Introduction
Essential thrombocythemia (ET) is the most common
subtype of classic Philadelphia chromosome negative bone marrow
proliferation tumor (MPN). Bone marrow fibrosis, thrombus and risk
of bleeding are the most common complications of ET. Approximately
10% of patients have acute leukemia transformation (1). In recent years, the ET rate has been on
the increase annually, and approximately 5–8/10 million individuals
were diagnosed with ET in 2005 (2).
ET may be the result of combined effects of gene and
environment. JAK2V617F mutations can occur in 50–60% of ET
patients, and the mutation-positive patients have a higher risk of
thromboembolism. Reverse positive mutation status was negatively
correlated with survival outcomes and serious adverse event
occurrence, suggesting that the JAK2V617F mutation plays an
important role in the pathogenesis of ET (3,4).
However, 50% of patients show a JAK2V617F-negative state. Recently,
gene sequencing identified that MPLW515L/K and CALR gene of
mutations can compensate for the deficiency of JAK2V617F
negativity, which can provide an important basis for explaining
genetic pathogenesis mechanism of ET (5,6).
The aim of the present study was to analyze the
mutation rate of JAK2V617F, MPLW515L/K and CALR genes in adult
patients with essential thrombocythemia (ET) and the accuracy of
the combined detection by the receiver operating curve.
Patients and methods
Patients
A total of 342 patients were selected consecutively
in the Second Affiliated Hospital of Wenzhou Medical University
with simple high platelet (≥300×109/l) from January 2013
to January 2016. The platelet count was continuously monitored on
weekly basis to take mean value. Exclusion criteria were: Patients
with a high or low level of white blood cells, red blood cells or
hemoglobin, history of recent surgery, bleeding, blood transfusion,
pregnancy, malignant tumor, blood diseases such as leukemia and
myeloproliferative disorders, severe hepatic and renal dysfunction,
or those combined with nerve or spirit system diseases.
Furthermore, patients with poor compliance, those who could not
undergo surgery following various examinations, e.g., bone marrow
biopsy and genetic testing, and those with incomplete data were
excluded. All the patients were tested for detection of blood, bone
marrow biopsy and the genes. The study was approved by the Ethics
Committee of the Wenzhou Medical University. Patients or their
family members provided written informed consent.
According to the hematopoietic and lymphoid tissue
tumor on ET classification standards, 2008 (7): i) platelet count ≥450×109/l;
ii) bone marrow biopsy showed that mature and volume increased
megakaryocyte increase dominated, with no obvious increase of
neutrophils or polycythemia; iii) excluded were those with other
myeloproliferative disorders, such as polycythemia vera, primary
myelofibrosis and chronic myeloid leukemia; and iv) acquired
JAK2V617F mutation or other clonal marker, or secondary
thrombocythemia with no clonal markers. One hundred and fifty-four
cases were diagnosed with ET (45.03%) and 188 patients were
diagnosed with secondary thrombocythemia. In the ET group, 86
patients were male and 68 female, aged 38–69 years and median age
of 53.4 years and platelet of 463–3,547×109/l with
average of 865.3±65.4×109/l. Ninety-eight cases were
male and 90 cases were female in the secondary thrombocythemia
group, aged 34–72 years and median age of 56.3 years, platelets of
357–2,451×109/l and average of
73.28±82.3×109/l. There were no significant differences
in the gender, age, and platelet count between the two groups
(p>0.05).
Gene detection method
Mutations in JAK2V617F and MPLW515L/K gene were
detected by allele-specific polymerase chain reaction (AS-PCR).
Reagents and equipment used were DNA extraction kit (Dynal, Oslo,
Norway), PTC-200™ PCR instrument (MJ Research, Inc., Waltham, MA,
USA), CS-6R centrifuge (Beckman Coulter, Inc., Brea, CA, USA), the
gel imaging analysis system (Transilluminator 202D; Cold Spring
Harbor Laboratory, Cold Spring Harbor, NY, USA).
The main steps are as follows: i) DNA extraction: a)
500 µl peripheral blood was extracted in a 2-ml EP tube. One
milliliter of red blood cell lysis was added at 4°C and centrifuged
at 8,000 × g for 1 min; b) the supernatant was discarded and
residual white lump in the pipe bottom was retained. Protease (125
µl) was added with gentle mixing for 5 sec. Subsequently, the tube
was placed in 65°C water bath for 10 min and gently mixed every 2
min until the precipitation was completely dissolved; iii) 275 µl
of protein clear solution was added with gentle mixing for 5 sec.
It was followed by incubation at −20°C for about 10 min and then
centrifuged at 10,000 × g for 5 min; d) the supernatant was
discarded, and 1 ml of 70% ethanol was added with gentle mixing for
5 sec. The tube contents were then centrifuged at 10,000 × g for 1
min; e) the ethanol was discarded, and the DNA containing rube was
placed at room temperature for 2 min for volatilization of extra
alcohol. Subsequently, 100 µl DNA dissolving solution was added and
then tube was incubated at 65°C for 15 min to fully dissolve DNA;
and f) DNA concentration and purity were measured with UV
spectrophotometer.
ii) Primer sequences were designed for two forward
primers and a reverse universal primer. PCR product was 203 bp and
the reference was 364 bp. Primers were produced by Shanghai
Biological Engineering Co., Ltd. (Shanghai, China). Forward
specific, 5′-AGCATTTGGTTTTAAATTATGGAGTATATT-3′; forward internal,
5′-ATCTATAGTCATGCTGAAAGTAGGAGAAAG-3′ and reverse,
5′-CTGAATAGTCCTACAGTGTTTTCAGTTTCA-3′.
iii) PCR reaction: 25 ng of DNA (10 mmol/l), 0.2 of
forward primer and 0.4 µl (10 mmol/l) reverse primer, 12.5 Master
mix and double ionized water were added for final volume of 25 µl.
PCR amplification cycle parameters were pre-denaturation at 95°C
for 4 min, denaturation at 95°C for 30 sec, annealing at 55°C for
30 sec, extension at 72°C for 1 min, 35 cycles, final extension at
72°C for 7 min. 1.5% agarose gel containing ethidium bromide was
prepared for the analysis. Five microliters of PCR products and 1X
TBE buffer was mixed and added for electrophoresis at 90 V for 40
min. The image was taken and analyzed with UV perspective image
instrument.
iv) Gene sequencing: positive specimens were
retrieved and purified after PCR amplification with the outside
primers. Product was sent to Shanghai Yingjun Biotechnology Co.
Ltd. (Shanghai, China) for sequencing. The result underwent
alignment with the wild-type gene sequence. Semi-quantitative
analysis results were shown as target gene copy
number/corresponding wild-type gene copy number. MPLW515 primer
sequences: forward primer M1, 5′-AGTAGGGGCTGGCTGGAT-3′ (409 bp) and
reverse M2, 5′-CTAGTCGCCGAGGTGAGC-3′ (409 bp); specific primer M3,
5′-CCTGCTGCTGCTGAGGTTGC-3′ (279 bp) and reverse M2,
5′-CTAGTCGCCGAGGTGAGC-3′ (279 bp).
PCR reaction mixture consists of DNA 25 ng, 12.5 µl
of 2X Taq PCR Master mix [KT201; Tiangen Biotech (Beijing) Co.,
Ltd., Beijing, China], 0.25 µl for each M1, M3 primers (10 mmol/l),
0.5 µl of M2 primer (10 mmol/l) and deionized water to make up
final volume of 25 µl. PCR parameters and gene sequencing were
similar to those above.
CALR gene mutation was detected by direct sequencing
method. QIAamp DNA Mini kit (Qiagen, Valencia, CA, USA), exon 9
primers (Invitrogen, Carlsbad, CA, USA) of 5.0 pmol/µl.
Primer sequence was: forward,
5′-CTGGTCCTGGTCCTGATGT-3′ and reverse,
5′-TCTCACAGAGACATTATTTGGC.
PCR reaction mixture consists of 50 ng DNA, 15 µl 2X
Taq PCR Master mix [KT201; Tiangen Biotech (Beijing) Co., Ltd.],
0.5 µl for each primer and deionized water was added to make up
final volume of 30 µl. PCR conditions were 98°C for 3 min, 98°C for
10 sec, 63°C for 30 sec, 72°C for 30 sec for 29 cycles, and final
extension at 72°C for 5 min. Gene sequencing was the same as
above.
Observation index
The expression of JAK2V617F, MPLW515L/K and CALR
gene positive mutation rate in two groups and the relative
expression levels were measured.
Statistical analysis
SPSS 19.0 statistical software (Chicago, IL, USA)
was used for data processing. Measurement data are presented as
mean ± standard deviation, the comparison between groups is shown
using t-test. Countable data are presented as percentage,
comparison between the groups were with χ2 test. The
diagnostic sensitivity, specificity and accuracy was analyzed using
the receiver operating curve (ROC). P<0.05 indicates a
statistically significant difference.
Results
Sequencing results of three gene
mutations
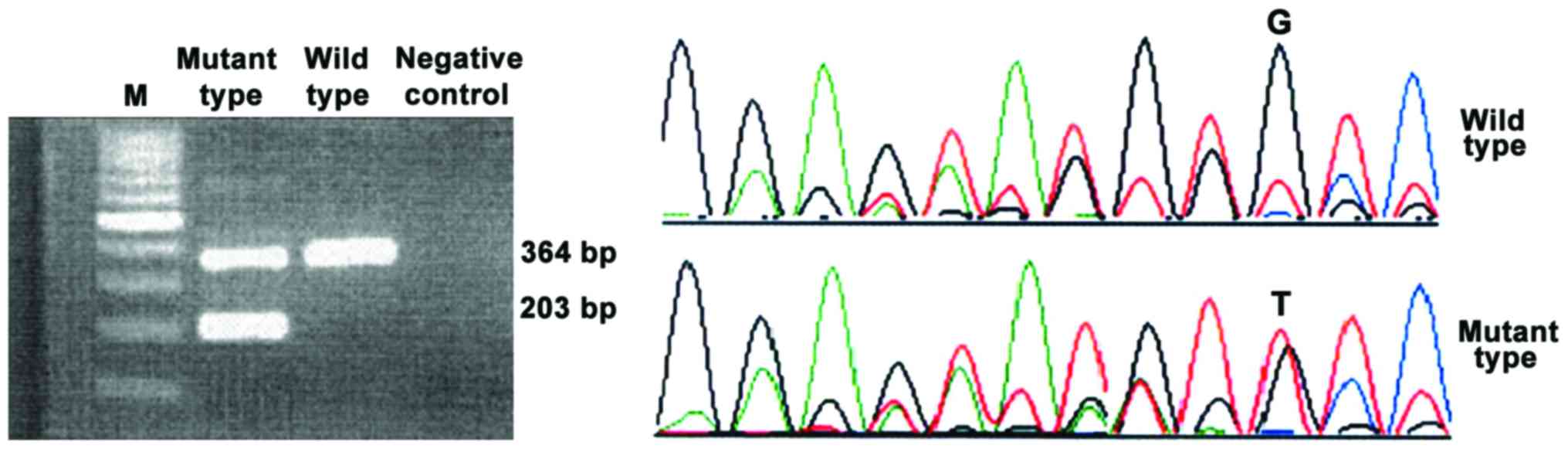
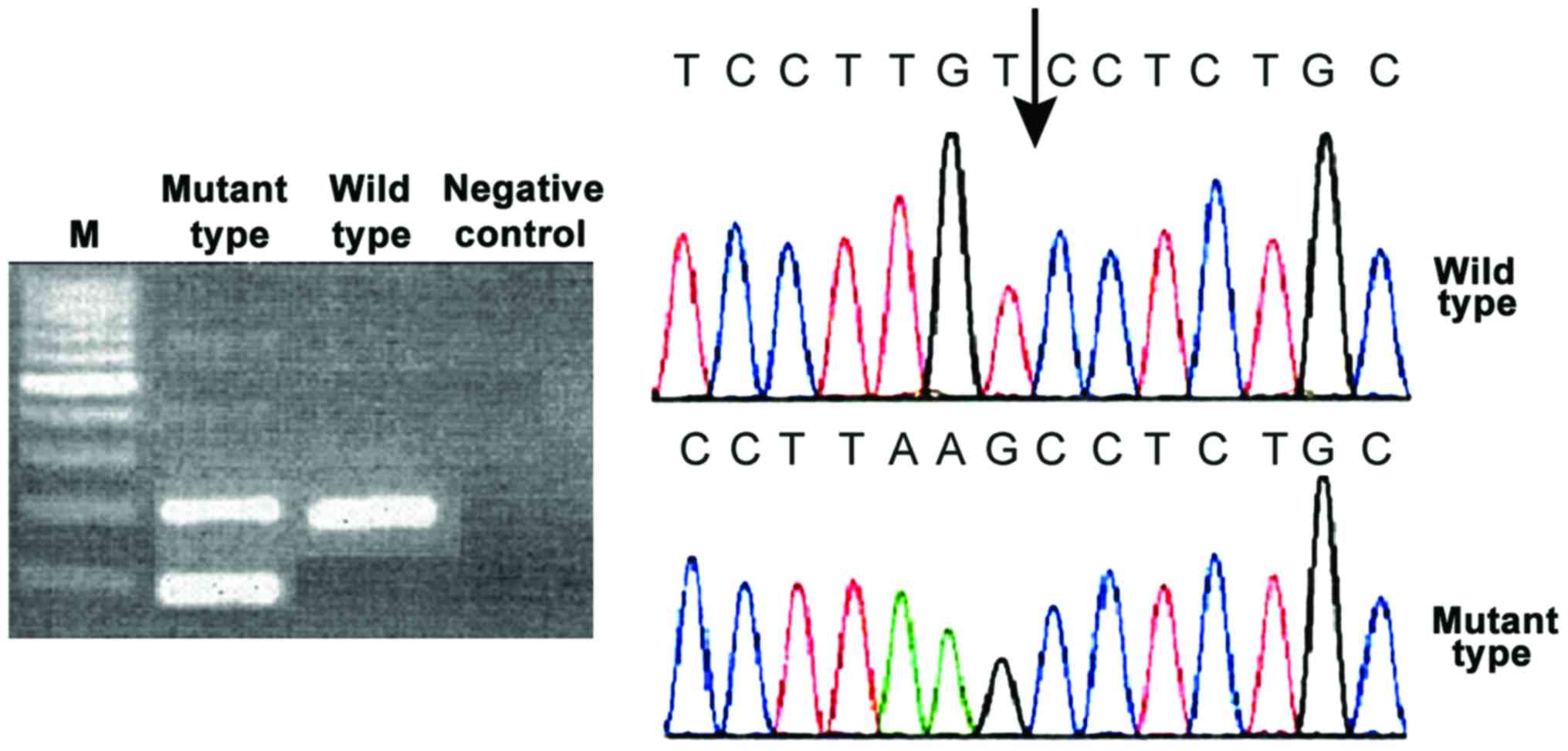
The mutant type of the three genes showed two
electrophoretic bands and the wild-type showed only one band.
JAK2V617F was located in exon 12. The first base G was replaced by
T and the codon was changed from V to F. Only V617 codon was
affected by the point mutation that causes missense mutation, which
did not change the open reading frame. Open reading frame of MPL
was not changed either. CALR gene mutations have both 52 bp
deletion and 5 bp insertion. All mutations can cause the change of
the open reading frame. C amino acid sequence of CALR protein was
changed (Figs. 1–3).
There were 89 (57.79%) cases of JAK2V617F mutations
in the ET group, 7 (4.55%) cases of MPL mutations and 48 (31.17%)
cases of CALR mutations. CALR and JAK2V617F mutations at the same
time were positive in 7 cases (4.55%) and negative in 24 cases
(15.58%). JAK2V617F positive with CALR negative were observed in 82
cases (53.25%), 41 cases (26.62%) were JAK2V617F negative and CALR
positive. In addition, 6 (3.19%) cases were detected with JAK2V617F
mutation in the secondary thrombocythemia group, 0 cases of MPL
mutation, and 1 case (0.53%) of CALR mutation. The positive
mutation rate of JAK2V617F and CALR in the ET group was
significantly higher than that in the secondary thrombocythemia
group, and the difference was statistically significant
(χ2=125.800, p<0.001; χ2=64.734,
p<0.001).
ROC analysis
The JAK2V617F average expression level in patients
with positive JAK2V617F and CALR mutation at the same time was
0.34±0.06. The CALR average expression level was 0.15±0.04. Also
the average expression levels in patients with negative mutations
at the same time were 0.07±0.01 and 0.03±0.01, respectively. The
average expression levels in patients with positive JAK2V617F
mutation and negative CALR mutation were 0.58±0.04 and 0.04±0.01,
0.08±0.02 and 0.22±0.03 in patients with negative JAK2V617F
mutation and positive CALR mutation.
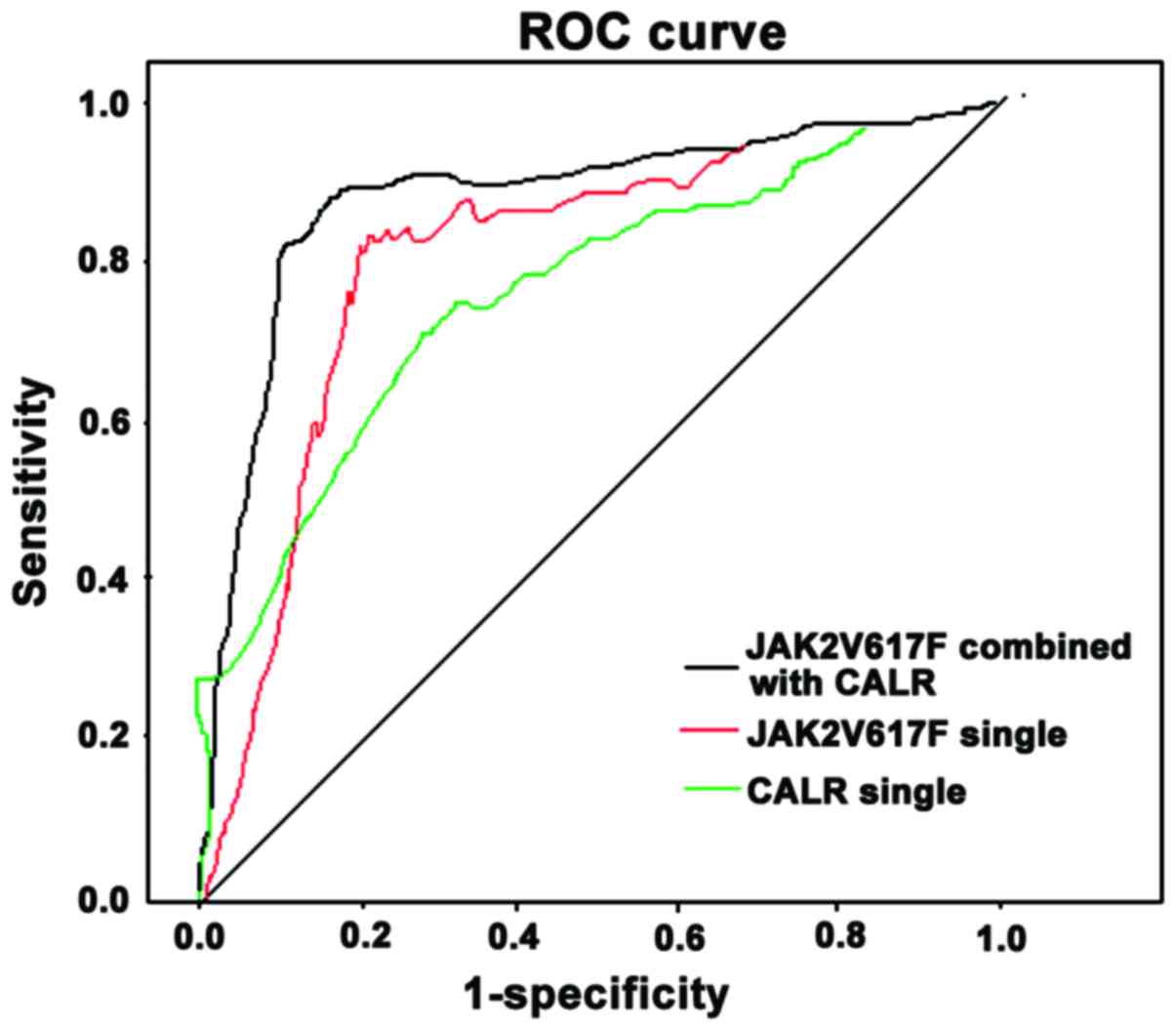
When JAK2V617F mutation positive was the only
diagnostic criteria, the area under the curve (AUC) was 0.721, 95%
confidence interval (CI) of 0.356–0.925, sensitivity of 72.4%,
specificity of 79.5% and the cut-off of 0.25. When CALR mutation
positive was the only diagnostic criteria, the AUC was 0.664, 95%
CI of 0.291–0.848 with 68.4% sensitivity, 82.4% specificity. The
cut-off was 0.09. When the combined positive CALR and JAK2V617F
mutation was used for diagnosis, AUC was 0.862. 95% CI was
0.467–0.963. The sensitivity was 85.9%. The specificity was 87.8%
and the cut-off was 0.21 and 0.07, respectively (Fig. 4).
Discussion
JAK is a class of non-receptor tyrosine protein
kinase. The gene encoding JAK2 is on chromosome 9 (9p24) and it is
expressed in almost all tissues. The most obvious structure of JAK
is characterized in that the C terminal has two sections of
catalytic and N terminal has three conserved structural sections.
The middle part has two sections. Downstream of JAK signal is
protein sub-family of signal transduction and transcriptional
activation (STAT). JAK can bind with the conserved BOX1 and BOX2 of
cytokine receptor and identify the motif in the cytokine receptor
juxtamembrane region. It occurs in a series of phosphorylation
under the stimulation of receptors and ligands and selective
activate downstream substrates STAT, which translocates to the
nucleus and binds with the nuclear specific DNA regulatory elements
so as to guide transcription, which is the JAK-STAT pathway
(8). The identification of JAK2V617F
provides an important clue to explore the pathogenesis of ET.
MPL is thrombopoietin receptor and its peptide
sequence has 633 amino acids. Two types of somatic cell mutations
MPLW515L/K were confirmed to exist in 5 and 1% of patients with
negative JAK2V617F bone marrow fibrosis (9). MPLW515 was located in the only
concurrent region of MPL, which can inhibit the MPL gene
spontaneous activation. MPLW515L mutation is located in the
suppressed sequence of cytoplasmic and transmembrane region of MPL
gene sequence of KWQFP. The mutations within this region can damage
the inhibitory function of KWQFP sequence and thus constitutively
activate the JAK-STAT signal transduction pathway (10). CALR is Ca2+ binding
protein complex, mainly located in the endoplasmic reticulum lumen
and the mediated Ca2+ homeostasis can regulate a variety
of cellular functions, including the differentiation and maturation
of megakaryocytes and platelet formation, integrin mediated signal
transduction, immune response, cell apoptosis, proliferation and
phagocytosis, wound healing and fibrosis (11). Kampfl et al (12) and Nangalia et al (13) detected CALR exon 9 mutation in most
of the JAK2 and MPL negative samples using sequencing method, while
CALR gene mutation was not found in a healthy population, lymphatic
system tumor, acute leukemia or solid tumors.
We concluded through the study that JAK2V617F and
MPL mutations do not cause frameshift, and CALR mutations can lead
to the change of open reading. The JAK2V617F and CALR gene mutation
rates in the ET group were significantly higher than those in the
group of secondary thrombocythemia and the positive MPL mutation
rate was only 4.55%. MPL lower positive mutation rate may be
related to racial factors (14) and
sample size. The innovation of the study is that the three kinds of
genes in MPN had certain positive mutation rates based on a
previous study (15). Gene analysis
for outpatient service with simple increased platelets indicates
that the CALR and JAK2V617F mutation can be a specific marker of ET
screening. The shortcoming of the study was that it did not involve
other types of MPN, which cannot compare the CALR and JAK2V617F
mutation in different types of MPN. In addition, the study detected
the relative expression levels of JAK2V617F and CALR through
quantitative analysis and set positive JAK2V617F, CALR mutation
alone and combination as diagnostic criteria respectively, showed
that combination use of those two can improve obviously the
diagnostic sensitivity, specificity and accuracy. It provides an
important reference basis for early clinical recognition.
In conclusion, with the increased JAK2V617F- and
CALR-positive mutation rate, the combination of both can
significantly improve the sensitivity, specificity and accuracy of
the diagnosis of ET.
Acknowledgements
This study was supported by the National Natural
Science Fund (81472651/H1604).
References
|
1
|
Barbui T, Thiele J, Vannucchi AM and
Tefferi A: Rationale for revision and proposed changes of the WHO
diagnostic criteria for polycythemia vera, essential
thrombocythemia and primary myelofibrosis. Blood Cancer J.
5:e3372015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haider M, Gangat N, Lasho T, Hussein AK
Abou, Elala YC, Hanson C and Tefferi A: Validation of the revised
international prognostic score of thrombosis for essential
thrombocythemia (IPSET-thrombosis) in 585 Mayo clinic patients. Am
J Hematol. 91:390–394. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liew EL, Araki M, Hironaka Y, Mori S, Tan
TZ, Morishita S, Edahiro Y, Ohsaka A and Komatsu N: Identification
of AIM2 as a downstream target of JAK2V617F. Exp Hematol Oncol.
5:22016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mahjoub S, Baccouche H, Sahnoun M, Kaabi
H, Manai Z, Slama H and Ben Romdhane N: The JAK2 mutation in
myeloproliferative disorders: A predictive factor of thrombosis.
Tunis Med. 93:474–477. 2015.(In French). PubMed/NCBI
|
|
5
|
Okabe M, Yamaguchi H, Usuki K, Kobayashi
Y, Kawata E, Kuroda J, Kimura S, Tajika K, Gomi S, Arima N, et al:
Clinical features of Japanese polycythemia vera and essential
thrombocythemia patients harboring CALR, JAK2V617F, JAK2Ex12del,
and MPLW515L/K mutations. Leuk Res. 40:68–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marty C, Pecquet C, Nivarthi H, Elkhoury
M, Chachoua I, Tulliez M, Villeval JL, Raslova H, Kralovics R,
Constantinescu SN, et al: Calreticulin mutants in mice induce an
MPL-dependent thrombocytosis with frequent progression to
myelofibrosis. Blood. 127:1317–1324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tefferi A, Thiele J and Vardiman JW: The
2008 World Health Organization classification system for
myeloproliferative neoplasms: Order out of chaos. Cancer.
115:3842–3847. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Čokić VP, Mitrović-Ajtić O, Beleslin-Čokić
BB, Marković D, Buač M, Diklić M, Kraguljac-Kurtović N, Damjanović
S, Milenković P, Gotić M, et al: Proinflammatory cytokine IL-6 and
JAK-STAT signaling pathway in myeloproliferative neoplasms.
Mediators Inflamm. 2015:4530202015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cabagnols X, Favale F, Pasquier F,
Messaoudi K, Defour JP, Ianotto JC, Marzac C, Le Couédic JP, Droin
N, Chachoua I, et al: Presence of atypical thrombopoietin receptor
(MPL) mutations in triple-negative essential thrombocythemia
patients. Blood. 127:333–342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feenstra JD Milosevic, Nivarthi H,
Gisslinger H, Leroy E, Rumi E, Chachoua I, Bagienski K, Kubesova B,
Pietra D, Gisslinger B, et al: Whole-exome sequencing identifies
novel MPL and JAK2 mutations in triple-negative myeloproliferative
neoplasms. Blood. 127:325–332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Verger E, Cassinat B, Chauveau A, Dosquet
C, Giraudier S, Schlageter MH, Ianotto JC, Yassin MA, Al-Dewik N,
Carillo S, et al: Clinical and molecular response to interferon-α
therapy in essential thrombocythemia patients with CALR mutations.
Blood. 126:2585–2591. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Klampfl T, Gisslinger H, Harutyunyan AS,
Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B,
Pietra D, et al: Somatic mutations of calreticulin in
myeloproliferative neoplasms. N Engl J Med. 369:2379–2390. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nangalia J, Massie CE, Baxter EJ, Nice FL,
Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, et al:
Somatic CALR mutations in myeloproliferative neoplasms with
nonmutated JAK2. N Engl J Med. 369:2391–2405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim SY, Im K, Park SN, Kwon J, Kim JA and
Lee DS: CALR, JAK2, and MPL mutation profiles in patients with four
different subtypes of myeloproliferative neoplasms: primary
myelofibrosis, essential thrombocythemia, polycythemia vera, and
myeloproliferative neoplasm, unclassifiable. Am J Clin Pathol.
143:635–644. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rumi E, Pietra D, Ferretti V, Klampfl T,
Harutyunyan AS, Milosevic JD, Them NC, Berg T, Elena C, Casetti IC,
et al: Associazione Italiana per la Ricerca sul Cancro Gruppo
Italiano Malattie Mieloproliferative Investigators: JAK2 or CALR
mutation status defines subtypes of essential thrombocythemia with
substantially different clinical course and outcomes. Blood.
123:1544–1551. 2014. View Article : Google Scholar : PubMed/NCBI
|