Introduction
Gastric cancer is among the most common types of
human cancer and has a poor 5-year survival rate, primarily due to
its high rates of recurrence and metastasis (1,2).
Although advances in the diagnosis and treatment of gastric cancer
have been made, gastric cancer remains to be among the top five
cancers worldwide in terms of cancer-related mortality (1–3). Thus,
elucidation of the molecular mechanisms underlying the development
and progression of gastric cancer is required to develop effective
strategies for its treatment (4,5).
MicroRNAs (miRs), as a class of small non-coding
RNAs of 18–25 nucleotides in length, serve as regulators of gene
expression by binding directly to the 3′-untranslational region
(UTR) of their target mRNAs, which leads to mRNA degradation and/or
translation inhibition (6,7). MiRs have been found to participate in
the regulation of various biological processes, including cell
proliferation, differentiation, apoptosis and motility and cell
cycle progression (7,8). Moreover, many oncogenes and tumor
suppressor genes are the targets of miRs, and thus miRs serve key
roles in various human cancers, including gastric cancer (9–12). For
instance, a previous study observed that miR-126 was significantly
downregulated in gastric cancer, and its downregulation was
associated with malignant progression (13). MiR-30a-5p, as a member of the miR-30
family, was recently observed to be significantly downregulated in
gastric cancer due to high promoter methylation induced by DNA
methyltransferase 1 (14). Moreover,
reduced expression of miR-30a-5p has been associated with a high
rate of lymph node metastasis (14).
These data suggest that miR-30a-5p may serve a suppressive role in
gastric cancer. However, data on the regulatory role of miR-30a-5p
in the growth and metastasis of gastric cancer are limited.
Insulin-like growth factor 1 receptor (IGF-1R) is a
key member of the IGF receptor family, and exhibits a high affinity
for insulin-like growth factor (15). Previous studies have indicated that
IGF-1R may be involved in malignant transformation events (16,17).
IGF-1R is expressed to a high level in various human cancer
tissues, where it promotes the survival and inhibits the apoptosis
of cancer cells, indicating that IGF-1R may be a key oncogene and
therapeutic target in the treatment of human cancers, including
gastric cancer (15–19). Recently, Ge et al (20) reported that inhibition of IGF-1R with
small interfering (si)-RNA inhibited the proliferation, cell cycle
progression, migration and invasion of gastric cancer cells, and
led to their apoptosis. However, the regulatory mechanism of IGF-1R
expression remains unknown, and thus insight into the regulation of
IGF-1R may aid to develop more effective strategies for the
treatment of gastric cancer.
Therefore, the present study aimed to elucidate the
molecular mechanism of miR-30a-5p in regulating the proliferation
and invasion of gastric cancer cells.
Materials and methods
Tissue specimen collection
The present study was approved by the Ethics
Committee of the Second Xiangya Hospital of Central South
University (Changsha, China). A total of 43 gastric cancer tissues
and 10 matched adjacent normal gastric tissues were obtained from
patients at the Second Xiangya Hospital of Central South University
from January 2013 to December 2013. Written informed consent was
obtained from all patients in the current study. The patients
included 18 females and 25 males with a mean age of 54.2±7.8 years.
Cancers were classified as T1 or T2-T4 stage (21). The clinical information of gastric
cancer patients is summarized in Table
I. All tissues were immediately snap-frozen in liquid nitrogen
following surgical removal and stored at −80°C until use.
 | Table I.Clinical characteristics of patients
with gastric cancer. |
Table I.
Clinical characteristics of patients
with gastric cancer.
| Variables | Number of patients
with T1 (from 13 patients) | Number of patients
with T2-T4 (from 30 patients) |
|---|
| Age |
|
|
| ≤55 | 4 | 11 |
|
>55 | 9 | 19 |
| Sex |
|
|
| Male | 8 | 17 |
|
Female | 5 | 13 |
Cell culture
Four common human gastric cancer cell lines, AGS,
HGC27, BGC823 and SGC7901, and the normal gastric mucosa epithelial
cell line GES-1 were purchased from the Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences (Shanghai, China).
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal
bovine serum (FBS; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified incubator containing 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from gastric tissue and
cells with TRIzol® reagent (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Total RNA was then
converted into cDNA using a High Capacity cDNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Reverse transcription was performed at
16°C for 30 min, followed by an incubation at 42°C for 30 min and
enzyme inactivation at 85°C for 5 min. PCR was subsequently
performed to measure the expression of miR-30a-5p using an miRNA
Q-PCR Detection kit and SYBR-Green (both from GeneCopoeia, Inc.,
Rockville, MD, USA) on an ABI 7500 thermocycler (Thermo Fisher
Scientific, Inc.) under the following conditions: 95°C for 10 min,
followed by 45 cycles of 95°C for 15 sec and 60°C for 15 sec. The
U6 gene (primer sequences not provided by manufacturer) was used as
an internal reference. The expression of IGF-1R mRNA was detected
by qPCR using a standard SYBR-Green RT-PCR kit (Takara Bio, Inc.,
Otsu, Japan), in accordance with the manufacturer's instructions.
GAPDH was used as an internal reference. The specific primers used
to detect IGF-1R were as follows: Forward,
5′-ATGCTGACCTCTGTTACCTCT-3′ and reverse,
5′-GGCTTATTCCCCACAATGTAGTT-3′. The specific primers used for GAPDH
were as follows: Forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse,
5′-AAGTGGTCGTTGAGGGCAATG-3′. The reaction was performed at 95°C for
5 min, followed by 40 cycles of denaturation at 95°C for 15 sec and
an annealing/elongation step at 60°C for 30 sec. The mRNA
expression levels of miR-30a-5p and IGF-1R were normalized to that
of U6 and GAPDH, respectively. Relative expression was analyzed by
the 2-ΔΔCq method (22).
PCR was performed in triplicate.
Western blot analysis
Cells were solubilized in cold
radioimmunoprecipitation assay lysis buffer (Thermo Fisher
Scientific, Inc.). The concentration of protein was determined
using a bicinchoninic acid (BCA) Protein Assay kit (Pierce; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. The protein was extracted by centrifugation at 12,000
× g for 20 min at 4°C. Proteins (50 µg) were loaded and separated
using 10% SDS-PAGE and transferred onto a polyvinylidene difluoride
membrane (Thermo Fisher Scientific, Inc.). The membrane was
incubated with phosphate-buffered saline (PBS) containing 5%
skimmed milk (China Mengniu Dairy Co., Ltd., Hong Kong, China)
overnight at 4°C, followed by incubation with rabbit anti-IGF-1R
monoclonal antibody (1:500; sc-7952) or rabbit anti-GAPDH
monoclonal antibody (1:500; sc-367714; both from Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at room temperature for 3 h.
Following three washes with PBS, the membrane was incubated with
mouse anti-rabbit IgG conjugated to alkaline phosphatase (1:5,000;
sc-2358; Santa Cruz Biotechnology, Inc.) at room temperature for 1
h. An enhanced chemiluminescence kit (Pierce; Thermo Fisher
Scientific, Inc.) was then used to perform chemiluminescent
detection, according to the manufacturer's instructions. Protein
expression was analyzed using Image-Pro Plus software 6.0 (Media
Cybernetics, Inc., Rockville, MD, USA). Relative protein expression
was represented as a density ratio compared to that of GAPDH. The
experiments were performed in triplicate.
Transfection
Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) was used to perform cell transfection according
to the manufacturer's instructions. For miR-30a-5p and IGF-1R
function analysis, AGS cells (1×105 cells) were cultured
to 70% confluence in DMEM supplemented with 10% FBS at 37°C in a
humidified incubator containing 5% CO2. Subsequently, cells were
transfected with 100 nm negative control miR (miR-NC group),
miR-30a-5p mimic (miR-30a-5p group; all from GeneCopoeia, Inc.) or
miR-30a-5p mimic and IGF-1R plasmid (miR-30a-5p+IGF-1R group;
Amspring, Changsha, China), respectively. AGS cells alone were
considered as the non-transfected control group.
MTT assay
Cell proliferation was determined using an MTT
assay. AGS cells were cultured in 96-well plates (104
cells/plate) with DMEM supplemented with 10% FBS at 37°C for 24 h.
A total of 100 µl DMEM containing 0.5 g/l MTT was added to each
well. Following incubation at 37°C for 12, 24, 48 and 72 h, the
medium was removed by aspiration and 50 µl dimethyl sulfoxide
(DMSO) was added. Following incubation at 37°C for 10 min, the
absorbance of each sample at 570 nm was measured using a microplate
reader (Tecan Infinite M200; Tecan Group Ltd., Männedorf,
Switzerland).
Cell invasion assay
A Transwell assay was conducted to evaluate cell
invasion using Transwell chambers pre-coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA). A cell suspension
(2×105 cells/ml) was prepared in serum-free DMEM, and
300 µl of the cell suspension was added into the upper chamber,
while 300 µl DMEM supplemented with 10% FBS was added into the
lower chamber. Following culture at 37°C for 24 h, a cotton-tipped
swab was used to wipe out the cells that did not migrate through
the filter. The filter was then fixed at room temperature for 20
min in 90% alcohol, and cells were stained with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The invaded cells
were photographed under an inverted light microscope (Olympus
Corporation, Tokyo, Japan). A total of 0.5 g/l MTT was then added
and incubated at 37°C for 4 h. The medium containing MTT was
subsequently removed, and 50 µl DMSO was added to each well.
Following incubation at 37°C for 10 min, the optical density (OD)
at 570 nm was measured using a microplate reader (Tecan Infinite
M200; Tecan Group Ltd.). The relative cell invasive capacity was
determined by the following equation: Relative cell invasive
capacity = OD value/OD value of the control group.
Bioinformatics prediction
TargetScan software 3.1 (www.targetscan.org) was used to predict the putative
target genes of miR-30a-5p, according to the manufacturers
instructions.
Dual luciferase reporter assay
A wild-type (WT) IGF-1R 3′UTR containing the binding
sequences of miR-30a-5p and a mutant type (MUT) IGF-1R 3′UTR
lacking the binding sequences of miR-33b were supplied by Yearthbio
(Changsha, China), and a Directed Mutagenesis kit (Stratagene;
Agilent Technologies, Inc., Santa Clara, CA, USA) was used in
accordance with the manufacturer's instructions. The WT or MUT
IGF-1R 3′UTR were subcloned into a psiCHECK-2 vector (Promega
Corporation, Madison, WI, USA) downstream of the Renilla
luciferase gene. AGS cells were cultured in DMEM with 10% FBS at
37°C in a humidified incubator containing 5% CO2 to 60–70%
confluence, then co-transfected with the luciferase reporter
vectors and miR-30a-5p mimic or miR-NC using Lipofectamine 2000
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. Luciferase activity was measured 48 h after
transfection using a Dual-Luciferase® Reporter Assay
System (Promega Corporation) on an Lmax Microplate Luminometer
(Molecular Devices, LLC, Sunnyvale, CA, UA), in accordance with the
manufacturer's instructions. Using Renilla fluorescence
activity as internal reference, the fluorescence values of each
group were measured.
Statistical analysis
Data were presented as mean ± standard deviation of
at least three independent experiments. SPSS 17.0 software (SPSS,
Inc., Chicago, IL, USA) was used to perform statistical analysis.
Differences were analyzed by one-way analysis of variance and
Tukey's post hoc test was performed. The relevance analysis between
miR-30a-5p expression and IGF-1R expression was conducted through
the Spearman's Correlation Coefficient. P<0.05 was considered to
indicate a statistically significant difference.
Results
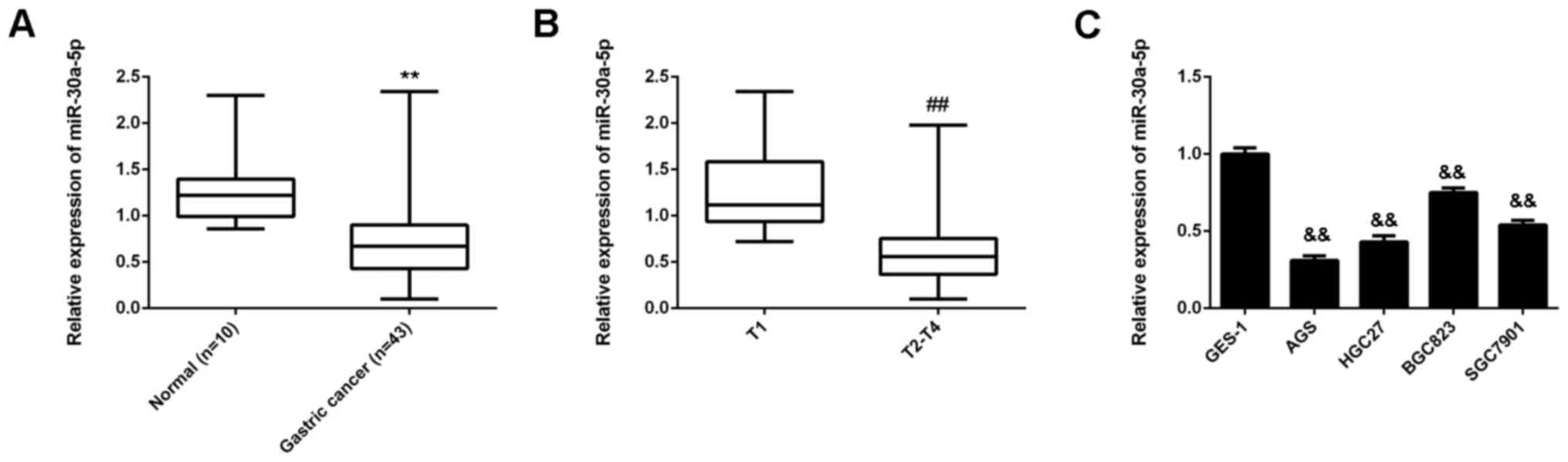
MiR-30a-5p is downregulated in gastric
cancer tissues and cell lines
To determine the role of miR-30a-5p in gastric
cancer, the levels of miR-30a-5p expression were evaluated in
gastric cancer tissues (n=43) and normal gastric tissues (n=10).
RT-qPCR data indicated that the expression of miR-30a-5p was
significantly decreased in gastric cancer tissues compared with
normal gastric tissues (P<0.01; Fig.
1A). In addition, miR-30a-5p expression was significantly lower
in T2-T4 stage cancer compared with that in T1 stage cancer
(P<0.01), suggesting that decreased levels of miR-30-5p were
associated with advanced malignancy (Fig. 1B). The expression of miR-30a-5p in
human gastric cancer cell lines was subsequently assessed. Similar
to gastric cancer tissues, it was observed that miR-30a-5p was
downregulated in the gastric cancer cell lines AGS, HGC27, BGC823
and SGC7901, when compared with the gastric mucosa epithelial cell
line GES-1 (P<0.01; Fig. 1C).
Thus, miR-30a-5p was downregulated in gastric cancer, suggesting
that reduced expression of miR-30a-5p may be involved in the
malignant progression of gastric cancer.
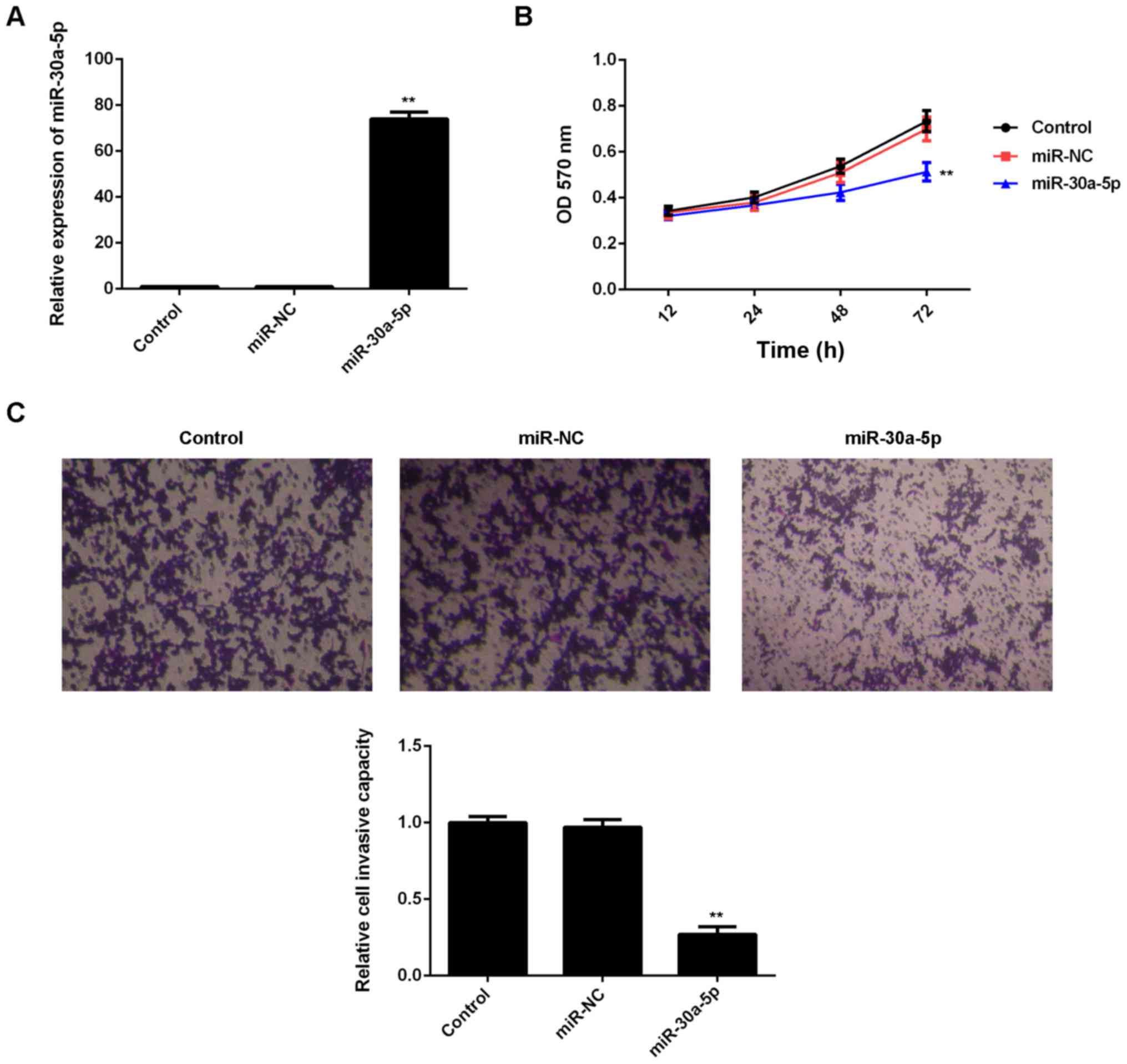
MiR-30a-5p inhibits the proliferation
and invasion of AGS cells
As AGS cells exhibited the lowest levels of
miR-30a-5p among the four gastric cancer cell lines, this cell line
was used in subsequent experiments. In addition, as miR-30a-5p was
downregulated in gastric cancer, miR-30-5p mimic was used to
upregulate its levels. Transfection with miR-30-5p mimic
significantly increased the expression of miR-30-5p in AGS cells
(P<0.01), while transfection with miR-NC had no affect on
miR-30-5p expression, relative to the control group (Fig. 2A). An MTT assay was subsequently
performed to evaluate cell proliferation, whereby it was observed
that overexpression of miR-30a-5p significantly inhibited the
proliferation of AGS cells compared with the control group
(P<0.01), indicating that miR-30a-5p may serve a suppressive
role in the proliferation of gastric cancer cells (Fig. 2B). Moreover, a Transwell assay was
conduced to assess the invasive capacities of AGS cells in each
transfection group. As depicted in Fig.
2C, transfection with miR-30a-5p mimic significantly decreased
the invasion of AGS cells, relative to the control group
(P<0.01). Collectively, these data suggest that miR-30a-5p
exerts inhibitory effects on the proliferation and invasion of
gastric cancer cells.
IGF-1R is a target gene of
miR-30a-5p
The putative targets of miR-30a-5p were investigated
by bioinformatical analysis. TargetScan software predicated that
IGF-1R was a potential target of miR-30a-5p (Fig. 3A), and that the targeting
relationship between miR-30a-5p and IGF-1R was evolutionally
conserved (Fig. 3B). To verify this
prediction, reporter vectors containing WT and MUT IGF-1R 3′UTR
were generated (Fig. 3C and 3D). A
luciferase reporter assay was subsequently conducted, and it was
observed that luciferase activity was significantly decreased in
AGS cells co-transfected with the WT IGF-1R 3′UTR vector and
miR-30a-5p mimic (P<0.01 vs. control; Fig. 3E). However, luciferase activity was
unaffected in cells co-transfected with the MUT IGF-1R 3′UTR vector
and miR-30a-5p mimic, relative to the control group (Fig. 3E). These data demonstrated that
miR-30a-5p may directly bind to the 3′UTR of IGF-1R mRNA. As miRs
negatively mediate the expression of their target genes, the
effects of miR-30a-5p on the expression of IGF-1R in AGS cells were
subsequently investigated. Data from western blot analysis
indicated that overexpression of miR-30-5p significantly decreased
the protein expression of IGF-1R in AGS cells compared with the
control group (P<0.01; Fig. 3F);
however, upregulation of miR-30-5p had no affect on the mRNA
expression of IGF-1R in AGS cells (Fig.
3G), indicating that miR-30a-5p suppressed the expression of
IGF-1R at the post-transcriptional level. Collectively, these
findings indicated that miR-30a-5p negatively mediated the protein
expression of IGF-1R in AGS cells by directly binding to the 3′UTR
of IGF-1R mRNA.
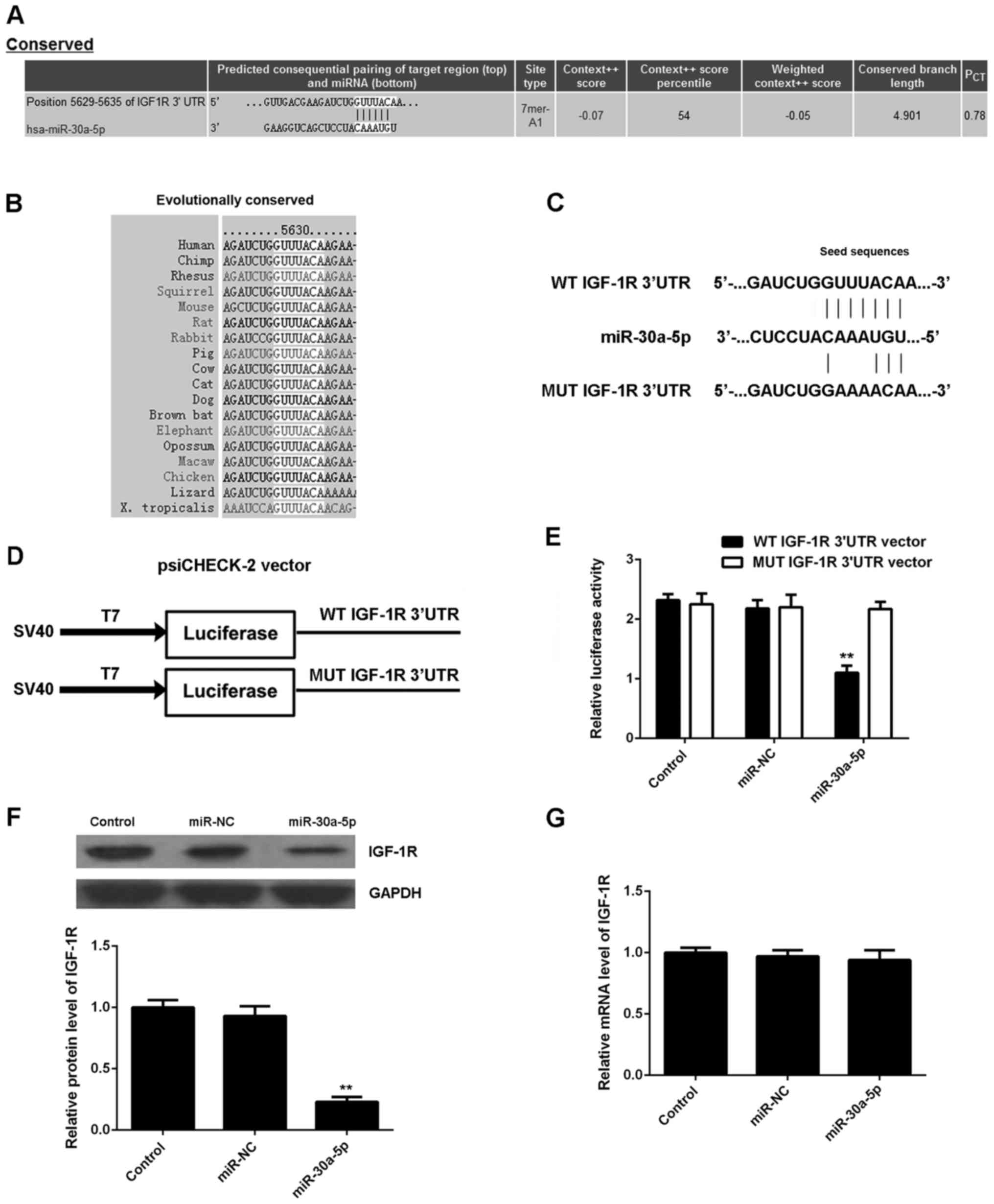 | Figure 3.Targeting of IGF-1R by miR-30a-5p. (A)
TargetScan software predicated that IGF-1R was a potential target
of miR-30a-5p, and that (B) their targeting relationship was
evolutionally conserved. (C and D) Luciferase reporter vectors
containing WT and MUT IGF-1R 3′UTR were constructed. (E) Luciferase
activity was significantly decreased in AGS cells co-transfected
with the WT IGF-1R 3′UTR vector and miR-30a-5p mimic, but was
unaffected in cells co-transfected with the MUT IGF-1R 3′UTR vector
and miR-30a-5p mimic, relative to the control group. (F) Western
blot analysis and (G) reverse transcription-quantitative polymerase
chain reaction were used to measure the expression of IGF-1R at the
protein and mRNA levels, respectively, in AGS cells transfected
with miR-NC or miR-30a-5p mimic. Data are presented as mean ±
standard deviation of at least three independent experiments
**P<0.01 vs. control. IGF-1R, insulin-like growth factor 1
receptor; miR, microRNA; WT, wild-type; MUT, mutant; 3′UTR, 3′
untranslated region; miR-NC, negative control miR; control,
non-transfected AGS cells. |
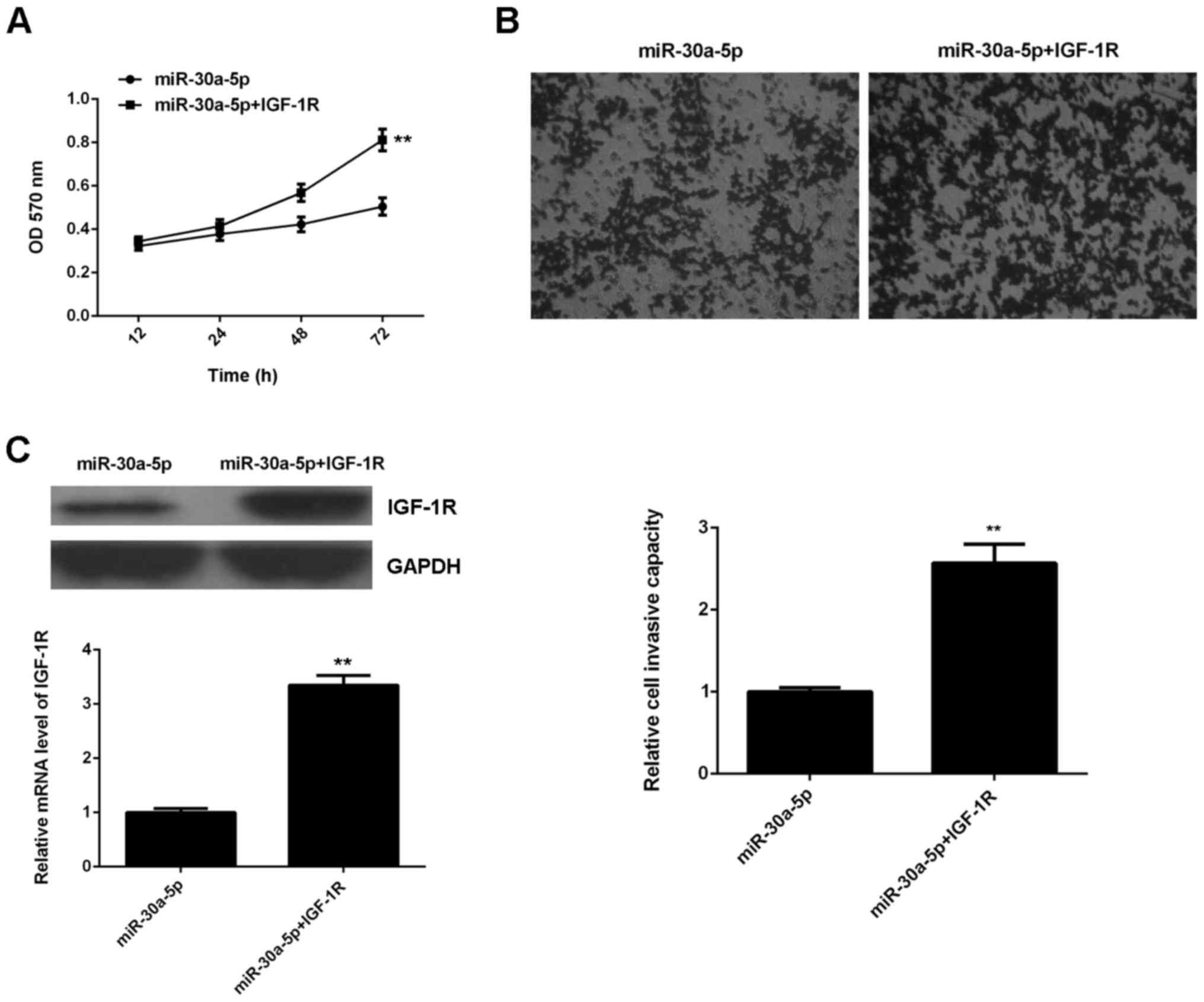
IGF-1R acts as a downstream effector
in miR-30a-5p-mediated suppression of gastric cancer proliferation
and invasion
It was subsequently determined whether IGF-1R was
involved in the inhibitory effects of miR-30a-5p on the
proliferation and invasion of gastric cancer cells. AGS cells were
transfected with miR-30a-5p mimic or co-transfected with miR-30a-5p
mimic and IGF-1R plasmid. An MTT assay was then conducted to assess
the effects on cell proliferation. The data indicated that cell
proliferation was significantly higher in the miR-30a-5p+IGF-1R
group, when compared with that in the miR-30a-5p group (P<0.01;
Fig. 4A), suggesting that miR-30a-5p
inhibited the proliferation of AGS cells through direct targeting
of IGF-1R. Similarly, it was observed that cell invasion was
significantly upregulated in the miR-30a-5p+IGF-1R group compared
to the miR-30a-5p group (P<0.01; Fig.
4B), suggesting that miR-30a-5p inhibited the invasion of AGS
cells by directly targeting IGF-1R. To confirm these findings,
levels of IGF-1R protein were measured in the miR-30a-5p and
miR-30a-5p+IGF-1R groups. Results indicated that IGF-1R expression
was significantly higher in the miR-30a-5p+IGF-1R group compared
with the miR-30a-5p group (P<0.01; Fig. 4C). The above data suggested that
IGF-1R was involved in the inhibitory effects of miR-30a-5p on the
proliferation and invasion of gastric cancer cells.
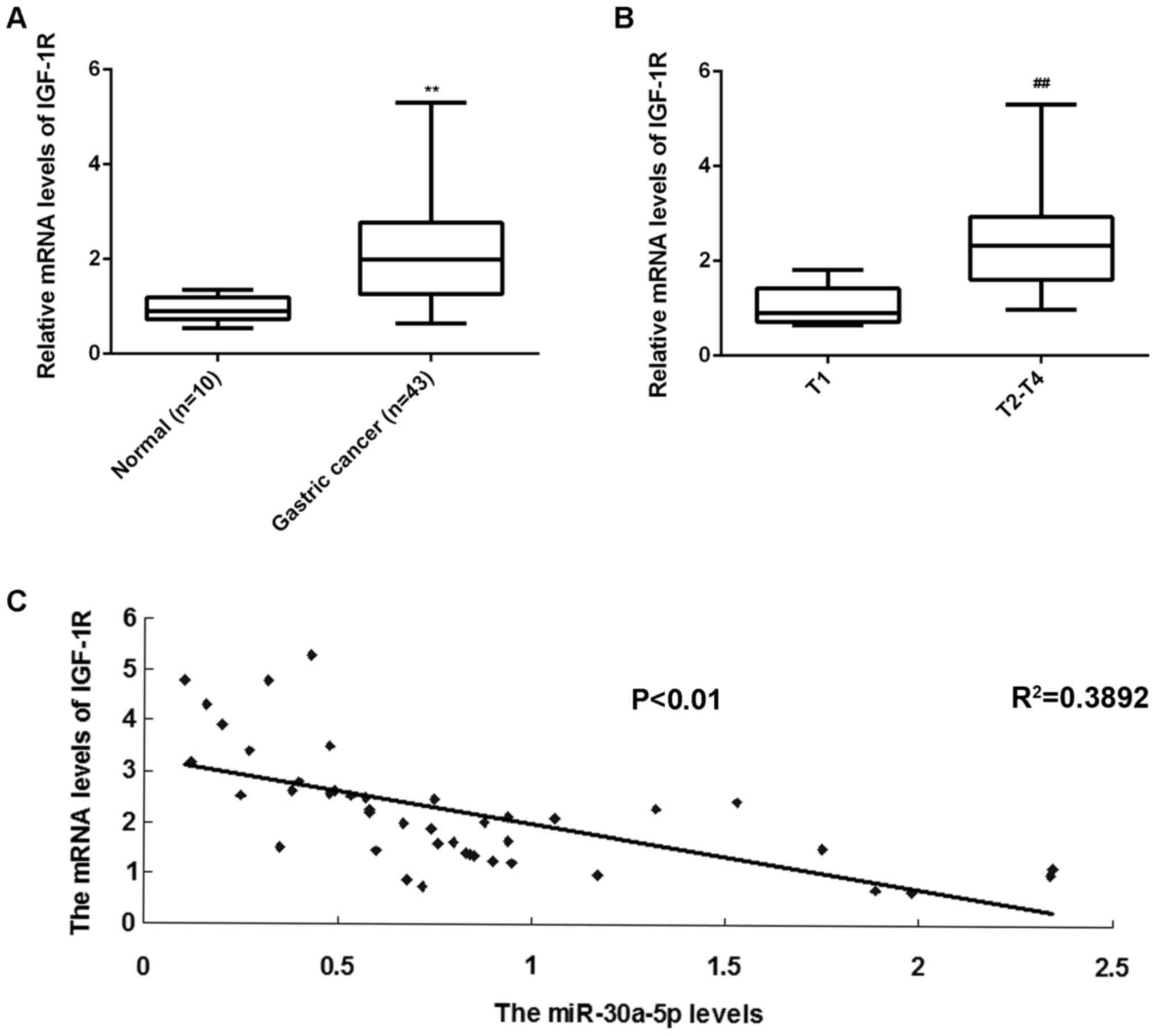
IGF-1R is upregulated in gastric
cancer tissues
RT-qPCR was performed to measure the expression of
IGF-1R in gastric cancer tissues and normal gastric tissues.
Results indicated that levels of IGF-1R mRNA were significantly
upregulated in gastric cancer tissues compared with normal gastric
tissues (P<0.01; Fig. 5A).
Furthermore, IGF-1R mRNA was significantly increased in T2-T4 stage
cancer compared with T1 stage cancer (P<0.01; Fig. 5B), suggesting that its upregulation
may be involved in the malignant progression of gastric cancer. In
addition, an inverse correlation between the expressions of
miR-30a-5p and IGF-1R was identified in gastric cancer tissues
(R2=0.3892, P<0.01; Fig.
5C). Therefore, increased levels of IGF-1R in gastric cancer
may be attributed to a downregulation in miR-30a-5p.
Discussion
A recent study identified a tumor suppressive role
of miR-30a-5p in gastric cancer (14). However, the underlying molecular
mechanism of miR-30a-5p in the regulation of gastric cancer
proliferation and invasion is not well understood. The present
study observed that miR-30a-5p was significantly downregulated in
gastric cancer tissues compared with non-tumor gastric tissues.
MiR-30a-5p was also downregulated in gastric cancer cell lines.
Furthermore, IGF-1R was identified as a novel target of miR-30a-5p,
and its levels of protein expression were negatively mediated by
miR-30a-5p in AGS cells. Overexpression of miR-30a-5p significantly
inhibited the proliferation and invasion of AGS cells, while
restoration of IGF-1R reversed the suppressive effects of
miR-30a-5p on the proliferation and invasion of AGS cells. In
addition, it was observed that IGF-1R was significantly upregulated
in gastric cancer tissues compared with non-tumor gastric tissues,
and its expression was reversely correlated with that of miR-30a-5p
in gastric cancer tissues.
MiR-30a-5p is deregulated and serves a role in many
human cancers. Significant downregulation of miR-30a-5p has been
identified in anaplastic thyroid carcinomas and non-small cell lung
cancer (23,24), while its upregulation has been
documented in glioma (25) and
squamous cell carcinoma of head and neck and esophagus (26), suggesting a dual role of miR-30a-5p
in different cancer types. For instance, knockdown of miR-30a-5p
suppressed glioma cell growth by targeting the septin-7 and PR
domain zinc finger protein 1 genes, indicating that miR-30a-5p
serves as an oncogene in glioma (27,28).
However, in colon cancer, overexpression of miR-30a-5p may inhibit
the proliferation of cancer cells, while also stimulating their
apoptosis and and cell cycle arrest at the G1 phase, at least in
part through targeting of denticleless protein homolog, indicating
that miR-30a-5p serves a suppressive role in colon cancer (29). Similarly, miR-30a-5p has been
reported to inhibit cell proliferation, induce cell apoptosis and
suppress the invasion and migration of hepatocellular carcinoma
(HCC) cells in vitro and in vivo (30).
More recently, Li et al (31) investigated the association of a
gastric cancer-specific miR signature (miR-10b, miR-21, miR-223,
miR-338, let-7a, miR-30a-5p and miR-126) with the prognosis of
gastric cancer patients, and observed that the miR signature was an
independent predictor of overall survival and relapse-free
survival, as validated in 50 patients with gastric cancer and 60
control patients. However, the exact role of miR-30a-5p in gastric
cancer remains unknown. In the present study, a significant
decrease in miR-30a-5p was observed in gastric cancer tissues and
cell lines, suggesting that its downregulation may serve a role in
the development and progression of gastric cancer. To the best of
our knowledge, it was also demonstrated for the first time that
overexpression of miR-30a-5p significantly inhibited the
proliferation and invasion of AGS cells, suggesting that miR-30a-5p
may also exert an inhibitory effect on the growth and metastasis of
gastric cancer.
As miRs negatively regulate the protein expression
of their target mRNAs, the current study investigated the putative
targets of miR-30a-5p in gastric cancer. Bioinformatical analysis
indicated that IGF-1R was a potential target of miR-30a-5p, and
their targeting relationship was evolutionally conserved. To verify
this predication, a luciferase reporter assay was performed.
Results indicated that miR-30a-5p directly bound to the 3′UTR of
IGF-1R mRNA, thus suggesting that IGF-1R is a direct target of
miR-30a-5p. It was further observed that the expression of IGF-1R
was negatively mediated by miR-503 at the post-transcriptional
level in gastric cancer cells. IGF-1R functions as a key oncogene
in the development and maintenance of cancer, even in the absence
of its cognate ligands and intrinsic kinase activity (17). Furthermore, IGF-1R has previously
been used as a therapeutic target in the treatment of cancer. For
instance, Yue et al (32)
generated a murine anti-IGF-1R antibody (4F2), and observed that
treatment with 4F2 decreased the proliferation and promoted the
apoptosis of HCC cells. In the current study, upregulation of
IGF-1R significantly reversed the inhibitory effects of miR-503
overexpression on the proliferation and invasion of AGS cells,
indicating that IGF-1R serves as a downstream effector in
miR-503-meditated suppression of gastric cancer proliferation and
invasion. Moreover, IGF-1R was significantly upregulated in gastric
cancer tissues compared with non-tumor gastric tissues, and its
expression levels were inversely correlated with that of miR-503 in
gastric cancer tissues, suggesting that high-level expression of
IGF-1R in gastric cancer may be attributed to a downregulation in
miR-503.
To the best of our knowledge, the present study was
the first to demonstrate that miR-30a-5p, as an miR that is
significantly downregulated in gastric cancer, had suppressive
effects on the proliferation and invasion of gastric cancer cells
by directly targeting IGF-1R. These data provide greater insight
into the regulatory mechanisms of miRs in gastric cancer, and
suggest that the miR-30a-5p/IGF-1R axis is a potential therapeutic
target in the treatment of gastric cancer.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Piazuelo MB and Correa P: Gastric cáncer:
Overview. Colomb Med (Cali). 44:192–201. 2013.PubMed/NCBI
|
|
4
|
Matuszcak C, Haier J, Hummel R and Lindner
K: MicroRNAs: Promising chemoresistance biomarkers in gastric
cancer with diagnostic and therapeutic potential. World J
Gastroenterol. 20:13658–13666. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi J, Qu YP and Hou P: Pathogenetic
mechanisms in gastric cancer. World J Gastroenterol.
20:13804–13819. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li PF, Chen SC, Xia T, Jiang XM, Shao YF,
Xiao BX and Guo JM: Non-coding RNAs and gastric cancer. World J
Gastroenterol. 20:5411–5419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pang Y, Young CY and Yuan H: MicroRNAs and
prostate cancer. Acta Biochim Biophys Sin (Shanghai). 42:363–369.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng R, Chen X, Yu Y, Su L, Yu B, Li J,
Cai Q, Yan M, Liu B and Zhu Z: miR-126 functions as a tumour
suppressor in human gastric cancer. Cancer Lett. 298:50–63. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qiao F, Zhang K, Gong P, Wang L, Hu J, Lu
S and Fan H: Decreased miR-30b-5p expression by DNMT1 methylation
regulation involved in gastric cancer metastasis. Mol Biol Rep.
41:5693–5700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Singh P, Alex JM and Bast F: Insulin
receptor (IR) and insulin-like growth factor receptor 1 (IGF-1R)
signaling systems: Novel treatment strategies for cancer. Med
Oncol. 31:8052014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
King H, Aleksic T, Haluska P and Macaulay
VM: Can we unlock the potential of IGF-1R inhibition in cancer
therapy? Cancer Treat Rev. 40:1096–1105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Crudden C, Girnita A and Girnita L:
Targeting the IGF-1R: The tale of the tortoise and the hare. Front
Endocrinol (Lausanne). 6:642015.PubMed/NCBI
|
|
18
|
Chen HX and Sharon E: IGF-1R as an
anti-cancer target-trials and tribulations. Chin J Cancer.
32:242–252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morishita A, Gong J and Masaki T:
Targeting receptor tyrosine kinases in gastric cancer. World J
Gastroenterol. 20:4536–4545. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ge J and Chen Z, Huang J, Yuan W, Den Z
and Chen Z: Silencing insulin-like growth factor-1 receptor
expression inhibits gastric cancer cell proliferation and invasion.
Mol Med Rep. 11:633–638. 2015.PubMed/NCBI
|
|
21
|
Wittekind C: The development of the TNM
classification of gastric cancer. Pathol Int. 65:399–403. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Visone R, Pallante P, Vecchione A,
Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V,
Borbone E, et al: Specific microRNAs are downregulated in human
thyroid anaplastic carcinomas. Oncogene. 26:7590–7595. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu J, Zeng Y, Xu C, Qin H, Lei Z, Shen D,
Liu Z and Huang JA: Expression profile analysis of microRNAs and
downregulated miR-486-5p and miR-30a-5p in non-small cell lung
cancer. Oncol Rep. 34:1779–1786. 2015.PubMed/NCBI
|
|
25
|
Wang K, Jia Z, Zou J, Zhang A, Wang G, Hao
J, Wang Y, Yang S and Pu P: Analysis of hsa-miR-30a-5p expression
in human gliomas. Pathol Oncol Res. 19:405–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kimura S, Naganuma S, Susuki D, Hirono Y,
Yamaguchi A, Fujieda S, Sano K and Itoh H: Expression of microRNAs
in squamous cell carcinoma of human head and neck and the
esophagus: miR-205 and miR-21 are specific markers for HNSCC and
ESCC. Oncol Rep. 23:1625–1633. 2010.PubMed/NCBI
|
|
27
|
Jia Z, Wang K, Wang G, Zhang A and Pu P:
MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth
by targeting SEPT7. PLoS One. 8:e550082013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X, Wang K, Han L, Zhang A, Shi Z,
Zhang K, Zhang H, Yang S, Pu P, Shen C, et al: PRDM1 is directly
targeted by miR-30a-5p and modulates the Wnt/β-catenin pathway in a
Dkk1-dependent manner during glioma growth. Cancer Lett.
331:211–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baraniskin A, Birkenkamp-Demtroder K,
Maghnouj A, Zöllner H, Munding J, Klein-Scory S, Reinacher-Schick
A, Schwarte-Waldhoff I, Schmiegel W and Hahn SA: MiR-30a-5p
suppresses tumor growth in colon carcinoma by targeting DTL.
Carcinogenesis. 33:732–739. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dai H, Kang B, Zuo D and Zuo G: Effect of
miR-30a-5p on the proliferation, apoptosis, invasion and migration
of SMCC-7721 human hepatocellular carcinoma cells. Zhonghua Gan
Zang Bing Za Zhi. 22:915–920. 2014.(In Chinese). PubMed/NCBI
|
|
31
|
Li X, Zhang Y, Ding J, Wu K and Fan D:
Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yue L, Wang Y, Wang H, Gao H, Liang J, Sui
A, Xiang J, Zhou F, Xu C, Zhao W, et al: Inhibition of
hepatocellular carcinoma cell growth by an anti-insulin-like growth
factor-I receptor monoclonal antibody. Oncol Rep. 28:1453–1460.
2012.PubMed/NCBI
|