Introduction
Prostate cancer is a common visceral cancer of men
worldwide (1). Patients in which the
disease is clinically localized at diagnosis typically receive a
radical prostatectomy or radiotherapy treatment (2,3).
However, there is a 20–40% recurrence rate within a year following
these treatments (4). Patients with
recurrent, metastatic or locally advanced prostate cancer are
primarily treated with androgen deprivation therapy (ADT) (5). However, all patients eventually develop
resistance to ADT, termed castration-resistant prostate cancer
(CRPC). Therefore, it is necessary to develop a more effective
treatment for CRPC.
Special AT-rich sequence-binding protein 1 (SATB1)
is a transcription factor that regulates histone modifications and
serves an important role in gene transcription (6). It has been previously demonstrated that
SATB1 is overexpressed in a variety of types of malignant cancer,
including nasopharyngeal carcinoma (7), cutaneous malignant melanoma (8), osteosarcoma (9) and small cell lung cancer (10). The authors of the present study have
previously demonstrated that SATB1 expression was positively
correlated with bone metastasis and the Gleason score of prostatic
carcinomas (11). SATB1
overexpression promoted prostate cancer cell proliferation and
invasion, whereas SATB1 knockdown inhibited it (11). These findings indicated that SAYB1
serves an oncogenic role in prostate cancer development (6–11).
RNA interference may downregulate target genes and
has been suggested as a potential therapeutic strategy in human
cancer therapy (7). RNA interference
involves post-transcriptional gene silencing via a process in which
double-stranded RNA inhibits gene expression in a
sequence-dependent manner through degradation of the corresponding
mRNA (12). Gene knockdown and its
inhibitory action on gene expression have been successfully
observed in rat (13) and human
cells cultured in vitro (14). The inhibition of several targets
using synthetic small interfering (si)RNA suppresses the growth of
various cancer cell lines and provides a potential nucleotide-based
approach for suppressing gene products for loss of function
analyses (7,8).
Based on previous studies, three siRNA against SATB1
were designed and the efficacy of SATB1 siRNA transfection was
investigated with regard to prostate cancer cells and the
suppression of SATB1 expression in vitro. The findings of
the present study suggest that SATB1 may be an important target
protein in prostate cancer cells.
Materials and methods
Cell culture
The prostate cancer cell line DU145 and normal human
lung fibroblast (NHLF) cells were purchased from the Shanghai Cell
Collection (Shanghai, China) (15).
The cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% heat-inactivated fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), 4 mM glutamine, 50 U/ml
penicillin and 50 µg/ml streptomycin at 37°C in a humidified
atmosphere with 5% CO2. The cells were screened
routinely to verify the absence of mycoplasma contamination in the
log phase of growth.
siRNA preparation
siRNA duplex sequences were synthesized, purified
and annealed by Ambion (Thermo Fisher Scientific, Inc.). The three
SATB1 siRNA targeting the region containing nucleotides 2147–2185
of the SATB1 complementary DNA (ncbi.nlm.nih.gov; accession no. AB6304) were as
follows: SATB1 siRNA1 (siRNA1), sense 5′-CGAGUCCUUAAACCAAACAATT-3′
and antisense 3′-UUGUUGGUUUAAGGACUGCTT-5′; SATB1 siRNA2 (siRNA2),
sense 5′-GAGGUGUCUUCCGAAAUCUTT-3′ and antisense
3′-AGAUUUCGGAAGACACCUCTT-5′; SATB1 siRNA3 (siRNA3), sense
5′-CCCAGUCUUUGCUGGUAAATT-3′ and antisense
3′-UUUACCAGCAAAGACUGGGTT-5′; and NC sense
5′-UUCUCCGAACGUGUCAGUTT-3′ and antisense
3′-TTAAGAGGCUUGCACAGUGCA-5′. The selected sequences were submitted
to the Basic Local Alignment Search Tool (ncbi.nlm.nih.gov/blast/) to ensure that the selected
genes were targeted specifically. A green fluorescein-labeled (FAM)
siRNA was purchased from Ambion (Thermo Fisher Scientific, Inc.)
and used as the negative control (NC).
siRNA transfection
siRNA transfection was performed using siPORT™ Lipid
Transfection agent (Ambion; Thermo Fisher Scientific, Inc.). A
total of 3 µl lipid and 3 µl siRNA were diluted separately in 250
µl Opti-MEM® I (Invitrogen; Thermo Fisher Scientific,
Inc.). The diluted lipid was mixed with the diluted siRNA and the
mixture was incubated for 20 min at room temperature for complex
formation. The Opti-MEM® I complex was added to each
well of a 6-well plate to a total volume of 200 µl and the entire
mixture was added to the cells in one well resulting in a final
concentration of 10, 50 or 100 nM siRNA. The blank group was not
transfected with anything. The cells were harvested and assayed at
24, 48 or 72 h following transfection.
Detecting transfection efficiency
under fluorescence microscopy
DU145 cells were plated at a density of
105 cells/6 cm dish. The NC (FAM siRNA) and Blank group
were used to transfect the cells for 24 h. The cell transfection
efficiency was detected under an Olympus fluorescence microscope
(Olympus Corporation, Tokyo, Japan) and images were captured in the
bright field at 24 h following treatment (magnification, ×200).
Cell viability assay
The viability of the DU145 and NHLF cells was
examined using a Cell Counting kit-8 (CCK-8; Shanghai Tongren
Pharmaceutical Co., Ltd., Shanghai, China) according to the
manufacturer's protocol. Briefly, 5×103 cells/well were
seeded in 96-well plates and cultured for 48 h at 37°C. A total of
10 µl CCK-8 solution was added to each well and the plates were
incubated for 1 h at 37°C. The absorbance at 450 nm was measured
using a microplate reader. Results were representative of three
individual experiments in triplicate.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from DU145 cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. RNA was reverse transcribed at 37°C
for 40 min using BeyoRT™ M-MuLV Reverse Transcriptase (Beyotime
Institute of Biotechnology, Haimen, China). The thermal protocol
for the PCR was 3 min at 94°C, followed by 30 cycles of 30 sec at
94°C, 30 sec at 58°C and 1 min at 72°C and 1 cycle of 10 min at
72°C. PCR amplifications were performed as described using
SYBR®Green and TaqMan™ (Beyotime Institute of
Biotechnology). The 2−ΔΔCq method of quantification was
used as previously described (16).
The following primers were used: β-actin, forward
5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse 5′-GGGCACGAAGGCTCATCATT-3′;
and SATB1, forward 5′-TGCAAAGGTTGCAGCAACCAAAAGC-3′ and reverse
5′-AACATGGATAATGTGGGGCGGCCT-3′. All primers were purchased from
Invitrogen (Thermo Fisher Scientific, Inc.).
Western blot analysis
The DU145 cells were harvested from the plates and
aliquots of cell extracts were separated on 12% SDS-PAGE. A
radioimmunoprecipitation protein lysis extraction buffer (Beyotime
Institute of Biotechnology) was used and the BCA method was used to
determination the protein content. For each individual sample 20 µl
protein was loaded per lane into a 4% (w/v) agarose gel and run in
an electrophoresis buffer for 2 h. The proteins were then
transferred onto a nitrocellulose membrane and blocked by 3% bovine
serum albumin (BSA; Gibco; Thermo Fisher Scientific, Inc.) at 37°C
for 2 h and incubated overnight at 4°C with rabbit polyclonal
antibodies (all dilution 1:1,000) directed against the following
proteins: SATB1 (cat. no. PA5-30163; GenHunter Corporation,
Nashville, TN, USA), matrix metalloproteinase 2 (MMP2; cat. no.
AF902; Cell Signaling Technology, Inc., Danvers, MA, USA) and
β-actin (cat. no. AB10024; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA). The membranes were then washed with a washing buffer (ph
7.5) consisting of 100 mM Tris, 1.21 g; 100 mM NaCl, 5.84 g; and
0.1% Tween-20 1 ml and incubated with horseradish
peroxidase-labeled goat anti-rabbit immunoglobulin G [(H+L); cat.
no. ZB2301; dilution 1:2,000; OriGene Technologies, Inc., Beijing,
China)] in Tris-buffered saline containing Tween-20 at 37°C for 2 h
and developed using the nitroblue tetrazolium
chloride/5-bromo-4-chloro-3-indolyl phosphate color substrate
(Promega Corporation, Madison, WI, USA). The band densities were
scanned and analyzed using ImageJ software version 1.48 u (National
Institutes of Health, Bethesda, MD, USA).
Cell adhesion, migration and invasion
assays
Polyvinyl chloride microtiter plates (96 wells) were
coated with 50 µl SATB1 siRNA and incubated at 4°C overnight.
Following washing with 0.9% sodium chloride, each well was filled
with DMEM supplemented with 1% BSA and containing the cells, for 2
h at 37°C and washed again. For the adhesion assay the cells were
detached using trypsin (Cell Signaling Technology, Inc.) digestion
at 37°C for 24 h, washed three times with 0.9% sodium chloride and
resuspended in incomplete DMEM; the cells were subsequently added
to the siRNA-coated plates at 2×104 cells/well in 100 µl
DMEM. Following 1 h incubation at 37°C in 5% CO2, the
unbound cells were removed by washing with incomplete DMEM.
Adherent cells were fixed with a solution of 4% paraformaldehyde in
PBS (pH 7.2) at 37°C for 30 min and stained with 0.5% crystal
violet at 37°C for 30 min, and subsequently observed using a light
microscope at magnification, ×100. The absorbance of the plates was
read on an ELX-800 spectrometer reader (BioTek Instruments, Inc.,
Winooski, VT, USA) at 490 nm.
The cell migration and invasion assays were
performed using a modified two-chamber migration apparatus with an
8-µm pore size (Corning Incorporated, Corning, NY, USA). For the
migration assays, the underside of a Transwell filter was coated
with 10 µg/ml human plasma fibronectin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) at 37°C overnight. Briefly, siRNA-transfected
DU145 cells (5×104) and DMEM without FBS were seeded
into the upper chambers and DMEM supplemented with 10% FBS was used
to fill the lower compartment. Following 24 h incubation at 37°C in
a humidified atmosphere containing 5% CO2, the cells in
the upper chamber were removed with a cotton swab. The filters were
fixed in 95% methanol at 37°C for 30 min and stained with crystal
violet at 37°C for 30 min. The cells that migrated to the lower
surface were counted in five light microscopic fields per filter at
magnification, ×100 in a minimum of three independent
experiments.
For the invasion assay, a similar procedure was
performed using Matrigel-coated (Sigma-Aldrich; Merck KGaA)
Transwell chambers. Cells at the bottom of the wells were
quantified following 48 h incubation at 37°C using a light
microscope. Cells that migrated to the bottom surface of the
membrane were observed under light microscopy and then photographed
in five random fields at magnification, ×100.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Data were analyzed by one-way analysis of variance followed by
Duncan's new multiple range method or the Newman-Keuls test.
P<0.05 was considered to indicate a statistically significant
difference. All statistical analyses were performed by using SPSS
version 13.0 for Windows (SPSS, Inc., Chicago, IL, USA) and each
experiment was repeated five times.
Results
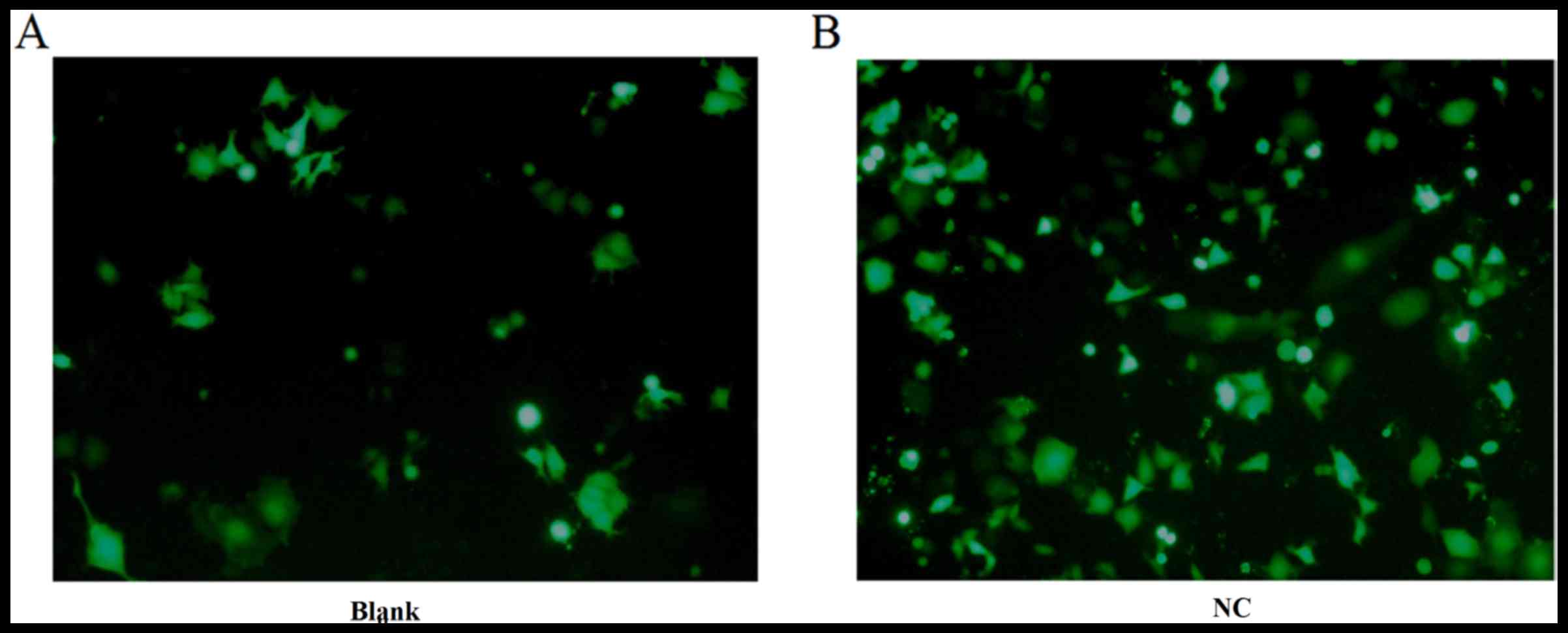
Detecting transfection efficiency
under fluorescence microscopy
The NC (FAM siRNA) and the Blank group were used to
transfect DU145 cells for 24 h as described above. The cell
transfection efficiency was detected under fluorescence microscopy
and images were captured in the bright field (Fig. 1). The green fluorescence within the
cytoplasm was clearer in the NC group than in the blank group.
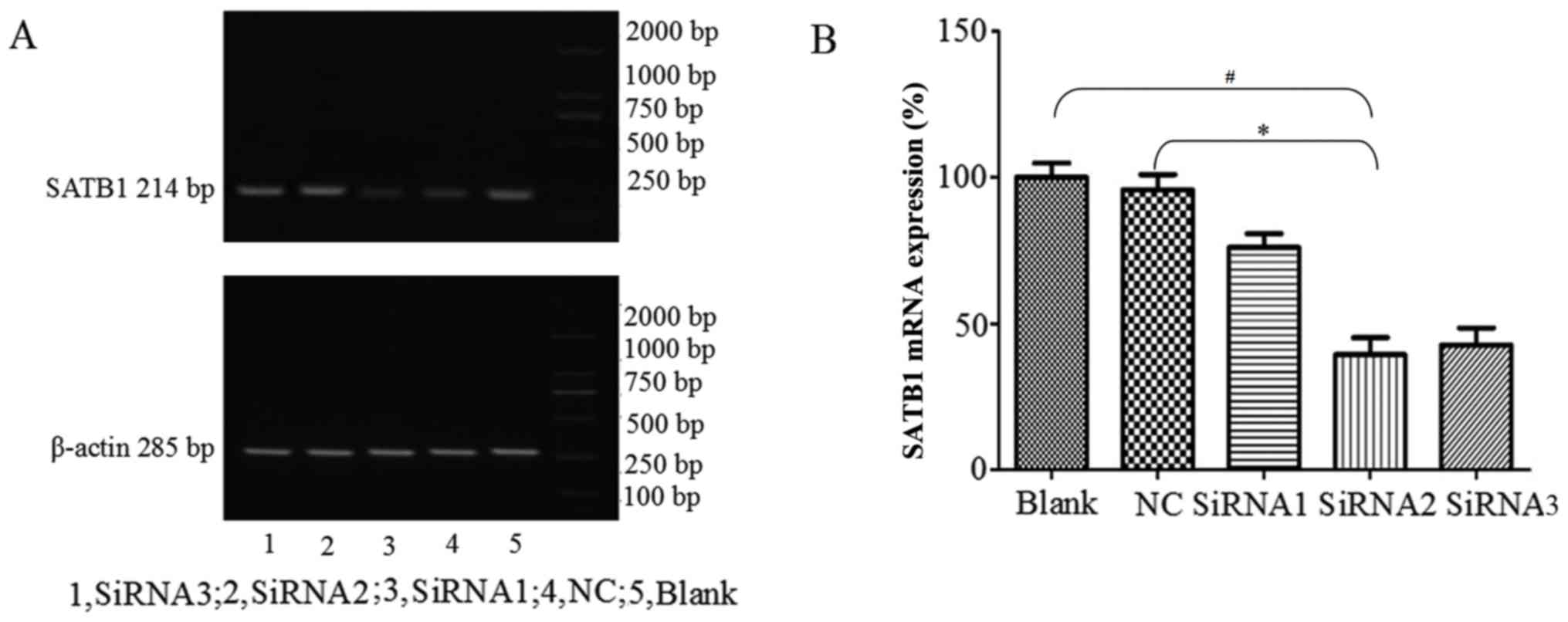
SATB1 and MMP2 expression following
siRNA transfection
Three siRNA were transfected into cells using
siPORT™ Lipid Transfection agent, and blank control (Blank) and NC
groups were also established. Following 48 h transfection, the
cells were collected and SATB1 mRNA and protein expression levels
were detected using RT-qPCR and western blot analysis,
respectively. The amplified fragment observed following RT-qPCR was
of the expected size (Fig. 2A). The
levels of SATB1 mRNA expression were significantly lower following
siRNA2 transfection compared with the NC and Blank groups
(P<0.05; Fig. 2B). siRNA2 had the
most notable inhibitory effect compared with the other siRNA; SATB1
mRNA expression following siRNA2 transfection was 39.39±5.78%.
Western blotting revealed that the levels of SATB1
protein expression following siRNA2 transfection were significantly
lower than the levels in the NC and Blank groups (P<0.05;
Fig. 3). SATB1 protein expression
was 0.69±0.03% following siRNA1 transfection, 0.28±0.05% following
siRNA2 transfection and 0.36±0.04% following siRNA3 transfection
(Fig. 3A and B). Similar to the mRNA
expression findings, siRNA2 had the most notable inhibitory effect
of the siRNA groups. As siRNA2 had the most notable inhibitory
effect of the siRNA groups, only siRNA2 transfection was
investigated in the following experiments.
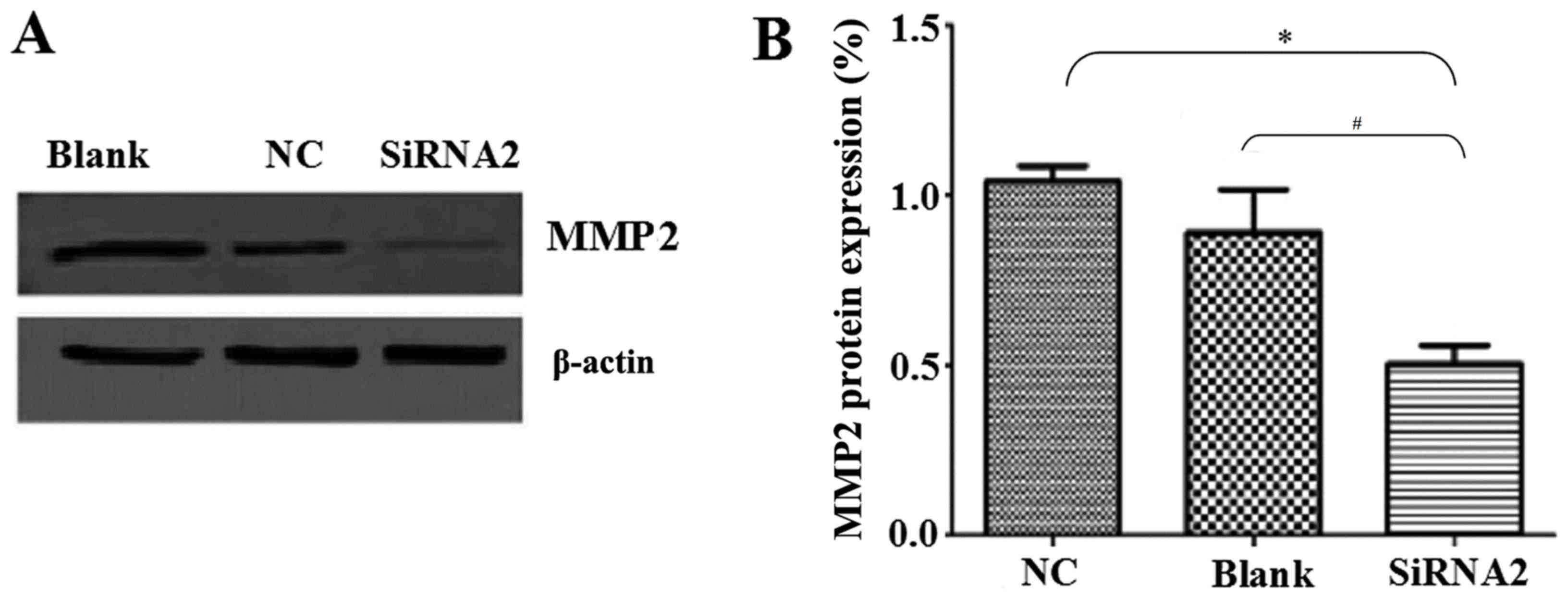
The levels of MMP2 protein expression were also
determined by western blot analysis. MMP2 is involved in radical
decomposition of the basement membrane and its overexpression is
closely associated with the metastasis of malignant tumors
(17). MMP2 protein levels were
significantly reduced by siRNA2 transfection compared with the
levels in the NC and Blank groups (P<0.05; Fig. 4A and B). MMP2 expression was
0.50±0.06% in the siRNA2 transfection group, 1.04±0.05% in the NC
group and 0.89±0.10% in the Blank group.
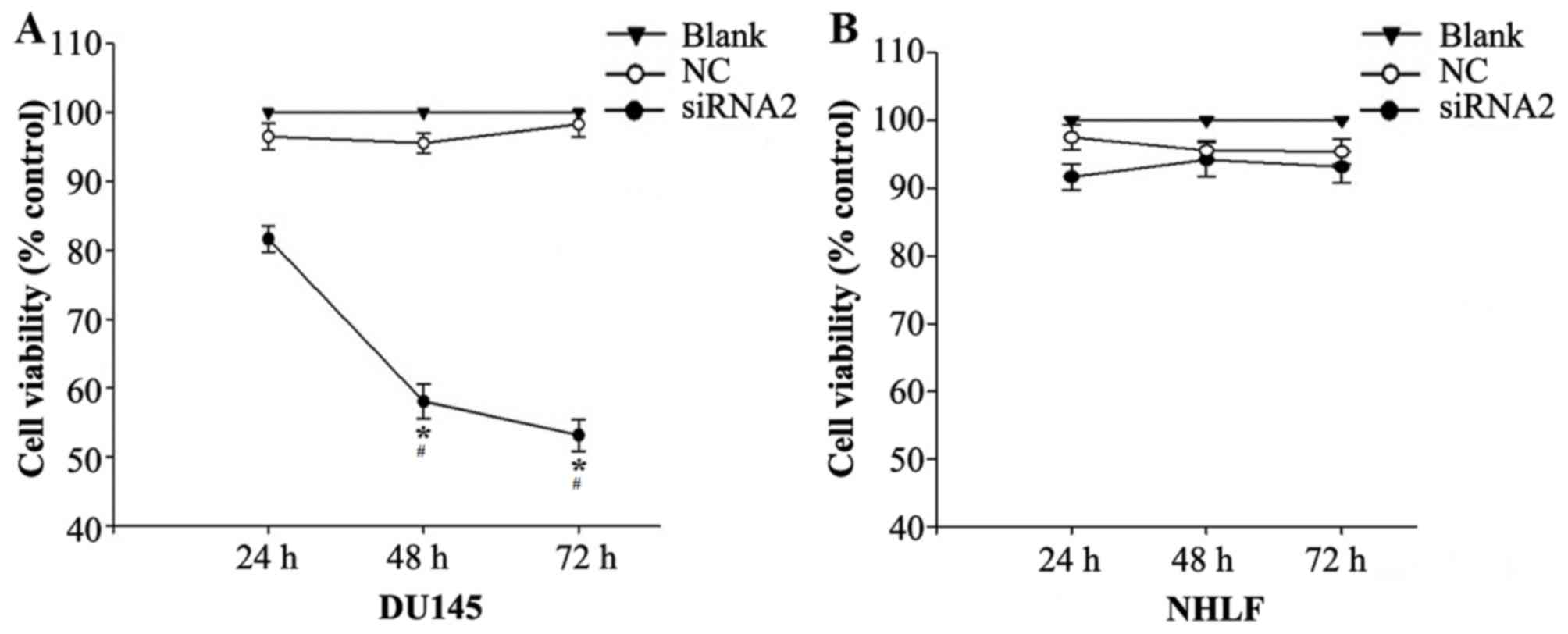
Silencing SATB1 inhibits the growth of
DU145 and NHLF cells
siRNA2 was transfected into DU145 and NHLF cells and
their viability was investigated using a CCK-8 assay. CCK-8
demonstrated that siRNA2 significantly inhibited DU145 cell
viability compared with the level in the NC and Blank groups
(P<0.05). The DU145 cell viability rate following siRNA2
transfection was 81.65±1.91%, 58.07±2.53% and 53.16±2.31% at 24, 48
and 72 h, respectively, following transfection (Fig. 5A). In the NHLF cells siRNA2
transfection had similar toxic effects compared with the NC and
Blank groups, no significant differences were observed (Fig. 5B).
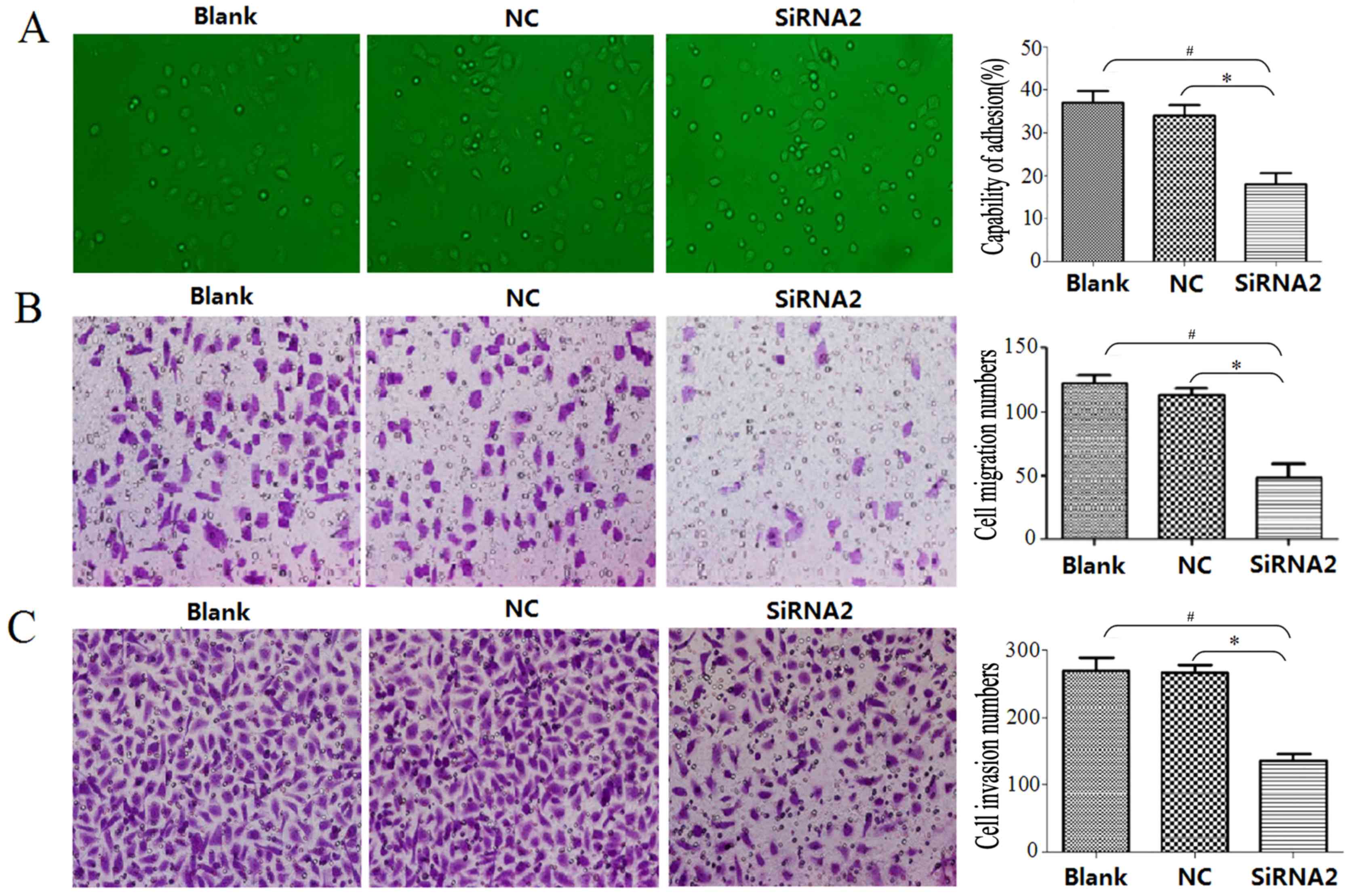
siRNA2 reduces cell adhesion,
migration and invasive capability in DU145 cells
DU145 cell adhesion capability was evaluated using a
cell adhesion assay, the migration capability was evaluated using a
Transwell migration assay and the invasive capability was evaluated
using a Transwell invasion assay. The adhesion assay demonstrated
that siRNA2 significantly inhibited adhesion between DU145 cells
and the cell-extracellular matrix compared with that observed in
the Blank and NC groups (P<0.05); the cell adhesion capability
was 18±6% in the siRNA2 transfection group, 34±4% in the NC group
and 37±6% in the Blank group (Fig.
6A). At 24 h, the number of migrated cells in the siRNA2
transfection group, NC group and Blank group was 49.08±10.64,
115.47±5.98 and 122.53±6.06, respectively (Fig. 6B). The siRNA2 transfection group had
a significantly reduced number of migrated cells compared with the
number in the NC and Blanks groups (P<0.05). DU145 cell invasive
ability was significantly inhibited 48 h following siRNA2
transfection compared with the number in the NC and Blanks groups
(P<0.05). The number of invading cells was 136.60±10.31 in the
siRNA2 transfection group, 270.19±11.70 in the NC group and
276.56±17.95 in the Blank group (Fig.
6C).
Discussion
Prostate cancer is the most frequently diagnosed
non-skin cancer among men and presents a global public health
problem (18). At present, there is
limited information available to determine which cases of prostate
cancer are likely to remain latent vs. those that are likely to
metastasize and require additional aggressive treatment (19). SATB1 has been proposed as a potential
oncogene, and siRNA-mediated knockdown of SATB1 in aggressive
cancer cells has been demonstrated to inhibit tumor growth and
metastasis (20). Previous studies
have suggested that SATB1 modulates cell proliferation and lineage
development and it has been implicated in breast cancer (21).
In the present study, it was demonstrated that SATB1
siRNA effectively inhibited the viability of prostate DU145 cancer
cells. The levels of SATB1 mRNA expression following transfection
with SATB1 siRNA2 were significantly lower than that in the NC
group. siRNA2 was transfected into DU145 and NHLF cells and a CCK-8
assay revealed that siRNA2 significantly inhibited DU145 cell
viability compared with the NC and Blank groups. siRNA2
transfection had similar effects in the NHLF cells.
Western blot analysis revealed that the levels of
SATB1 and MMP2 protein expression following siRNA2 transfection
were significantly lower than the NC group. The adhesion assay
demonstrated that siRNA2 significantly inhibited the adhesion
between DU145 cells and the cell-extracellular matrix.
In a previous study, it was observed that SATB1
staining was stronger in prostatic carcinoma with metastasis than
in prostatic carcinoma without metastasis and was absent in benign
prostate hyperplasia (11). These
results suggest that SATB1 is crucially implicated in the
metastasis of prostate cancer (11).
In the present study, SATB1 siRNA inhibited prostate cancer cell
invasion effectively in vitro. These results are consistent
with a previous study that reported that pSilencer-SATB1-short
hairpin (sh)RNA was markedly efficacious against prostate cancer
xenografts in nude mice (20).
Mao et al (22) reported that the toxicity of the
oncolytic adenovirus (OAds) carrying shRNA targeting SATB1 was
reduced by generating E1B 55-kDa-deleted OAds. However, others have
reported that the oncolytic anti-tumor activity of OAds, including
reducing the circulation time, inducing immunogenicity and
increasing the accumulation and toxicity in the liver, are
insufficient for effectively eliminating tumors (23). Therefore, there is an urgent
requirement for the development of novel treatments.
In the present study, it was demonstrated that
silencing SATB1 significantly inhibited DU145 cell viability,
adhesion, migration and invasion in vitro, indicating that a
SATB1-targeting siRNA was successfully engineered. The approach
exhibited clear anti-cancer cell efficacy. The results of the
present study suggest that SATB1 siRNA may be a promising agent for
the treatment of human prostate cancer, although further studies
are required in vivo to confirm this.
References
|
1
|
Suh YS, Joung JY, Kim SH, Seo HK, Chung J
and Lee KH: Establishment and application of prostate cancer
circulating tumor cells in the Era of precision medicine. Biomed
Res Int. 2017:72063072017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yamoah K, Beecham K, Hegarty SE, Hyslop T,
Showalter T and Yarney J: Early results of prostate cancer
radiation therapy: An analysis with emphasis on research strategies
to improve treatment delivery and outcomes. BMC Cancer. 13:232013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pinkawa M, Schoth F, Böhmer D, Hatiboglu
G, Sharabi A, Song D and Eble MJ: Current standards and future
directions for prostate cancer radiation therapy. Expert Rev
Anticancer Ther. 13:75–88. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barret E, Harvey-Bryan KA, Sanchez-Salas
R, Rozet F, Galiano M and Cathelineau X: How to diagnose and treat
focal therapy failure and recurrence? Curr Opin Urol. 24:241–246.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qin J, Lee HJ, Wu SP, Lin SC, Lanz RB,
Creighton CJ, DeMayo FJ, Tsai SY and Tsai MJ: Androgen
deprivation-induced NCoA2 promotes metastatic and
castration-resistant prostate cancer. J Clin Invest. 124:5013–5026.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tattermusch A and Brockdorff N: A scaffold
for X chromosome inactivation. Hum Genet. 130:247–253. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ye CS, Zhou DN, Yang QQ and Deng YF:
Silencing SATB1 influences cell invasion, migration, proliferation,
and drug resistance in nasopharyngeal carcinoma. Int J Clin Exp
Pathol. 7:914–922. 2014.PubMed/NCBI
|
|
8
|
Zhang L, Cheng F, He R, Chen H, Liu Y and
Sun J: Inhibition of SATB1 by shRNA suppresses the proliferation of
cutaneous malignant melanoma. Cancer Biother Radiopharm. 29:77–82.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Su X, Guo L, Zhong L, Li W, Yue
Z, Wang X, Mu Y, Li X, Li R and Wang Z: Silencing SATB1 inhibits
the malignant phenotype and increases sensitivity of human
osteosarcoma U2OS cells to arsenic trioxide. Int J Med Sci.
11:1262–1269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang B, Zhou H, Wang X and Liu Z:
Silencing SATB1 with siRNA inhibits the proliferation and invasion
of small cell lung cancer cells. Cancer Cell Int. 13(8)2013.
|
|
11
|
Mao L, Yang C, Wang J, Li W, Wen R, Chen J
and Zheng J: SATB1 is overexpressed in metastatic prostate cancer
and promotes prostate cancer cell growth and invasion. J Transl
Med. 11:1112013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Betáková T and Svančarová P: Role and
application of RNA interference in replication of influenza
viruses. Acta Virol. 57:97–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song Y, Liu C, Liu X, Trottier J, Beaudoin
M, Zhang L, Pope C, Peng G, Barbier O, Zhong X, et al: H19 promotes
cholestatic liver fibrosis by preventing ZEB1-mediated inhibition
of epithelial cell adhesion molecule. Hepatology. 66:1183–1196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li X, Chen H, Xue L, Pang X, Zhang X, Zhu
Z, Zhu W, Wang Z and Wu H: p53 performs an essential role in
mediating the oncogenic stimulus triggered by loss of expression of
neurofibromatosis type 2 during in vitro tumor progression. Oncol
Lett. 14:2223–2231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao L, Gu J, Dong A, Zhang Y, Zhong L, He
L, Wang Y, Zhang J, Zhang Z, Huiwang J, et al: Potent antitumour
activity of oncolytic adenovirus expressing mda-7/IL-24 for
colorectal cancer. Hum Gene Ther. 16:845–858. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng Y, Liu W, Guo L and Yang X: The
expression level of miR-203 in patients with gastric cancer and its
clinical significance. Pathol Res Pract. 213:1515–1518. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Roomi MW, Kalinovsky T, Rath M and
Niedzwiecki A: A nutrient mixture inhibits glioblastoma xenograft
U-87 MG growth in male nude mice. Exp Oncol. 38:54–56.
2016.PubMed/NCBI
|
|
18
|
Chen CB, Eurich DT, Majumdar SR and
Johnson JA: Risk of prostate cancer across different racial/ethnic
groups in men with diabetes: A retrospective cohort study. Diabet
Med. 35:107–111. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H,
He J, Han P and Tian D: Upregulation of SATB1 promotes tumor growth
and metastasis in liver cancer. Liver Int. 32:1064–1078. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Hu SC, Yang CS, Chen JC, Zheng JN,
Sun XQ and Wang JQ: Inhibition of prostate cancer cell growth in
vivo with short hairpin RNA targeting SATB1. Oncol Lett.
14:6592–6596. 2017.PubMed/NCBI
|
|
21
|
Han HJ, Russo J, Kohwi Y and
Kohwi-Shigematsu T: SATB1 reprogrammes gene expression to promote
breast tumor growth and metastasis. Nature. 452:187–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mao LJ, Zhang J, Liu N, Fan L, Yang DR,
Xue BX, Shan YX and Zheng JN: Oncolytic virus carrying shRNA
targeting SATB1 inhibits prostate cancer growth and metastasis.
Tumour Biol. 36:9073–9081. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Choi JW, Lee JS, Kim SW and Yun CO:
Evolution of oncolytic adenovirus for cancer treatment. Adv Drug
Deliv Rev. 64:720–729. 2012. View Article : Google Scholar : PubMed/NCBI
|