Introduction
Glioma is the most common primary brain malignant
tumor, accounting for ~30% of central nervous system tumors and 80%
of all malignant tumors in the brain (1,2). Despite
great improvements in the diagnosis and treatment of this tumor,
the prognosis of glioma patients at a late stage remains poor
(1,2). Deregulations of oncogenes or tumor
suppressors have been identified in glioma in the past decade
(3). Therefore, understanding the
molecular mechanism underlying glioma progression is beneficial for
the development of novel diagnostic and therapeutic strategies for
this disease.
MicroRNAs (miRs), a class of non-coding RNAs with a
length of 18–25 nucleotides, function as important regulators of
gene expression by binding to the complementary regions of their
target mRNAs, leading to protein translation inhibition or mRNA
degradation (4,5). Through inhibition of their target
genes, miRs serve key roles in a variety of cellular biological
processes, including cell survival, proliferation, cell cycle
progression, differentiation, apoptosis, migration, as well as
tumorigenesis (5–7). In recent years, numerous miRs have been
reported to function as oncogenes or tumor suppressors in glioma
(8,9). For instance, miR-30a-5p promotes glioma
cell growth by targeting septin 7 (9), while miR-124 inhibits the migration and
invasion of glioma cells by directly inhibiting the expression of
Rho-associated protein kinase 1 (8).
However, the molecular mechanism underlying the role of miRs in
glioma progression remains largely unclear.
miR-219 has been demonstrated to generally serve a
tumor suppressive role in human cancer. For instance, Lei et
al (10) demonstrated that
miR-219 played anti-proliferative, pro-apoptotic and
anti-metastatic roles, as well as reduced the levels of
phosphorylated extracellular signal-regulated kinases 1/2 in
gastric cancer cells. In addition, Huang et al (11) reported that miR-219 inhibited
hepatocellular carcinoma cell proliferation by targeting
glypican-3. Recently, miR-219-5p has been identified to exert a
tumor suppressor function by targeting roundabout guidance receptor
1 and epidermal growth factor receptor (EGFR) in glioblastoma, the
most malignant type of glioma (12,13).
However, certain other target genes of miR-219 may also serve key
roles in glioma, and the regulatory mechanism of miR-219 underlying
glioma growth and metastasis needs to be fully uncovered.
Therefore, the present study aimed to investigate
the clinical significance of miR-219 expression in glioma through
the examination of glioma tissues and various glioma cell lines.
Furthermore, the molecular mechanism of miR-219 underlying glioma
malignant progression was investigated.
Materials and methods
Clinical tissue samples
The study was approved by the Ethics Committee of
the First Hospital of Changsha (Changsha, China). A total of 63
glioma tissues and 12 normal brain tissues were obtained from the
hospital between July 2010 and March 2012. The tissues were
resected during surgery. Written informed consents were obtained
from all patients. The clinical information of glioma patients
included in the current study is summarized in Table I. The glioma patients were diagnosed
according to the World Health Organization (WHO) stage and grading
system (14). The Karnofsky
performance scale (KPS) runs from 100 to 0, where 100 is ‘perfect
health’ and 0 is death. Its purpose is to allow physicians to
evaluate a patient's ability to survive chemotherapy for cancer
(15). All patients involved in this
study received no treatment prior to surgical resection. All tissue
samples were immediately snap-frozen and stored in liquid nitrogen
until further use.
 | Table I.Association between miR-219 levels and
clinicopathological characteristics of glioma patients. |
Table I.
Association between miR-219 levels and
clinicopathological characteristics of glioma patients.
|
|
| miR-219
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases, n | Low (n=36) | High (n=27) | P-value |
|---|
| Age, years |
|
|
| >0.999 |
|
<50 | 25 | 20 | 15 |
|
| ≥50 | 38 | 16 | 12 |
|
| Sex |
|
|
| 0.430 |
| Male | 40 | 21 | 19 |
|
|
Female | 23 | 15 | 8 |
|
| WHO grade |
|
|
| 0.010 |
|
I–II | 27 | 10 | 17 |
|
|
III–IV | 36 | 26 | 10 |
|
| KPS |
|
|
| 0.004 |
|
>90 | 24 | 8 | 16 |
|
|
≤90 | 39 | 28 | 11 |
|
Cell culture
Normal human astrocytes were purchased from the IBS
Cell Bank of Fudan University (Shanghai, China), and cultured in
astrocyte media (ScienCell Research Laboratories, Inc., Carlsbad,
CA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified
incubator containing 5% CO2. In addition, human glioma
cell lines, including A172, U87, U251 and U373, were purchased from
the Cell Bank of Chinese Academy of Sciences (Shanghai, China).
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS in a 37°C
humidified atmosphere with 5% CO2.
Cell transfection
Cell transfection was conducted in the U87 glioma
cell lines using Lipofectamine 2000 (Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. Briefly, U87
cells were cultured to 70% confluence, and resuspended in
serum-free DMEM. Scramble miR (miR-NC), miR-219 mimic for miR-219
upregulation, negative control (NC) inhibitor, miR-219 inhibitor
for miR-219 knockdown, and pc-DNA3.1-Sal-like protein 4 (SALL4)
open reading frame plasmid for SALL4 upregulation were diluted in
OPTI-MEM (Thermo Fisher Scientific, Inc.), which was then added
with diluted Lipofectamine 2000. All mimics, inhibitors and
plasmids were purchased from Yearthbio (Changsha, China). Following
incubation for 20 min at room temperature, the mixture was added
into the U87 cell suspension. Following incubation at 37°C and 5%
CO2 for 6 h, the transfection mixture was replaced with
DMEM with 10% FBS. MiR-NC was the corresponding control for miR-219
mimic, and NC inhibitor was the corresponding control for miR-219
inhibitor.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from tissues and cell lines
using TRIzol reagent (Thermo Fisher Scientific, Inc.). RNA quality
and concentration was measured using a Nanodrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.) RNA was
converted into cDNA using PrimeScript 1st Strand cDNA Synthesis kit
(Takara Bio, Inc., Tokyo, Japan). For miR-219 expression detection,
the miRNA Q-PCR Detection kit (GeneCopoeia, Rockville, MD, USA) was
used to conduct qPCR on an ABI 7300 Plus thermal cycler (Applied
Biosystems; Thermo Fisher Scientific, Inc.). U6 gene was used as an
internal control. For mRNA expression detection, SYBR Green I
Real-Time PCR kit (Biomics Biopharma, Nantong, China) was used to
conduct qPCR on an ABI 7300 Plus system. The primers for SALL4
were: Forward, 5′-AGCACATCAACTCGGAGGAG-3′ and reverse,
5′-CATTCCCTGGGTGGTTCACTG-3′. The primers for the internal control
gene GAD PH were: Forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse,
5′-AAGTGGTCGTTGAGGGCAATG-3′. The reaction conditions were as
follows: 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 30 sec. The relative expression levels were analyzed
by the 2−ΔΔCq method (16). According to the mean value of miR-219
levels that served as the cutoff value, glioma tissues were divided
into the high and low miR-219 expression groups.
Western blotting
U87 cells were lysed in ice-cold buffer containing
0.5 mol/l Tris-HCl, pH 7.4, 1.5 mol/l NaCl, 2.5% deoxycholic acid,
10% NP-40 and 10 mmol/l EDTA (Sigma-Aldrich; Merck, Darmstadt,
Germany) with cocktail protease inhibitors (Thermo Fisher
Scientific, Inc.). Cell lysates were then obtained by
centrifugation at 16,000 × g for 20 min at 4°C. Protein
concentrations were determined by Bio-Rad Protein Assay kit
(Bio-Rad Laboratories, Inc., Hercules, CA, USA), and 50 µg protein
was subjected to 12% SDS-PAGE, followed by immunoblotting onto a
polyvinylidene difluoride membrane (Thermo Fisher Scientific,
Inc.). The membrane was blocked for 1 h in phosphate-buffered
saline with Tween 20 (PBST) containing 5% nonfat dry milk (Yili,
Beijing, China). Subsequently, the membrane was incubated at room
temperature for 3 h with primary antibodies: Rabbit polyclonal to
SALL4 antibody (1:50, ab29112, Abcam, Cambridge, MA, USA) and
rabbit polyclonal to GAPDH antibody (1:50, ab9485, Abcam). The
membrane was then washed three times with PBST, then immersed in
PBST containing horseradish peroxidase-conjugated goat anti-rabbit
secondary antibody (1:5,000, ab7090, Abcam) at room temperature for
1 h. After washing in PBST four times (10 min/wash), an enhanced
chemiluminescence kit (Thermo Fisher Scientific, Inc.) was used to
perform chemiluminescence detection. The relative protein
expression was determined using Image J software v.1.48 (National
Institutes of Health, Bethesda, MD, USA), represented as the
density ratio vs. GAPDH.
Cell proliferation assay
Cell proliferation was measured by MTT assay. U87
cells in 200 µl serum-free DMEM were plated into 96-well plates at
a density of 5,000 cells per well. After cells were adhered, the
medium was replaced with DMEM containing 10% FBS, and incubated at
37°C for 12, 24, 48 and 72 h, respectively. Next, 20 µl MTT
solution (5 mg/ml) was added into each well. Following incubation
at 37°C for 4 h, the MTT solution was replaced with 150 µl dimethyl
sulfoxide to dissolve the tetrazolium crystals for 10 min.
Absorbance at 570 nm was detected with a microplate reader (Thermo
Fisher Scientific, Inc.).
Cell migration assay
U87 cells were plated at 1×105 cells/well
in 24-well plates with 1 ml DMEM and incubated for 24 h. Scratch
wounds were generated using a 200 µl pipette tip. The medium was
then replaced with fresh DMEM with 10% FBS. The wound size was
recorded by camera at 0 and 24 h, respectively. Quantification was
implemented by Image J v1.48 software.
Cell invasion assay
In this assay, 24-well-plate-matched Bio-coat cell
invasion chambers (BD Biosciences, Franklin Lakes, NJ, USA) were
used to measure the invasion of glioma cells. Briefly,
1.0×105 U87 cells were seeded in the upper transwell
chamber in serum-free medium, with 500 µl DMEM containing 10% FBS
in the lower chamber. Following incubation at 37°C for 24 h, the
cells that did not migrate through the pores were carefully wiped
out with a cotton-tipped swab. The cells that had migrated were
then fixed in 90% alcohol, followed by staining with 0.1% crystal
violet (Sigma-Aldrich; Merck). Subsequent to washing with PBS for
three times, invading cells were observed and photographed using an
inverted microscope (Olympus Corp., Tokyo, Japan).
Bioinformatics analysis
Bioinformatics analysis was performed to predict the
potential target genes of miR-219 using TargetScan online software
(version 7.1; www.targetscan.org), according to the manufacturer's
instructions. SALL4 was shown as the putative target gene of
miR-219, and this association was then confirmed using a luciferase
reporter gene assay.
Luciferase reporter gene assay
Luciferase reporter gene assay was conducted to
confirm the association between miR-219 and its potential target,
SALL4. Initially, the mutant type (MT) of SALL4 3′-untranslated
region (UTR) lacking complementarity with the miR-219 binding
sequence was constructed using QuickChange Site-Directed
Mutagenesis kit (Stratagene; Agilent Technologies, Inc., Santa
Clara, CA, USA), according to the manufacturer's instructions. The
wild type (WT) or mutant type (WT) 3′UTRs of SALL4 were also
constructed and inserted into the multiple cloning sites in the
psiCHECK2 luciferase reporter vector (Promega Corp., Madison, WI,
USA). Lipofectamine 2000 was used to co-transfected U87 cells with
WT-SALL4-3′UTR or MT-SALL4-3′UTR reporter plasmids, and miR-219
mimics or miR-NC. At 48 h after co-transfection, the luciferase
activities were examined using a dual-luciferase reporter assay
system (Promega Corp.), according to the manufacturer's protocol.
The Renilla luciferase activity was normalized to the
firefly luciferase activity.
Statistical analysis
Data in the present study are expressed as the mean
± standard deviation of three independent experiments. The
difference between two groups was analyzed using Student's t-test,
while differences among more than two groups were analyzed using
analysis of variance. Associations between SALL4 expression and the
clinicopathological characteristics in glioma patients were
assessed using the χ2 test. Statistical analysis was
performed using SPSS version 19.0 (IBM Corp., Armonk, NY, USA). A
P-value that was <0.05 was considered to indicate a
statistically significant difference.
Results
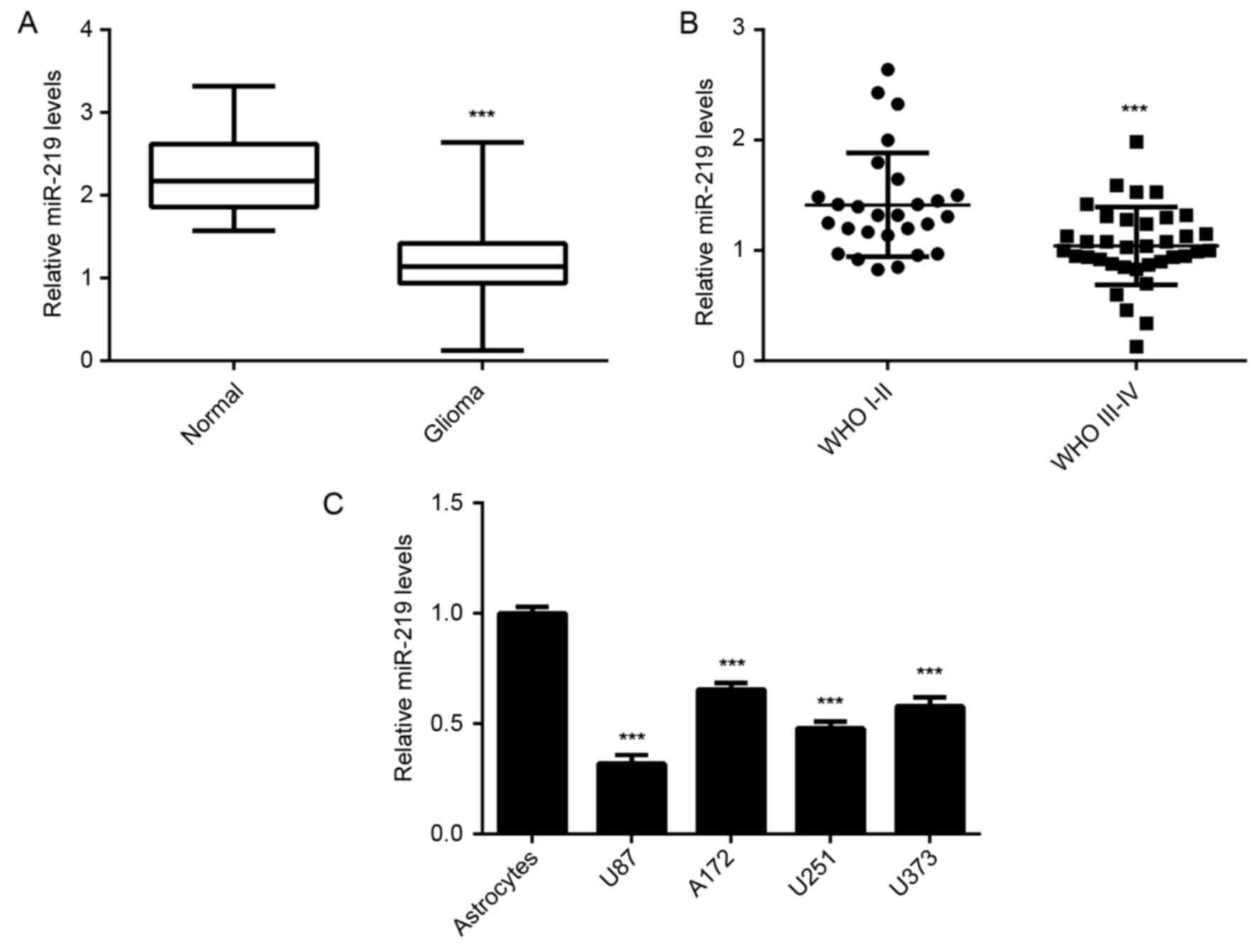
miR-219 is downregulated in
glioma
In the current study, the expression of miR-219 was
examined in a total of 63 glioma tissues and 12 normal brain
tissues. The RT-qPCR data revealed that the miR-219 levels were
significantly decreased in glioma tissues, when compared with the
normal brain tissues (Fig. 1A;
P<0.001). In addition, high-grade gliomas (WHO grade III–IV)
presented significantly lower levels of miR-219 as compared with
those in low-grade gliomas (WHO grade I–II) (Fig. 1B; P<0.001). Furthermore, miR-219
was significantly downregulated in several common human glioma cell
lines (U87, A172, U373 and U251) compared with the normal
astrocytes (P<0.001; Fig. 1C).
Therefore, miR-219 is concluded to be downregulated in glioma, and
may contribute to glioma progression.
Subsequently, the clinical significance of miR-219
expression in glioma was investigated. Glioma tissues were divided
into the high and low miR-219 expression groups, according to the
mean value of miR-219 levels that served as the cutoff value. It
was observed that low miR-219 expression was significantly
associated with an advanced WHO grade and KPS index, but not with
the age or sex of patients (Table
I). Accordingly, these results suggested that decreased
expression of miR-219 is significantly associated with glioma
progression in glioma patients.
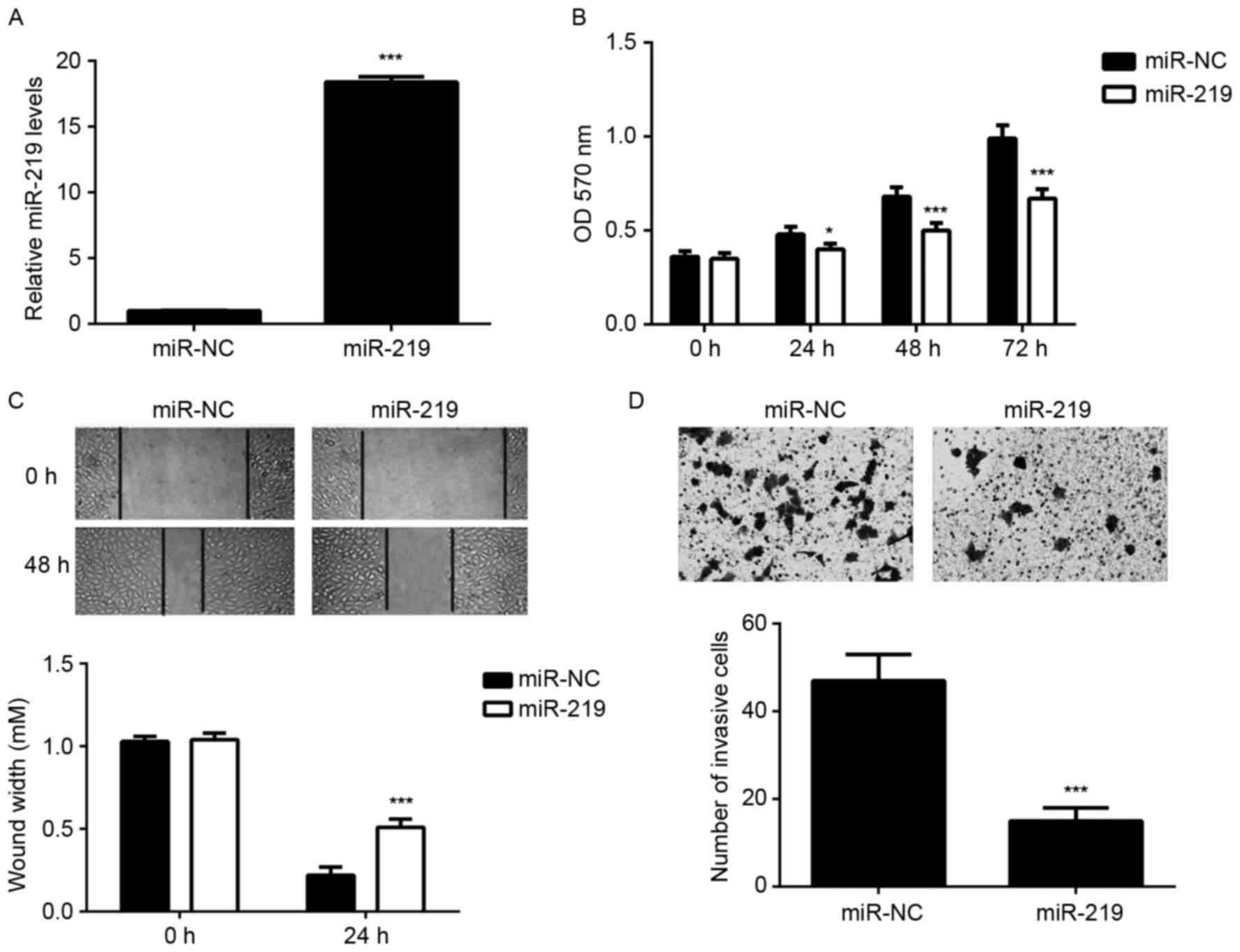
Ectopic expression of miR-219
suppresses U87 cell proliferation, migration and invasion
In order to upregulate the expression of miR-219,
U87 cells were transfected with miR-219 mimic. Transfection with
miR-NC was used as the control group. Following transfection, the
miR-219 levels were significantly higher in the miR-219 group
compared with those in the miR-NC group (Fig. 2A). MTT, wound healing and transwell
assays were then conducted to determine the cell proliferation,
migration and invasion, respectively. As indicated in Fig. 2B-D, ectopic expression of miR-219
significantly decreased the proliferation, migration and invasion
of U87 cells, when compared with the miR-NC group. These findings
suggest that miR-219 may function as a tumor suppressor in
glioma.
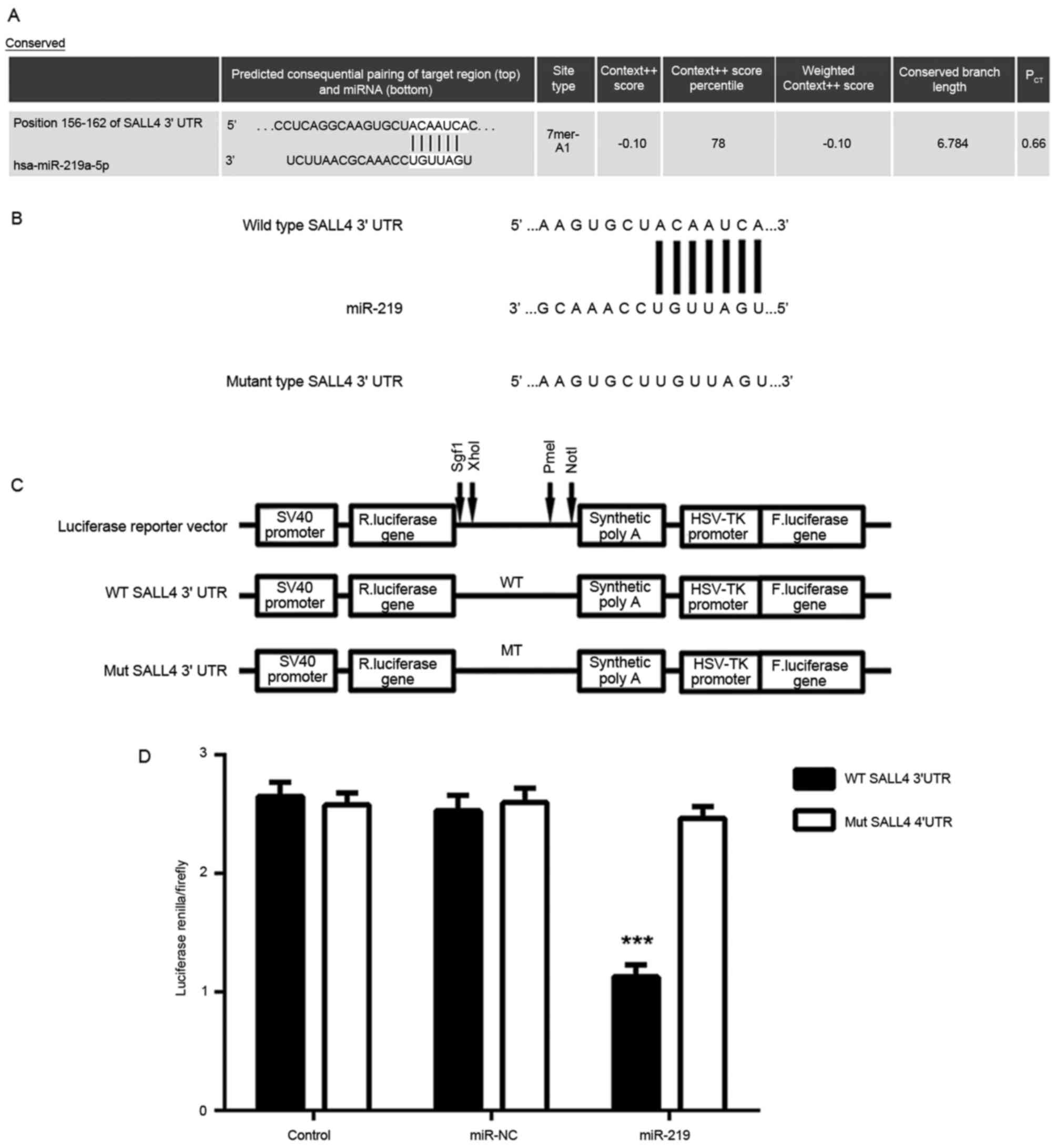
miR-219 negatively regulates its
target gene SALL4 in U87 cells
Next, bioinformatics analysis was performed to
predict the potential target genes of miR-219. As shown in Fig. 3A, SALL4 was observed to be a putative
target of miR-219. To verify the targeting association between
miR-219 and SALL4, luciferase reporter plasmids containing WT or MT
SALL4 3′UTRs were constructed (Fig. 3B
and C). The results of luciferase reporter gene assay
demonstrated that the luciferase activity was significantly
inhibited in U87 cells co-transfected with the WT-SALL4-3′UTR
luciferase reporter plasmid and miR-219 mimic, when compared with
the control group, but the luciferase activity was unchanged
compare with the controlwhen transfected with MT-SALL4-3′UTR
luciferase reporter plasmid and miR-219 mimic (Fig. 3D). These findings indicate that SALL4
is a target gene of miR-219, and that miR-219 directly binds to the
3′UTR of SALL4 mRNA in U87 cells.
As miRs generally inhibit the expression of their
target genes at the post-transcriptional level, the present study
then investigated the effects of miR-219 on the protein expression
of SALL4 in U87 cells. As shown in Fig.
4A, upregulation of miR-219 significantly decreased the protein
expression of SALL4. Next, U87 cells were transfected with miR-219
inhibitor to knockdown its expression, with NC inhibitor
transfection used as the control group. Following transfection, the
miR-219 levels were significantly reduced in the miR-219 inhibitor
group, when compared with the NC inhibitor group (Fig. 4B). Western blot analysis data
revealed that downregulation of miR-219 significantly increased the
protein expression of SALL4 in U87 cells as compared with that in
the NC inhibitor group (Fig. 4C).
Therefore, these findings suggest that miR-219 negatively regulated
the protein expression of its target gene SALL4 in U87 cells.
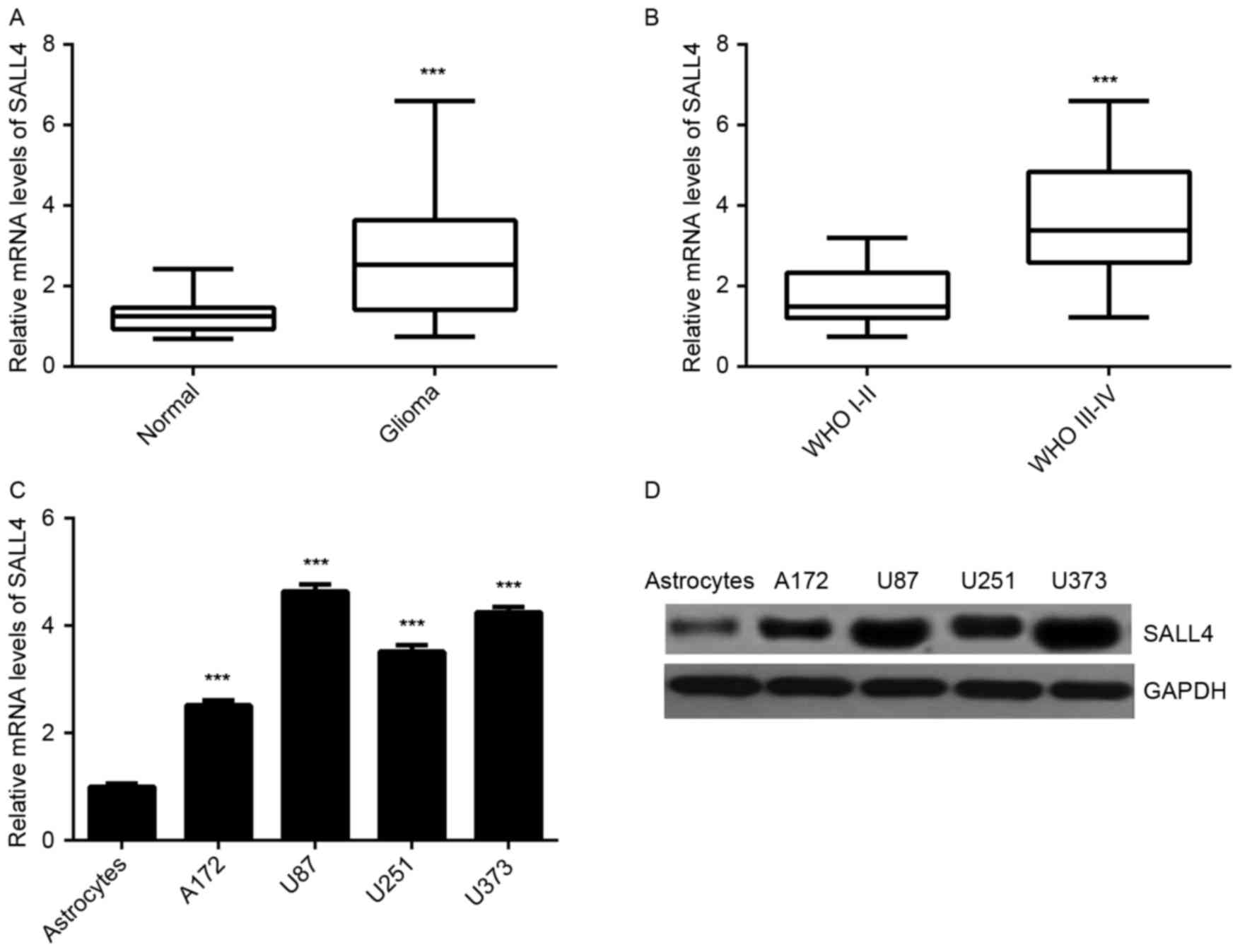
SALL4 is upregulated in glioma
cells
As the earlier experiments observed that miR-219 was
downregulated in glioma tissues and that it negatively regulated
SALL4 in glioma cells, the expression of SALL4 in glioma tissues
and cell lines was subsequently investigated. The results of
RT-qPCR indicated that SALL4 was significantly upregulated in
glioma tissues as compared with the normal brain tissues (Fig. 5A). Furthermore, the high-grade
gliomas (WHO grade III–IV) exhibited higher SALL4 levels in
comparison with those in low-grade gliomas (WHO grade I–II;
Fig. 5B). Similarly, SALL4
expression levels were also higher in glioma cell lines compared
with normal astrocytes (Fig. 5C and
D). These observations reveal that the increased expression of
SALL4 in glioma may be caused by the downregulation of miR-219.
Overexpression of SALL4 attenuated the
suppressive effects of miR-219 on the malignant phenotypes of U87
cells
The current study further examined whether SALL4 was
involved in the miR-219-mediated proliferation, migration and
invasion of U87 cells. miR-219-overexpressing U87 cells were
transfected with SALL4 expression plasmid, in order to upregulate
its expression. Following transfection, the mRNA and protein levels
of SALL4 were significantly higher in the miR-219 + SALL4 group
compared with those in the miR-219 group (Fig. 6A and B). MTT, wound healing and
transwell assays were subsequently conducted to determine cell
proliferation, migration and invasion, respectively. As indicated
in Fig. 6C-E, the proliferation,
migration and invasion of U87 cells were significantly upregulated
in the miR-219 + SALL4 group when compared with those in the
miR-219 group. Accordingly, overexpression of SALL4 attenuated the
suppressive effects of miR-219 upregulation on the malignant
phenotypes of U87 cells, suggesting that miR-219 serves a
suppressive role in glioma cells via directly targeting SALL4.
Discussion
MiR-219 was recently suggested to function as a
tumor suppressor in glioma, however, the underlying mechanism
remains largely unknown. In the present study, it was observed that
miR-219 was significantly downregulated in glioma tissues and cell
lines, and that reduced expression of miR-219 was significantly
associated with advanced pathological grade in glioma tissues.
Furthermore, ectopic expression of miR-219 inhibited the
proliferation, migration and invasion of U87 cells. SALL4, which
was markedly upregulated in glioma, was further identified as a
target gene of miR-219 in U87 cells, while overexpression of SALL4
significantly eliminated the suppressive effects of miR-219 on U87
cell proliferation, migration and invasion.
Recently, miR-219 has been reported to serve a
suppressive role in malignant tumors in the central nervous system
(17). For instance, Shi et
al reported that miR-219 inhibited the proliferation, migration
and invasion of medulloblastoma cells by targeting CD164 (17). In the present study, it was observed
that miR-219 was downregulated in glioma tissues compared with
normal brain tissues, as well as in glioma cell lines as compared
with normal astrocytes. The findings of the present study are
consistent with a previous study (12). In addition, low expression of miR-219
was identified to be significantly associated with high tumor
grade, suggesting that downregulation of miR-219 may contribute to
the malignant progression of glioma. Furthermore, it was
demonstrated that ectopic expression of miR-219 decreased the
proliferation, migration and invasion of U87 cells. Consistent with
the present study data, Jiang et al (12) also reported that miR-219 inhibited
cell proliferation and invasion, induced the apoptosis of
glioblastoma cells in vitro, and inhibited xenograft
formation in vivo. Rao et al (13) demonstrated that overexpression of
miR-219 in glioma cell lines inhibited the proliferation, growth
and migration via inhibition of receptor tyrosine kinase pathway
through directly targeting EGFR. Rao et al (18) also reported that overexpression of
miR-219-5p caused decreased soft agar colony formation of
glioblastoma cells.
The present study further identified SALL4 as a
target gene of miR-219, and the protein levels of SALL4 were
negatively mediated by miR-219 in U87 cells. SALL4, a zinc finger
transcription factor, is considered as an important marker for stem
cells and participates in the maintenance of embryonic stem cell
self-renewal (19). Recently, SALL4
has been demonstrated to be significantly upregulated in certain
types of human cancer, including hepatocellular carcinoma (20), intrahepatic cholangiocarcinoma
(21), esophageal squamous cell
carcinoma (22), gastric cancer
(23) and lung cancer (24). For instance, Deng et al
(21) showed that the increased
expression of SALL4 contributed to the malignant progression in
intrahepatic cholangiocarcinoma, as well as to the poor prognosis
of patients. It has been demonstrated that SALL4 can promote tumor
cell growth, metastasis and angiogenesis, as well as induce drug
resistance (21–23). Oikawa et al (25) reported that overexpression of SALL4
increased hepatocellular carcinoma cell proliferation in
vitro, while knockdown of SALL4 led to a significant decrease
in growth inhibition in vitro and in vivo. Kobayashi
et al (24) observed that
inhibition of SALL4 inhibited the proliferation of lung cancer
cells through induction of a cell cycle arrest at the G1 phase. In
the present study, SALL4 was significantly upregulated in glioma
tissues and cell lines, while high-grade gliomas (WHO grade III–IV)
presented higher SALL4 levels when compared with those in low-grade
tumors (WHO grade I–II). These findings are consistent with a
previous study reporting that increased expression of SALL4 was
associated with advanced pathological grade in glioma (26). Therefore, the increased expression of
SALL4 may be caused by the reduced miR-219 levels in glioma
tissues.
As SALL4 was negatively regulated by miR-219 in U87
cells, SALL4 may be involved in the miR-219-mediated malignant
phenotypes of U87 cells. To verify this hypothesis,
miR-219-overexpressing U87 cells were then transfected with SALL4
expression plasmid to upregulate its expression. The results in the
current study indicated that overexpression of SALL4 significantly
attenuated the inhibitory effects of miR-219 upregulation on U87
cell proliferation, migration and invasion. These findings support
that the tumor suppressive role of miR-219 in glioma cells is, at
least partly, via inhibition of SALL4 expression. Similarly, a
previous study reported that miR-107 inhibited glioma cell
proliferation and induced cell apoptosis via directly targeting
SALL4 (27). Therefore, the present
study further highlights the importance of the miR/SALL4 axis in
glioma.
In conclusion, to the best of our knowledge, the
current study is the first to demonstrate that miR-219 exerts
suppressive effects on the malignant phenotypes of glioma cells,
which was, at least partly, through inhibition of SALL4 expression.
These findings suggest that the miR-219/SALL4 association may be a
potential therapeutic target for glioma.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marumoto T and Saya H: Molecular biology
of glioma. Adv Exp Med Biol. 746:2–11. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:pp. 2999–3004. 2004, View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
An L, Liu Y, Wu A and Guan Y: microRNA-124
inhibits migration and invasion by down-regulating ROCK1 in glioma.
PLoS One. 8:e694782013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jia Z, Wang K, Wang G, Zhang A and Pu P:
MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth
by targeting SEPT7. PLoS One. 8:e550082013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lei H, Zou D, Li Z, Luo M, Dong L, Wang B,
Yin H, Ma Y, Liu C, Wang F, et al: MicroRNA-219-2-3p functions as a
tumor suppressor in gastric cancer and is regulated by DNA
methylation. PLoS One. 8:e603692013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang N, Lin J, Ruan J, Su N, Qing R, Liu
F, He B, Lv C, Zheng D and Luo R: MiR-219-5p inhibits
hepatocellular carcinoma cell proliferation by targeting
glypican-3. FEBS Lett. 586:884–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang Y, Yin L, Jing H and Zhang H:
MicroRNA-219-5p exerts tumor suppressor function by targeting ROBO1
in glioblastoma. Tumour Biol. 36:8943–8951. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rao SA, Arimappamagan A, Pandey P, Santosh
V, Hegde AS, Chandramouli BA and Somasundaram K: miR-219-5p
inhibits receptor tyrosine kinase pathway by targeting EGFR in
glioblastoma. PLoS One. 8:e631642013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Komori T: The 2016 WHO classification of
tumours of the central nervous system: The major points of
revision. Neurol Med Chir (Tokyo). 57:301–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Terret C, Albrand G, Moncenix G and Droz
JP: Karnofsky performance scale (KPS) or physical performance test
(PPT)? That is the question. Crit Rev Oncol Hematol. 77:142–147.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi JA, Lu DL, Huang X and Tan W: miR-219
inhibits the proliferation, migration and invasion of
medulloblastoma cells by targeting CD164. Int J Mol Med.
34:237–243. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rao SA, Santosh V and Somasundaram K:
Genome-wide expression profiling identifies deregulated miRNAs in
malignant astrocytoma. Mod Pathol. 23:1404–1417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Vega VB and Ng HH: Transcriptional
regulatory networks in embryonic stem cells. Cold Spring Harb Symp
Quant Biol. 73:203–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Han SX, Wang JL, Guo XJ, He CC, Ying X, Ma
JL, Zhang YY, Zhao Q and Zhu Q: Serum SALL4 is a novel prognosis
biomarker with tumor recurrence and poor survival of patients in
hepatocellular carcinoma. J Immunol Res. 2014:2623852014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng G, Zhu L, Huang F, Nie W, Huang W, Xu
H, Zheng S, Yi Z and Wan T: SALL4 is a novel therapeutic target in
intrahepatic cholangiocarcinoma. Oncotarget. 6:27416–27426. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Forghanifard MM, Khales Ardalan S,
Javdani-Mallak A, Rad A, Farshchian M and Abbaszadegan MR: Stemness
state regulators SALL4 and SOX2 are involved in progression and
invasiveness of esophageal squamous cell carcinoma. Med Oncol.
31:9222014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Xu Z, Xu X, Zhang B, Wu H, Wang
M, Zhang X, Yang T, Cai J, Yan Y, et al: SALL4, a novel marker for
human gastric carcinogenesis and metastasis. Oncogene.
33:5491–5500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kobayashi D, Kuribayashi K, Tanaka M and
Watanabe N: Overexpression of SALL4 in lung cancer and its
importance in cell proliferation. Oncol Rep. 26:965–970.
2011.PubMed/NCBI
|
|
25
|
Oikawa T, Kamiya A, Zeniya M, Chikada H,
Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW, et
al: Sal-like protein 4 (SALL4), a stem cell biomarker in liver
cancers. Hepatology. 57:1469–1483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang L, Yan Y, Jiang Y, Cui Y, Zou Y,
Qian J, Luo C, Lu Y and Wu X: The expression of SALL4 in patients
with gliomas: High level of SALL4 expression is correlated with
poor outcome. J Neurooncol. 121:261–268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|