approval no. IRB-2019-01-378). All the samples were
anonymized to protect patient confidentiality and clinical and
therapeutic data were collected from the hospital laboratory
information system in a predesigned format. Written informed
consent was obtained from all patients.
DNA isolation and quantification
Paraffin-embedded sections (20-µm-thick) were
prepared from the respective FFPE tissues using a microtome. A
total of two sections were used for genomic DNA extraction using
the QIAMP DNA FFPE Tissue kit (Qiagen GmbH). Briefly, the procedure
included the deparaffinization of paraffin-embedded sections by the
addition of xylene (Merck KGaA) followed by centrifugation at
19,283 x g for 2 min at 25˚C to remove the supernatant. Absolute
ethanol (Merck KGaA) was added to the pellet and centrifuged at
19,283 x g for 2 min at 25˚C to remove the supernatant and the
residual ethanol was dried. For the pellet, tissue lysis buffer and
proteinase K was added and incubated at 56˚C until complete sample
lysis, followed by incubation at 90˚C for 1 h. Another lysis buffer
along with absolute ethanol was added to the existing lysate and
the solution was transferred to a QIAmp minielute spin column
(Qiagen GmbH) and centrifuged at 6,297 x g for 1 min at 25˚C,
followed by two washing steps with wash buffer 1 and 2. Finally, an
elution buffer was added to the QIAmp minielute spin column to
obtain DNA.
The quantity and purity of the isolated DNA was
examined using a Nanodrop spectrophotometer (Thermo Fisher
Scientific, Inc.). The absorbance at 260 nm was determined to
calculate the concentration and purity at a 260/280 ratio. The mean
DNA yield was 137.62±85.75 ng/µl and the mean 260/280 ratio was
2.03±0.16.
Capillary sequencing
A total of 100 ng DNA was used to amplify the
PIK3CA exons 9 and 20 in separate tubes using primers
(25) and 2X Phusion U Green
Multiplex PCR Master Mix (Thermo Fisher Scientific, Inc.). The
master mix was subjected to PCR using an annealing temperature of
54˚C. The resulting PCR amplicons for exon 9 (195 bp) and 20 (338
bp) were confirmed by 2% agarose gel electrophoresis.
The same PCR product was purified using the
ExoSAP-IT express PCR product cleanup kit (cat. no. 75001.200.UL;
Thermo Fisher Scientific, Inc.) followed by sequencing PCR using
forward or reverse primers and BigDye terminator v3.1 cycle
sequencing master mix (Thermo Fisher Scientific, Inc.). PGEM DNA
was used as the control for the sanger sequencing reaction. The
product was purified using BigDye XTerminator purification kit
(Thermo Fisher Scientific, Inc.). The purified product was
subjected to capillary electrophoresis using Quantstudio sequencer
(Thermo Fisher Scientific, Inc.). Primary analysis and quality
control of the output sequence was performed using sequence
analysis software (Thermo Fisher Scientific, Inc.). Secondary
analysis to compare the DNA sequence with the reference
PIK3CA exon 9 and 20 DNA sequence (NM_006218.4) was achieved
by CodonCode aligner software, version 10.0.2 (CodonCode
Corporation). Finally, two novel mutation sequence data were
submitted to a data repository of the NCBI GenBank with accession
numbers PQ785769 and PQ785770.
Statistical analysis
All data, including demographic, histological,
clinical, therapeutic and mutation details were organized in MS
excel format. The statistical analysis was carried out using SPSS
version 22 (IBM Corp.). The Kolmogorov-Smirnov test was used to
determine the normality of the data, which revealed a normal
distribution. For categorical data, the Chi-squared test and
Fisher's exact test were used and for continuous variable data, the
Student's t-test was used. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
Demographic data
Among the 71 tissue samples collected, 58 samples
were from patients with colon cancer, and 5.2% of the cases were
cecum cancer, 12% were rectosigmoid cancer, 19% were sigmoid
cancer, and the remaining cases were from different parts of the
colon. The remaining 13 tissue samples were from patients with
rectal cancer. The diagnosis was confirmed using the hematoxylin
and eosin staining technique. The overall frequency for colon
(43.1%) and rectal (46.1%) cancer was lower among males. In rectal
cancer, the overall mean age at diagnosis was 50.69 years and
stratification based on sex revealed 53.17 and 48.57 years in male
and female patients, respectively. Similarly, the overall mean age
at diagnosis among the patients with colon cancer was 55.24 years.
The mean age at diagnosis of the male and female patients with
colon cancer was 58.68 and 52.64 years, respectively. The patient
demographics are presented in Table
I.
 | Table IDemographics of the patients in the
present study. |
Table I
Demographics of the patients in the
present study.
| Parameter | Colon cancer | Rectal cancer | P-value |
|---|
| Cases, n (%) | 58(100) | 13(100) | NA |
| Sex (M: F), n
(%) | 25 (43.1):33
(56.9) | 6 (46.15):7
(53.85) | 0.841 |
| Age at diagnosis,
years (mean ± SD) | 55.24±12.48 | 50.69±13.19 | 0.243 |
| PIK3CA
mutation | 6 (10.34) | 0 | 0.584 |
| Tumor type, n
(%) | | NA | NA |
|
Cecum
cancer | 3 (5.2) | | |
|
Rectosigmoid
cancer | 7(12) | | |
|
Sigmoid
cancer | 11(19) | | |
|
Different
parts of colon | 37 (63.8) | | |
| Tumor grade, n
(%) | | | 0.160 |
|
Well-differentiated
(grade 1) | 28 (48.27) | 3(23) | |
|
Moderately
differentiated (grade 2) | 23 (39.65) | 9 (69.23) | |
|
Poorly
differentiated (grade 3) | 7 (12.08) | 1 (7.69) | |
| Tumor stage, n
(%) | | | 0.0007 |
|
Stage I | 3 (5.17) | 5 (38.5) | |
|
Stage
II | 24 (41.37) | 8 (61.5) | |
|
Stage
III | 17 (29.31) | 0 (0) | |
|
Stage
IV | 14 (24.13) | 0 (0) | |
| Hospital, n
(%) | | | NA |
|
KFHU | 41 (70.6) | 2 (15.38) | |
|
Other | 17 (29.4) | 11 (84.62) | |
| Treatment strategy
(only KFHU) | | | 0.999 |
|
Surgical
resection | 20 (48.7) | 1(50) | |
|
Surgical
resection and chemotherapy/radiotherapy | 21 (51.2) | 1(50) | |
| Recurrence at 5
years | 13 (33.33) | No Data | NA |
Tumor classification
Tumor stage was classified based on the tumor, node
and metastasis staging system, which is calculated by assessing the
tumor size and growth, number of regional lymph nodes involved and
metastasis. Tumor grade is identified based on the morphology of
the tumor by comparing healthy and malignant cells under the
microscope. In the colon cancer group, well-differentiated tumors
were found in 48.27% of the cases, followed by moderately
differentiated tumors (39.65%) and poorly differentiated tumors
(12%). However, the majority of the tumors in the rectal cancer
group were moderately differentiated tumors (69.23%), followed by
23% of the tumors being well-differentiated and only 7.69% of the
cases exhibiting poorly differentiated tumors. The cancer stages
revealed that the majority of the tumors in the colon cancer group
were stage II tumors (41.37%), followed by stage III tumors
(29.31%), stage IV tumors (24.13%) and stage I tumors (5.17%). In
the rectal cancer group, the highest percentage of tumors (61.5%)
were observed in stage II and the remaining (38.4%) cases were in
stage I. There were no cases observed in the rectal cancer group
with stage III and stage IV disease (Table I).
Regional lymph node involvement was observed in only
the colon cancer cases. An average of 3.6 regional lymph nodes were
involved in patients with stage III and stage IV tumors. However,
in the rectal cancer cases, there was no involvement of the
regional lymph nodes as all the cases presented with stage I and
stage II disease.
Treatment and follow-up
Out of the 58 colon cancer cases, 70.6% of the
patients underwent treatment at KFHU and the remaining cases were
diagnosed at KFHU, but treated at other hospitals. Hence, the
therapeutic and follow-up data for the patients treated outside
KFHU was not obtained. Among the 70.6% of the cases of colon
cancer, 48.7% of the patients underwent surgical resection alone
and the remaining 51.2% of the patients underwent surgical
resection followed by chemotherapy. Out of the 13 rectal cancer
cases, only 15.38% of the cases were treated at KFHU and the
remaining patients were referred to King Fahd Specialist Hospital,
Dammam, Saudi Arabia, which is the major center for cancer
treatment in the Eastern Province of Saudi Arabia. Of the rectal
cancer cases who were treated at KFHU, 50% underwent surgical
resection and 50% were treated by surgical resection followed by
chemotherapy and radiotherapy. In the colon cancer cases, 67.24%
were followed up after treatment. Among these patients, recurrence
within 5 years was observed in 33.33% of the cases. The majority of
follow-up data from the rectal cancer group were not available as
the patients were treated at another center.
PIK3CA mutation
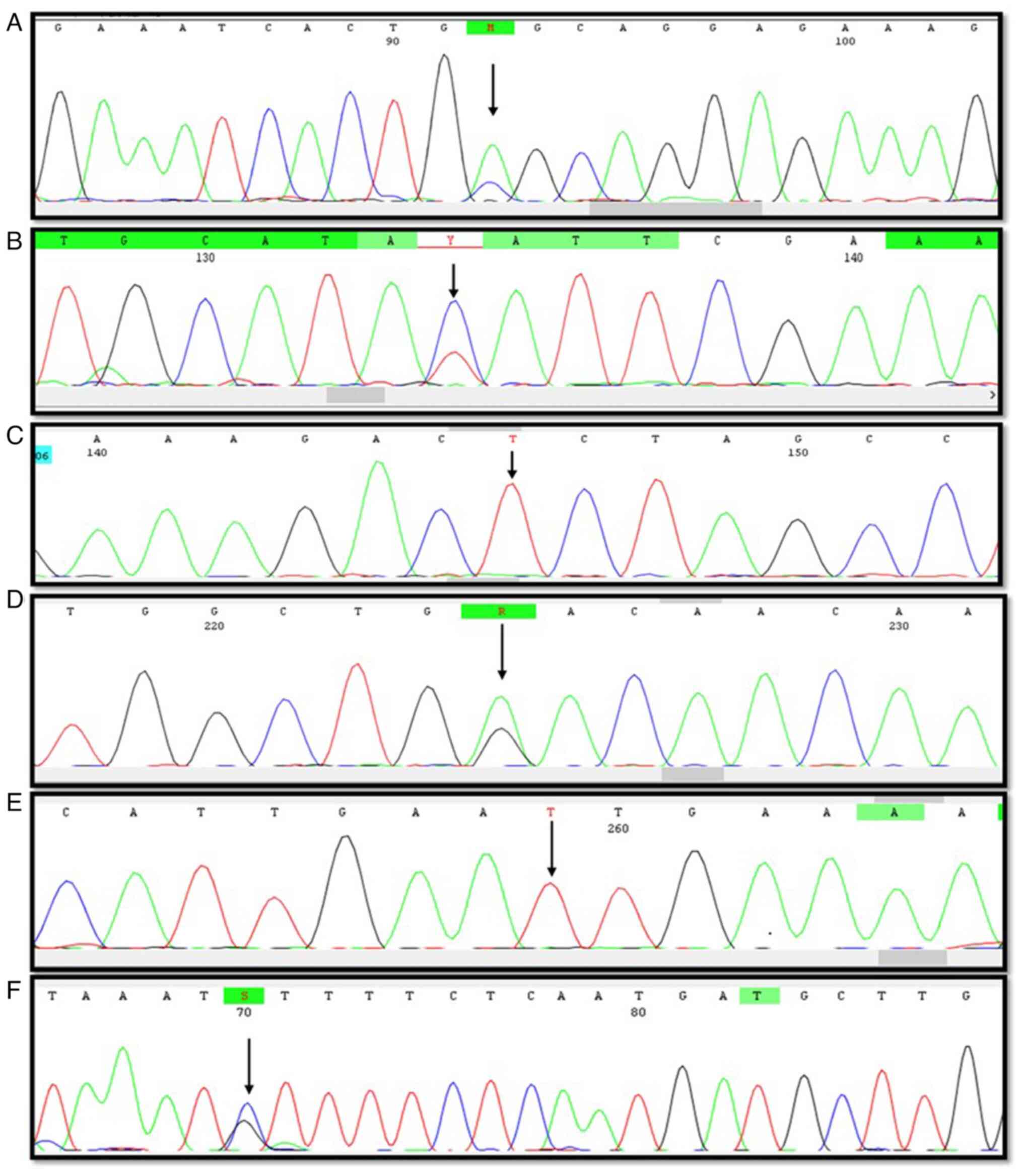
PIK3CA exon 9 and 20 genes were amplified by
Sanger Sequencing to determine the known and unknown mutations.
From the entire cohort, including mutation analysis of colon and
rectal tumors, it was revealed that 6 patients (8.4%) had mutations
in either exon 9 or 20. These mutations were observed only in colon
cancer tumors (10.3%) with no mutations in rectal cancer tumors. A
total of six different types of mutations were identified, with one
mutation in exon 9 and the remaining five mutations in exon 20
(Table II and Fig. 1). Out of the total mutations, 50%
of the mutations were silent mutations, followed by missense and
nonsense mutations (33.3 and 16.6%, respectively). Only one silent
mutation was observed in the 4 patients with colon cancer. Mutation
status comparison with tumor grade and stage revealed that 66.6% of
the mutations were observed in grade II tumors and 50% of the
mutations were observed in stage III tumors.
 | Table IIExon 9 and 20 mutation details,
clinical and histopathological details in colon cancer cases. |
Table II
Exon 9 and 20 mutation details,
clinical and histopathological details in colon cancer cases.
| PIK3CA
exon | Type of
mutation | Nucleotide
change | Amino acid
change | Age at diagnosis
(years) | Sex | Grade | Stage | Recurrence |
|---|
| 9 | Missense
mutation | c.1634 A>C | E545A | 64 | Female | 2 | III | No |
| 20 | Silent
mutation | c.3063 C>T | Y1021Y | 83 | Male | 1 | I | No |
| | | c.3063 C>T | Y1021Y | 55 | Male | 1 | III | No |
| | | c.3063 C>T | Y1021Y | 52 | Female | 2 | III | No |
| | | c.3063 C>T | Y1021Y | 41 | Female | 2 | IV | Yes |
| | | c.3075 C>T | T1025T | | | | | |
| | | c.3204 C>T | N1068N | | | | | |
| | Missense
mutationa | c.3001 C>G | L1001V | 57 | Female | 2 | IV | Yes |
| | Nonsense
mutationa | c.3153 G>A | W1051Ter | | | | | |
Exon 9 mutation c. 1634A>C was identified in a
patient with colon cancer with the primary tumor located in the
cecum. This mutation was not observed in the rectal group of
tumors. This was a missense mutation leading to an amino acid
change at codon 545 from glutamic acid to alanine (E545K). The
other hotspot mutations, such as c.1624G>A (E542K) and
c.1633G>A (E545K) in exon 9 were absent in the cohort.
Exon 20 mutations were observed in five patients,
with the silent mutation c.3063C>T observed in 4 patients who
all presented with colon cancer. One patient had the silent
mutation c.3063C>T along with two other silent mutations
(c.3075C>T and c.3204C>T). The missense mutation c.
3001C>G, which was also observed in a patient with colon cancer,
leading to an amino acid change at codon 1001 from leucine to
valine (L1001V). In the same patient, downstream of the missense
mutation, a nonsense mutation (c.3153G>A) was observed, which
created a termination codon sequence at codon 1051 from tryptophan
amino acid (W1051Ter). The two mutations, L1001V and W1051Ter, were
novel mutations and have not been previously reported. The
PIK3CA mutation rate was observed more frequently in female
patients (66.6%) compared with male patients and the missense
mutations were seen only in female patients. The mutation
comparison with the prognosis data revealed four patients (66.6%)
who had no recurrence of the disease with two patients (33.3%)
having a recurrence. The 2 patients who had a recurrence presented
with multiple mutations including silent, missense and nonsense
mutations.
Discussion
CRC represents the third most common type of cancer
globally and in Saudi Arabia it is ranked second following breast
cancer. Universally, among CRC subtypes, colon cancers are the most
predominant (60.2%) followed by rectal cancer (39.8%). A similar
trend has been observed in Saudi Arabia, with colon and rectal
cancers (61 and 39%, respectively) (3,4).
However, in the present study, 81.7% of the patients had colon
cancer and the remaining had rectal cancer. It was hypothesized
that this discrepancy may be due to the single center approach for
sample collection and the fact that the majority of the patients
were from the Eastern Province of Saudi Arabia. The age range at
diagnosis for the majority of male patients with CRC is between
60-64 years and in female patients this is 55-59 years (4). The present study observed a similar
pattern in the age range at diagnosis among male (57.61 years) and
female (51.93 years) patients with CRC, indicating that females
present with the disease at an earlier age than males. The
stratified mean age at diagnosis of the patients with colon cancer
(55.24 years) and rectal cancer (50.69 years) was in line with the
Saudi Cancer Incidence Report 2018(4). As age is one of the influencing
factors for the risk of developing CRC, the majority of CRC cases
are diagnosed after the age of 50 years. Therefore, after the age
of 40 years, CRC screening is recommended.
CRC is typically an asymptomatic disease and when
symptoms such as anemia, rectal bleeding or abdominal pain
manifest, the majority of the patients present at an advanced stage
of the disease (26). In the
present study, ~50% of patients with colon cancer presented at an
advanced stage but none of the patients with rectal cancer
presented at an advanced stage. Several studies have revealed that
the incidence of early-onset CRC is increasing globally (27,28).
Improved survival outcomes have been noted in patients whose
disease was diagnosed in the early onset of rectal cancer (29). Therefore, patients with rectal
cancer in the present study cohort may have improved survival rates
compared with the patients with colon cancer. Disease recurrence
data within 5 years from the diagnosis date were available for the
cases followed-up at KFHU and the data revealed that 33.35% of the
patients with colon cancer had disease recurrence. The majority
(77%) of the patients with colon cancer and disease recurrence
presented with stage IV of the disease. Diagnosis at various stages
of the disease influences the treatment options and survival rates.
Improvements in the understanding of the pathophysiology of CRC
enables physicians to select the optimum treatment options and can
increase the survival rate to 3 years (30).
CRC is a heterogenous disease with different genetic
and epigenetic backgrounds. The development of CRC is caused by an
accumulation of somatic mutations in multiple genes and epigenetic
events (31). The PI3K/Akt/mTOR
pathway plays a crucial role in these processes, such as cell
growth, cell cycle progression and survival. In this pathway, PI3K
is composed of a catalytic subunit and a regulatory subunit, and is
initially encoded by the PIK3CA gene. The PIK3CA gene
mutations were observed frequently in several cancers (32,33).
The majority of somatic mutations in PIK3CA are confined to
exon 9 and 20(34). Hence, the
present study focused on the hotspot mutations situated in exon 9
and 20 of the PIK3CA gene in the CRC cohort. The frequency
of PIK3CA somatic mutations in the CRC cohort were 5.7-32%.
Studies with a large CRC sample size, such as The Cancer Genome
Atlas study (11), Dana Farber
Cancer Institute study (35) and
The Memorial Sloan Kettering Cancer Center study (36), reported PIK3CA mutations to
be 24.7, 21.3 and 20.8%, respectively (37). Other studies with smaller sample
sizes reported varying mutation rates. For example, a study
conducted in the USA reported a mutation rate of 32% (38), followed by another USA study with a
rate of 18.1% (39), an Indian
study with a rate of 5.7% (40),
an Italian study with 8% (41) and
a Chinese study with 18.94% (42).
In the present study, the majority of the patients
were from the Eastern Province of Saudi Arabia, and it was observed
that 8.4% of these patients presented with the PIK3CA
mutation. These variations among different studies are possibly due
to the difference in the ethnicity of population and different
methodologies used to detect the mutations. PIK3CA mutation
status association with sex revealed mixed results. Of note, one
study reported that the PIK3CA mutation frequency in colon
cancer was higher in female patients (43) and another study reported there was
no association between sex and the PIK3CA mutation (44). In the present study, the overall
CRC frequency was higher in female patients (66.6%). Among the
patients who presented with the mutation, all the missense and
novel mutations were observed in female patients.
Various studies have found an association between
the PIK3CA mutation status with the clinicopathological
parameters, such as disease stage, survival, recurrence and
therapeutic response (45-49).
The present study revealed that the majority (83.3%) of colon
tumors with the PIK3CA mutation presented with an advanced
stage of the disease (stage III or IV). These results are in line
with those of another study, which reported that all PIK3CA
mutations were observed in high pathological stages and poorly
differentiated CRC tumors (45).
PIK3CA mutations have been shown to be associated with a
poor prognosis of patients with the disease in previous studies
(46-48);
in the present study, the patient who presented with the
PIK3CA missense mutation/nonsense mutation had a poor
prognosis. The present study reported on two novel mutations in the
PIK3CA gene, namely one missense mutation leading to an
L1001V amino acid change and a nonsense mutation leading to
termination (W1051Ter). Both of these novel mutations are situated
on exon 20 which is the kinase domain of the protein. The kinase
region mutations help to achieve gain of function and change the
transforming capacity of the tumor (49).
In conclusion, PIK3CA somatic mutations are
critical in understanding the pathogenicity of solid cancers,
particularly CRC. The novel mutations identified in the kinase
domain of PIK3CA may induce the hyperactivation of the protein in
the PI3K pathway leading to poor prognosis. Based on the present
study findings, CRC screening is recommended at an early age
(>40 years) for all patients, but particularly for female
patients who carry these mutations. The majority of the tumors if
detected early may present at a low grade and early stage, and the
response to treatment options in terms of prognosis may be
improved. However, the small sample size of the present study is a
limitation to a strong genetic association. Hence, larger sample
studies are warranted to understand the association between
PIK3CA mutations with disease prognosis and therapeutic
response.
Acknowledgements
The authors would like to thank Mr. Shakir Ahmed,
King Fahd Hospital of the University, Imam Abdulrahman Bin Faisal
University, Al Khobar, Saudi Arabia, for providing technical
support (he performed the staining process at the KFHU hospital and
also assisted the authors with locating the selected FFPE tissues
from the archive).
Funding
Funding: The present study was supported by the Deanship of
Scientific Research, Imam Abdulrahman Bin Faisal University (Grant
no. 2019-356-Med).
Availability of data and materials
The sequence data for the two novel mutations
generated in the present study may be found in NCBI GenBank under
accession numbers PQ785769 and PQ785770. The data generated in the
present study are available in the National Centre for
Biotechnology information database (https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA1230587)
with the accession no. PRJNA1230587.
Authors' contributions
CV was involved in the conceptualization of the
study, in the writing of the original draft of the manuscript, in
the study methodology (sequencing), in the writing, reviewing and
editing of the manuscript and in the formal analysis. AMAA, AA,
NJA, HMA, MAA and RAA were involved in the conception and design of
the study, provision of resources, data collection, analysis,
interpretation, drafting the article and critical revision of the
article. CC and SC were involved in the study methodology
(sequencing), in the formal analysis, and in the drafting and
critical revision of the article. SC and CC confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethical approval and consent to
participate
Institutional review board approval
(IRB-2019-01-378) was received from Imam Abdulrahman Bin Faisal
University. The procedures in the present study adhere to the
principles of the Declaration of Helsinki. Written informed consent
was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kuipers EJ, Grady WM, Lieberman D,
Seufferlein T, Sung JJ, Boelens PG, van de Velde CJH and Watanabe
T: Colorectal cancer. Nat Rev Dis Primers. 1(15065)2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hossain MS, Karuniawati H, Jairoun AA,
Urbi Z, Ooi J, John A, Lim YC, Kibria KMK, Mohiuddin AKM, Ming LC,
et al: Colorectal cancer: A review of carcinogenesis, global
epidemiology, current challenges, risk factors, preventive and
treatment strategies. Cancers (Basel). 14(1732)2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Saudi Health Council, National Cancer
Center and Saudi Cancer Registry: Cancer incidence report from
Saudi Arabia 2018. Saudi Health Council, 2022. https://shc.gov.sa/Arabic/NCC/Activities/AnnualReports/2018.pdf.
Accessed July 23, 2023.
|
|
5
|
World Health Organization (WHO): Cancer
Saudi Arabia 2020 country profile. WHO, Geneva, 2020. https://cdn.who.int/media/docs/default-source/country-profiles/cancer/sau-2020.pdf?sfvrsn=37936f36_2&download=true.
Accessed July 23, 2023.
|
|
6
|
Messersmith WA: NCCN guidelines updates:
Management of metastatic colorectal cancer. J Natl Compr Canc Netw.
17:599–601. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732.
2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Xie YH, Chen YX and Fang JY: Comprehensive
review of targeted therapy for colorectal cancer. Signal Transduct
Target Ther. 5(22)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Triantafillidis JK, Nasioulas G and
Kosmidis PA: Colorectal cancer and inflammatory bowel disease:
Epidemiology, risk factors, mechanisms of carcinogenesis and
prevention strategies. Anticancer Res. 29:2727–2737.
2009.PubMed/NCBI
|
|
10
|
Edwards BK, Ward E, Kohler BA, Eheman C,
Zauber AG, Anderson RN, Jemal A, Schymura MJ, Lansdorp-Vogelaar I,
Seeff LC, et al: Annual report to the nation on the status of
cancer, 1975-2006, featuring colorectal cancer trends and impact of
interventions (risk factors, screening, and treatment) to reduce
future rates. Cancer. 116:544–573. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cancer Genome Atlas Network. Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sepulveda AR, Hamilton SR, Allegra CJ,
Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C,
Lindor NM, Minsky BD, et al: Molecular biomarkers for the
evaluation of colorectal cancer: Guideline from the American
society for clinical pathology, college of American pathologists,
association for molecular pathology, and American society of
clinical oncology. Arch Pathol Lab Med. 141:625–657.
2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Krasinskas AM: EGFR signaling in
colorectal carcinoma. Patholog Res Int. 2011(932932)2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Novellasdemunt L, Antas P and Li VS:
Targeting Wnt signaling in colorectal cancer. A review in the
theme: Cell signaling: Proteins, pathways and mechanisms. Am J
Physiol Cell Physiol. 309:C511–C521. 2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Malapelle U, Pisapia P, Sgariglia R,
Vigliar E, Biglietto M, Carlomagno C, Giuffrè G, Bellevicine C and
Troncone G: Less frequently mutated genes in colorectal cancer:
Evidences from next-generation sequencing of 653 routine cases. J
Clin Pathol. 69:767–771. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Inamura K: Colorectal Cancers: An update
on their molecular pathology. Cancers (Basel).
10(26)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou R, Huang Y, Cheng B, Wang Y and Xiong
B: TGFBR1*6A is a potential modifier of migration and invasion in
colorectal cancer cells. Oncol Lett. 15:3971–3976. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Koveitypour Z, Panahi F, Vakilian M,
Peymani M, Seyed Forootan F, Nasr Esfahani MH and Ghaedi K:
Signaling pathways involved in colorectal cancer progression. Cell
Biosci. 9(97)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kwak MS, Cha JM, Yoon JY, Jeon JW, Shin
HP, Chang HJ, Kim HK, Joo KR and Lee JI: Prognostic value of KRAS
codon 13 gene mutation for overall survival in colorectal cancer:
Direct and indirect comparison meta-analysis. Medicine (Baltimore).
96(e7882)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hamada T, Nowak JA and Ogino S: PIK3CA
mutation and colorectal cancer precision medicine. Oncotarget.
8:22305–22306. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Samuels Y and Waldman T: Oncogenic
mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol.
347:21–41. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Al-Amri AM, Vatte C, Cyrus C, Chathoth S,
Hashim TM, Mohamed YS, Al Ali R, Alsaid A and Al Ali A: Novel
mutations of PIK3CA gene in head and neck squamous cell carcinoma.
Cancer Biomark. 16:377–383. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vatte C, Al Amri AM, Cyrus C, Chathoth S,
Alsayyah A, Ahmad A, Akhtar MS, Alrashidi NF, Jayaseeli N, Al
Wadani H, et al: Helical and kinase domain mutations of PIK3CA, and
their association with hormone receptor expression in breast
cancer. Oncol Lett. 18:2427–2433. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Qiu W, Tong GX, Manolidis S, Close LG,
Assaad AM and Su GH: Novel mutant-enriched sequencing identified
high frequency of PIK3CA mutations in pharyngeal cancer. Int J
Cancer. 122:1189–1194. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xi Y and Xu P: Global colorectal cancer
burden in 2020 and projections to 2040. Transl Oncol.
14(101174)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lu XQ, Li Y, Wang W, Feng WT, Shi OM and
Wang Q: International incidence trends in early- and late-onset
colorectal cancer: A population-based study. Int J Colorectal Dis.
35:1077–1086. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Saad El Din K, Loree JM, Sayre EC, Gill S,
Brown CJ, Dau H and De Vera MA: Trends in the epidemiology of
young-onset colorectal cancer: A worldwide systematic review. BMC
Cancer. 20(288)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zaborowski AM, Murphy B, Creavin B, Rogers
AC, Kennelly R, Hanly A, Martin ST, O'Connell PR, Sheahan K and
Winter DC: Clinicopathological features and oncological outcomes of
patients with young-onset rectal cancer. Br J Surg. 107:606–612.
2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ogino S, Chan AT, Fuchs CS and Giovannucci
E: Molecular pathological epidemiology of colorectal neoplasia: An
emerging transdisciplinary and interdisciplinary field. Gut.
60:397–411. 2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Reddy D, Ghosh P and Kumavath R:
Strophanthidin attenuates MAPK, PI3K/AKT/mTOR, and Wnt/β-catenin
signaling pathways in human cancers. Front Oncol.
9(1469)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang Y, Li J and Che G: Clinical
significance of PIK3CA gene in non-small-cell lung cancer: A
systematic review and meta-analysis. Biomed Res Int.
2020(3608241)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kumar R, Kumar R, Goel H, Kumar S,
Ningombam SS, Haider I, Agrawal U, Deo S, Gogia A, Batra A, et al:
Whole exome sequencing identifies novel variants of PIK3CA and
validation of hotspot mutation by droplet digital PCR in breast
cancer among Indian population. Cancer Cell Int.
23(236)2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Giannakis M, Mu XJ, Shukla SA, Qian ZR,
Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, et
al: Genomic correlates of immune-cell infiltrates in colorectal
carcinoma. Cell Rep. 15:857–865. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yaeger R, Chatila WK, Lipsyc MD, Hechtman
JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A,
Donoghue MTA, et al: Clinical sequencing defines the genomic
landscape of metastatic colorectal cancer. Cancer Cell.
33:125–136.e3. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Voutsadakis IA: The landscape of PIK3CA
mutations in colorectal cancer. Clin Colorectal Cancer. 20:201–215.
2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Samuels Y, Wang Z, Bardelli A, Silliman N,
Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al:
High frequency of mutations of the PIK3CA gene in human cancers.
Science. 304(554)2004.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gong J, Cho M, Sy M, Salgia R and Fakih M:
Molecular profiling of metastatic colorectal tumors using
next-generation sequencing: A single-institution experience.
Oncotarget. 8:42198–42213. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Shetty O, Vengurlekar V, Kapoor A, Kamble
V, Gurav M, Bhargava P, Srinivas S, Ramaswamy A, Ramadwar M,
Saklani AP, et al: The prevalence of BRAF, PIK3CA, and RAS
mutations in indian patients with colorectal cancer. South Asian J
Cancer. 11:190–194. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Reggiani Bonetti L, Barresi V, Maiorana A,
Manfredini S, Caprera C and Bettelli S: Clinical impact and
prognostic role of KRAS/BRAF/PIK3CA mutations in stage I colorectal
cancer. Dis Markers. 2018(2959801)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li X, Yang T, Li CS, Song Y, Lou H, Guan D
and Jin L: Surface enhanced raman spectroscopy (SERS) for the
multiplex detection of Braf, Kras, and Pik3ca mutations in plasma
of colorectal cancer patients. Theranostics. 8:1678–1689.
2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Jhawer M, Goel S, Wilson AJ, Montagna C,
Ling YH, Byun DS, Nasser S, Arango D, Shin J, Klampfer L, et al:
PIK3CA mutation/PTEN expression status predicts response of colon
cancer cells to the epidermal growth factor receptor inhibitor
cetuximab. Cancer Res. 68:1953–1961. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Sartore-Bianchi A, Martini M, Molinari F,
Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P,
De Dosso S, Mazzucchelli L, et al: PIK3CA mutations in colorectal
cancer are associated with clinical resistance to EGFR-targeted
monoclonal antibodies. Cancer Res. 69:1851–1857. 2009.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Barault L, Veyrie N, Jooste V, Lecorre D,
Chapusot C, Ferraz JM, Lièvre A, Cortet M, Bouvier AM, Rat P, et
al: Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH
kinase) signaling network correlate with poor survival in a
population-based series of colon cancers. Int J Cancer.
122:2255–2259. 2008.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Ogino S, Nosho K, Kirkner GJ, Shima K,
Irahara N, Kure S, Chan AT, Engelman JA, Kraft P, Cantley LC, et
al: PIK3CA mutation is associated with poor prognosis among
patients with curatively resected colon cancer. J Clin Oncol.
27:1477–1484. 2009.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Foltran L, De Maglio G, Pella N, Ermacora
P, Aprile G, Masiero E, Giovannoni M, Iaiza E, Cardellino GG,
Lutrino SE, et al: Prognostic role of KRAS, NRAS, BRAF and PIK3CA
mutations in advanced colorectal cancer. Future Oncol. 11:629–640.
2015.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Phipps AI, Ahnen DJ, Cheng I, Newcomb PA,
Win AK and Burnett T: PIK3CA somatic mutation status in relation to
patient and tumor factors in racial/ethnic minorities with
colorectal cancer. Cancer Epidemiol Biomarkers Prev. 24:1046–1051.
2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Dirican E, Akkiprik M and Özer A: Mutation
distributions and clinical correlations of PIK3CA gene mutations in
breast cancer. Tumour Biol. 37:7033–7045. 2016.PubMed/NCBI View Article : Google Scholar
|