Introduction
Lung cancer is the leading cause of
cancer-associated mortality globally (1). Non-small cell lung cancer (NSCLC),
accounts for ~85% of all types of lung cancer and patient prognosis
is poor (2). Despite marked
improvements in cancer detection and treatment, the 5-year overall
survival rate for NSCLC is only ~16% (3). It is thus important to understand the
molecular mechanisms involved in the growth of NSCLC to facilitate
the development of novel anticancer agents to improve patient
outcomes.
The chromatin assembly factor 1 (CAF-1) complex,
composed of chromatin assembly factor 1 subunit A (CHAF1A; also
known as p150), p60 and p48 subunits, has been implicated in the
assembly of nucleosomes on newly replicated DNA (4). CAF-1-mediated chromatin assembly serves
a pivotal role in DNA repair, protecting DNA from excessive
degradation (5). The interaction
between CAF-1 and proliferating cell nuclear antigen aids in
recruiting and stabilizing CAF-1 at the site of DNA replication
(6). It has been reported that
expression of a dominant-negative mutant of CHAF1A induces S-phase
cell cycle arrest, which is accompanied by DNA damage (7). A loss-of-function mutation in
CAF-1/p150 impairs the organization of heterochromatin in
pluripotent embryonic cells (8).
CAF-1 is an important regulator of somatic cell identity and its
absence leads to a more accessible chromatin structure, increasing
the expression of pluripotency-specific genes (9). Several previous studies have suggested
that there is an association between CAF-1 overexpression and
aggressive parameters of human cancer (10–12). For
instance, CAF-1/p60 overexpression is an independent poor
prognostic indicator of laryngeal carcinoma (10). In high-grade gliomas, CAF-1/p60
overexpression contributes to tumor aggressiveness and shorter
overall survival (11). Barbieri
et al (13) indicated that
CHAF1A overexpression predicts poor prognosis and stimulates the
development of neuroblastoma. Another study demonstrated that
upregulation of CHAF1A promotes the proliferation of colon cancer
cells (14). In glioblastoma
(15) and ovarian cancer (16) cells, CHAF1A also offers a growth
advantage. However, the expression and biological significance of
CHAF1A in NSCLC remains unclear. Therefore, the present study
examined the expression of CHAF1A in NSCLC and adjacent normal
tissues and investigated its function in the growth of NSCLC
cells.
Materials and methods
Tissue samples
Tumor and corresponding normal tissue specimens were
collected from 22 patients with NSCLC who underwent surgical
resection between October 2014 and January 2015 at the Department
of Thoracic Surgery of the Second Affiliated Hospital of Shanxi
Medical University (Taiyuan, China). There were 20 males and 2
females, with a median age of 67 years (range, 47–78 years).
Patients who received radiotherapy or chemotherapy prior to surgery
were excluded. Written informed consent was obtained from each
patient and the study was approved by the local Institutional
Review Board of the Second Affiliated Hospital of Shanxi Medical
University (Taiyuan, China).
Cell culture
The human lung cancer cell lines A549, H1299, H1688
and H1975, human embryonic kidney 293T cells and normal human lung
epithelial BEAS-2B cells were purchased from the American Type
Culture Collection (Manassas, VA, USA). All cells were routinely
maintained in RPMI 1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum, 100 units/ml penicillin and 100 mg/ml streptomycin at
37°C in an atmosphere of humidified air with 5% CO2.
Upon reaching 90% confluence, cells were subcultured every three
days.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues or cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reversely transcribed to first-strand cDNA using the
Reverse Transcription system (catalog no. A3500; Promega
Corporation, Madison, WI, USA). DNase (Tiangen Biotech Co., Ltd.,
Beijing, China) was employed to eliminate DNA prior to reverse
transcription. PCR amplifications were performed using the SYBR
Green Real-Time PCR Master mix (catalog no. 4309155; Invitrogen;
Thermo Fisher Scientific, Inc.). The cycling conditions consisted
of an initial denaturation at 95°C for 5 min and 40 cycles of
denaturation at 95°C for 20 sec, annealing at 59°C for 20 sec and
elongation at 72°C for 40 sec. The PCR primers were as follows:
Human CHAF1A forward, 5′-AGGGAAGGTGCCTATGGTG-3′ and reverse,
5′-CAGGGACGAATGGCTGAGTA-3′; human glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) forward, 5′-CAGGGACGAATGGCTGAGTA-3′ and
reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. Relative CHAF1A mRNA
expression was determined according to the 2−ΔΔCq method
following normalization to GAPDH (17). Each assay was repeated three
times.
Construction of recombinant lentivirus
encoding CHAF1A small hairpin (sh)RNA
Knockdown of CHAF1A expression in H1299 cells was
performed using lentivirus-mediated RNA interfering technology, as
previously described (18). In
brief, CHAF1A-targeting and negative control shRNAs were
synthesized by GeneChem Co., Ltd (Shanghai, China). The sequence of
CHAF1A shRNA was as follows:
5′-CCGGCCGACTCAATTCCTGTGTAAATTCAAGAGATTTACACAGGAATTGAGTCGGTTTTTG-3′.
Annealed DNA oligonucleotides were inserted into the GV115
lentivirus vector (GeneChem Co., Ltd.) and the construct was
verified by DNA sequencing, performed by Sangon Biotech Co., Ltd.
(Shanghai, China). For production of recombinant lentivirus, the
GV115-CHAF1A shRNA-expressing plasmid and lentivirus packaging
vectors were co-transfected into 293T cells using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h
post-transfection, lentivirus-containing supernatant was collected,
purified and titered as previously described (18). A non-specific shRNA-expressing
lentivirus was also prepared and used as a negative control.
Infection of cells with
CHAF1A-expressing lentivirus
H1299 cells were seeded at 4×105
cells/well onto 6-well plates and incubated in RPMI 1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C overnight
prior to transduction. Cells were infected with lentiviral
particles expressing control or CHAF1A shRNA at a multiplicity of
infection of 5. As the recombinant lentivirus is able to co-express
intended shRNAs and green fluorescent protein (GFP), transduction
efficiency was monitored by measuring GFP-positive cells under a
fluorescence microscope.
Measurement of cell proliferation
using the Cellomics method
H1299 cells were seeded onto 96-well plates at a
density of 2×103 cells/well and cultured for 5 days at
37°C. Plates were scanned and analyzed daily using the Cellomics
ArrayScan VTI (Thermo Fisher Scientific Inc.) (19). The system is an automated
fluorescence-imaging microscope that analyzes the intensity and
distribution of fluorescence in each individual cell. Cells with
green fluorescence were counted at five random fields at ×20
magnification.
Colony formation assay
H1299 cells were seeded onto 6-well plates (400
cells/well) and cultured for 14 days. The wells were stained with
Giemsa and the number of colonies containing >50 cells were
counted. The assay was repeated three times in triplicate.
Cell cycle analysis by flow
cytometry
H1299 cells (8×104 in RPMI 1640 medium)
were fixed with ice-cold 70% ethanol and suspended in staining
solution containing 0.5 mg/ml RNase, 0.05% Triton X-100 and 10
µg/ml propidium iodide (PI; Sigma Aldrich; Merck KGaA, Darmstadt,
Germany). Following 1 h incubation at 37°C in the dark, cells were
immediately analyzed using a flow cytometer with CellQuest v3.3
software (BD Biosciences, San Jose, CA, USA).
Western blot analysis
Protein samples were prepared in ice-cold
radioimmunoprecipitation assay buffer supplemented with
protease/phosphatase inhibitors (Sigma Aldrich; Merck KGaA) for 30
min. Lysates were cleared by centrifugation at 10,000 × g for 10
min at 4°C. Protein samples (50 µg in 10 µl loading buffer per
lane) were resolved by 12% SDS-PAGE and transferred to
nitrocellulose membranes. Membranes were blocked with 5% fat-free
milk at 37°C for 1 h and incubated with primary antibodies at 4°C
overnight. The primary antibodies used are summarized as follows:
Against cyclin D1 (sc-70899; 1:500 dilution), cyclin-dependent
kinase 2 (CDK2) (sc-6248; 1:500 dilution), S-phase
kinase-associated protein (SKP)2 (sc-74477; 1:800 dilution), p21
(sc-6246; 1:1,000 dilution), p27 (sc-53906; 1:1,000 dilution), and
β-actin (sc-47778; 1:2,000 dilution) (all Santa Cruz Biotechnology,
Inc., Dallas, TX, USA). The membranes were then incubated with
horseradish peroxidase-conjugated anti-mouse IgG (1:5,000 dilution;
A9044; Sigma-Aldrich; Merck KGaA) at room temperature for 1 h.
Immunoreactive signals were visualized by chemifluorescence using
ECL Plus Western Blotting Detection reagents (GE Healthcare
Bio-Sciences, Pittsburgh, PA, USA). Protein signals were quantified
using Quantity One software (version 4.6.2; Bio-Rad Laboratories,
Hercules, CA, USA). Each assay was repeated three times.
Statistical analysis
Data are presented as mean ± standard deviation. All
statistical calculations were performed using SPSS 11.7 software
(SPSS, Chicago, IL, USA). Differences between the means were
analyzed using Student's t-test or one-way analysis of variance
followed by Tukey's multiple comparison test. P<0.05 was
considered to indicate a statistically significant difference.
Results
CHAF1A is upregulated in NSCLC tissues
relative to adjacent normal tissues
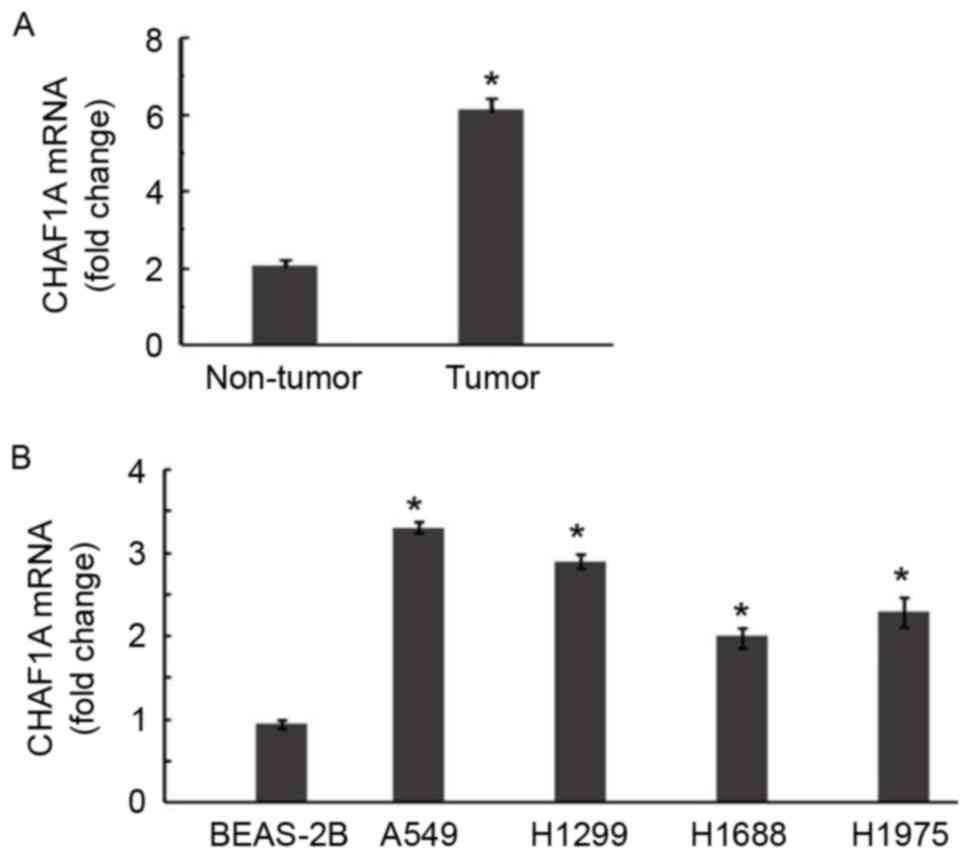
The expression of CHAF1A was examined in 22 samples
of NSCLC and corresponding normal lung tissue. RT-qPCR demonstrated
that CHAF1A mRNA levels were 2.9-fold greater in NSCLC samples
compared with adjacent normal tissues (P<0.05; Fig. 1A). Significantly increased expression
of endogenous CHAF1A was further confirmed in all 4 of the NSCLC
cell lines by RT-qPCR (P<0.05; Fig.
1B).
Silencing of endogenous CHAF1A in
NSCLC cells via infection with CHAF1A shRNA-expressing
lentivirus
To determine the biological roles of CHAF1A in NSCLC
growth, its expression was knocked down in NSCLC cells using
lentivirus-mediated shRNA technology. Microscopic examination of
GFP expression following lentivirus infection revealed a similar
transduction efficiency of ~80% for control and CHAF1A shRNA
(Fig. 2A). However, compared with
the delivery of control shRNA, CHAF1A shRNA transduction
significantly decreased levels of CHAF1A mRNA in H1299 cells by
84.7±1.2% (P<0.05; Fig. 2B).
Knockdown of CHAF1A impairs the growth
and colony formation of NSCLC cells
The effect of CHAF1A downregulation on the growth of
NSCLC cells was subsequently assessed. During a 5-day culture
period, H1299 cells transduced with CHAF1A-shRNA lentivirus and
control lentivirus were counted daily using the Cellomics method.
The number of cells was significantly lower in the CHAF1A shRNA
group compared with the control group on day 5 (162±10 vs. 523±8
cells/field; P<0.05; Fig. 3). The
colony formation assay further demonstrated that the delivery of
CHAF1A shRNA significantly reduced the number of colonies formed by
H1299 cells following 14-day incubation (21±9 vs. 84±12
colonies/dish; P<0.05; Fig. 4).
These results indicate that the suppression of NSCLC cell growth is
mediated by CHAF1A silencing.
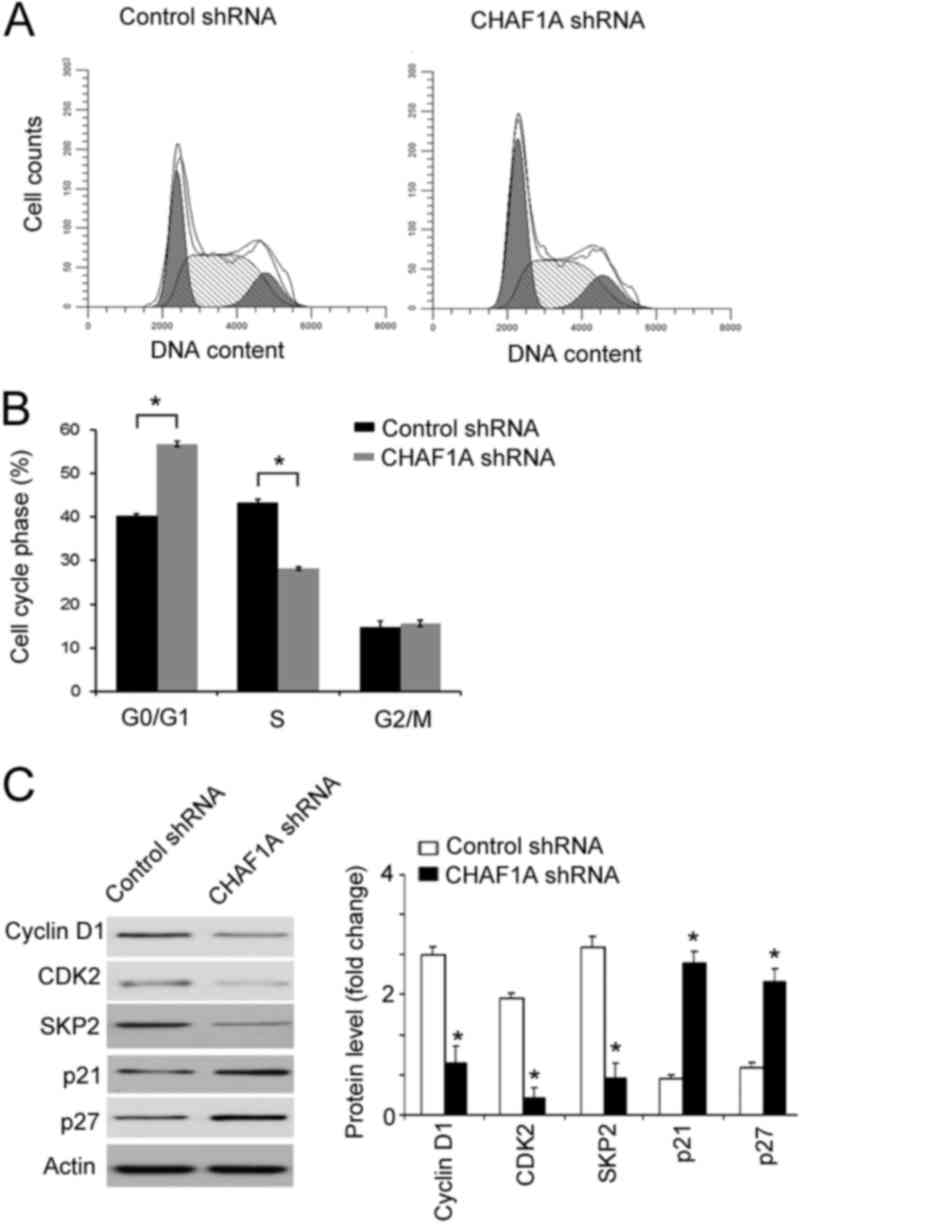
CHAF1A silencing induces cell cycle
arrest at the S-phase
Finally, it was assessed whether the anti-growth
effect of CHAF1A downregulation is associated with the induction of
cell cycle arrest. Cell cycle progression was analyzed by flow
cytometry following PI staining. As presented in Fig. 5A and B, the percentage of cells
entering the S-phase was significantly decreased in CHAF1A
shRNA-transduced cells relative to control cells (28±0.9 vs.
43±1.2%; P<0.05). By contrast, the percentage of cells at the
G0/G1 phase was significantly increased in CHAF1A shRNA-transduced
cells compared with the control (57±0.8 vs. 40±0.9%; P<0.05).
Western blot analysis revealed that CHAF1A silencing significantly
downregulated the expression of cyclin D1, CDK2, and SKP2
(P<0.05, but significantly increased the expression of p21 and
p27 (P<0.05; Fig. 5C). This
indicates that knockdown of CHAF1A leads to cell cycle arrest
during S-phase.
Discussion
The CAF-1 complex serves a critical role in DNA
replication and repair and its deregulation contributes to the
pathogenesis of cancer (5,8,9). The
CAF-1 subunit p60 is frequently upregulated in human cancer and
contributes to aggressive phenotypes (10,11).
Mascolo et al (20) reported
that overexpression of CAF-1/p60 is significantly associated with
node and/or distant metastases in cutaneous melanoma. The current
study examined the expression of CHAF1A, the largest subunit of
CAF-1, in NSCLC. Data from the present study indicated that CHAF1A
expression was significantly elevated in NSCLC tissues compared
with adjacent normal lung tissues. To the best of our knowledge,
this is the first report confirming the overexpression of CHAF1A in
NSCLC, although upregulation of CHAF1A has been identified in
aggressive neuroblastoma (13) and
colon cancer (14), suggesting its
involvement in the pathogenesis of cancer.
Having identified that CHAF1A is upregulated in
NSCLC, the current study sought to determine its biological roles
in this disease. Lentivirus-mediated delivery of CHAF1A shRNA
resulted in efficient knockdown of endogenous CHAF1A expression in
H1299 cells. Notably, it was determined in the current study using
the Cellomics method that CHAF1A silencing significantly suppressed
the proliferation of H1299 cells. Furthermore, CHAF1A
downregulation impaired the colony formation capacity of H1299
cells, resulting in the formation of fewer colonies following 14
day culture. In accordance with the results of the present study,
it has been determined that CHAF1A silencing inhibits the
tumorigenicity and growth of colon cancer cells (14). The results of the present study
indicate that CHAF1A is required for the growth of NSCLC cells.
This provides a rationale for targeting CHAF1A as a potential
therapeutic strategy for NSCLC.
Compelling evidence indicates that CAF-1 activity is
required for efficient replication of euchromatic DNA (21). Loss of CAF-1 results in cell cycle
arrest at the S-phase (21,22). Takami et al (23) reported that the depletion of each
subunit of the CAF-1 complex delays S-phase progression, followed
by accumulation in late S/G2 phase and aberrant mitosis. Quivy
et al (24) demonstrated that
the interaction between CHAF1A and heterochromatin protein 1 is
indispensable for pericentric heterochromatin replication and
S-phase progression in mouse cells. In agreement with these
studies, the current study revealed that a knockdown of CHAF1A
significantly increased the percentage of S-phase cells and
decreased the percentages of cells at the G0/G1 and G2/M phases,
indicating that it induced cell cycle arrest at the S-phase. At the
molecular level, both p21 and p27 are well-defined CDK inhibitors
that serve an important role in controlling cell cycle progression
(25). SKP2 functions as a positive
regulator of cell cycle progression by promoting p27 degradation
(26). These results provide an
explanation for the reduced proliferation observed in
CHAF1A-depleted H1299 cells.
Certain limitations of the present study should be
noted. Firstly, no information is available concerning the
association of CHAF1A expression with the clinicopathological
parameters and prognosis of NSCLC. Secondly, validation of the
in vivo effect of CHAF1A knockdown on the growth of NSCLC in
animal models is required. Finally, the exact mechanism for the
regulation of cell cycle arrest by CHAF1A in NSCLC cells remains
unclear and should be investigated.
In conclusion, the results of the current study
demonstrate that CHAF1A is upregulated in NSCLC and that its
depletion hinders the growth of NSCLC cells. This potentially
occurs through the induction of cell cycle arrest at the S-phase.
The results of the current study warrants further investigations
into the effect of depletion of CHAF1A on the tumorigenicity and
growth of NSCLC cells in animal models.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
NSCLC Meta-analysis Collaborative Group, .
Preoperative chemotherapy for non-small-cell lung cancer: A
systematic review and meta-analysis of individual participant data.
Lancet. 383:1561–1571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng YW, Li RM, Zhang XW and Ren XB:
Current adoptive immunotherapy in non-small cell lung cancer and
potential influence of therapy outcome. Cancer Invest. 31:197–205.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Groth A, Rocha W, Verreault A and Almouzni
G: Chromatin challenges during DNA replication and repair. Cell.
128:721–733. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mjelle R, Hegre SA, Aas PA, Slupphaug G,
Drabløs F, Saetrom P and Krokan HE: Cell cycle regulation of human
DNA repair and chromatin remodeling genes. DNA Repair (Amst).
30:53–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang K, Gao Y, Li J, Burgess R, Han J,
Liang H, Zhang Z and Liu Y: A DNA binding winged helix domain in
CAF-1 functions with PCNA to stabilize CAF-1 at replication forks.
Nucleic Acids Res. 44:5083–5094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ye X, Franco AA, Santos H, Nelson DM,
Kaufman PD and Adams PD: Defective S phase chromatin assembly
causes DNA damage, activation of the S phase checkpoint, and S
phase arrest. Mol Cell. 11:341–351. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Houlard M, Berlivet S, Probst AV, Quivy
JP, Héry P, Almouzni G and Gérard M: CAF-1 is essential for
heterochromatin organization in pluripotent embryonic cells. PLoS
Genet. 2:e1812006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheloufi S, Elling U, Hopfgartner B, Jung
YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Ang C Euong, Tenen D,
et al: The histone chaperone CAF-1 safeguards somatic cell
identity. Nature. 528:218–224. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mesolella M, Iorio B, Landi M, Cimmino M,
Ilardi G, Iengo M and Mascolo M: Overexpression of chromatin
assembly factor-1/p60 predicts biological behaviour of laryngeal
carcinomas. Acta Otorhinolaryngol Ital. 37:17–24. 2017.PubMed/NCBI
|
|
11
|
de Tayrac M, Saikali S, Aubry M, Bellaud
P, Boniface R, Quillien V and Mosser J: Prognostic significance of
EDN/RB, HJURP, p60/CAF-1 and PDLI4, four new markers in high-grade
gliomas. PLoS One. 8:e733322013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Polo SE, Theocharis SE, Grandin L,
Gambotti L, Antoni G, Savignoni A, Asselain B, Patsouris E and
Almouzni G: Clinical significance and prognostic value of chromatin
assembly factor-1 overexpression in human solid tumours.
Histopathology. 57:716–724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barbieri E, De Preter K, Capasso M, Chen
Z, Hsu DM, Tonini GP, Lefever S, Hicks J, Versteeg R, Pession A, et
al: Histone chaperone CHAF1A inhibits differentiation and promotes
aggressive neuroblastoma. Cancer Res. 74:765–774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu Z, Cui F, Yu F, Peng X, Jiang T, Chen
D, Lu S, Tang H and Peng Z: Up-regulation of CHAF1A, a poor
prognostic factor, facilitates cell proliferation of colon cancer.
Biochem Biophys Res Commun. 449:208–215. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng H, Du B, Jiang H and Gao J:
Over-expression of CHAF1A promotes cell proliferation and apoptosis
resistance in glioblastoma cells via AKT/FOXO3a/Bim pathway.
Biochem Biophys Res Commun. 469:1111–1116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia D, Yang X, Liu W, Shen F, Pan J, Lin
Y, Du N, Sun Y and Xi X: Over-expression of CHAF1A in Epithelial
Ovarian Cancer can promote cell proliferation and inhibit cell
apoptosis. Biochem Biophys Res Commun. 486:191–197. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen J, Liu WB, Jia WD, Xu GL, Ma JL, Ren
Y, Chen H, Sun SN, Huang M and Li JS: Embryonic morphogen nodal is
associated with progression and poor prognosis of hepatocellular
carcinoma. PLoS One. 9:e858402014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buchser WJ, Laskow TC, Pavlik PJ, Lin HM
and Lotze MT: Cell-mediated autophagy promotes cancer cell
survival. Cancer Res. 72:2970–2979. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mascolo M, Vecchione ML, Ilardi G,
Scalvenzi M, Molea G, Di Benedetto M, Nugnes L, Siano M, De Rosa G
and Staibano S: Overexpression of Chromatin Assembly Factor-1/p60
helps to predict the prognosis of melanoma patients. BMC Cancer.
10:632010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klapholz B, Dietrich BH, Schaffner C,
Hérédia F, Quivy JP, Almouzni G and Dostatni N: CAF-1 is required
for efficient replication of euchromatic DNA in Drosophila larval
endocycling cells. Chromosoma. 118:235–248. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen Z, Tan JL, Ingouff M, Sundaresan V
and Berger F: Chromatin assembly factor 1 regulates the cell cycle
but not cell fate during male gametogenesis in Arabidopsis
thaliana. Development. 135:65–73. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takami Y, Ono T, Fukagawa T, Shibahara K
and Nakayama T: Essential role of chromatin assembly
factor-1-mediated rapid nucleosome assembly for DNA replication and
cell division in vertebrate cells. Mol Biol Cell. 18:129–141. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Quivy JP, Gérard A, Cook AJ, Roche D and
Almouzni G: The HP1-p150/CAF-1 interaction is required for
pericentric heterochromatin replication and S-phase progression in
mouse cells. Nat Struct Mol Biol. 15:972–979. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fillies T, Woltering M, Brandt B, Van
Diest JP, Werkmeister R, Joos U and Buerger H: Cell cycle
regulating proteins p21 and p27 in prognosis of oral squamous cell
carcinomas. Oncol Rep. 17:355–359. 2007.PubMed/NCBI
|
|
26
|
Shin JS, Hong SW, Lee SL, Kim TH, Park IC,
An SK, Lee WK, Lim JS, Kim KI, Yang Y, et al: Serum starvation
induces G1 arrest through suppression of Skp2-CDK2 and CDK4 in
SK-OV-3 cells. Int J Oncol. 32:435–439. 2008.PubMed/NCBI
|