Introduction
Acute promyelocytic leukemia (APL) with
promyelocytic leukemia (PML)-retinoic acid receptor α (RARA) gene
fusion, a distinct subtype of acute myeloid leukemia (AML)
accounting for ~10% of cases (1).
The PML-RARA fusion is hypothesized to serve a vital role in the
pathogenesis of APL. Usually, patients with APL with the PML-RARA
fusion gene are sensitive to molecular target-based agents,
including all-trans retinoic acid (ATRA) and arsenic trioxide
(ATO).
The fusion gene usually results from classical
t(15;17)(q24;q12) rearrangements. However, a minority of APL cases
actually lacks this classical chromosomal aberration and is
additionally associated with the formation of the PML-RARA fusion
gene. APL cases lacking classical t(15;17) arise from insertion
events or more complex rearrangements, and the latter account for
10% of APL lacking classical t(15;17) (2). Due to its rarity, the outcome of
complex translocations has remained to be characterized.
Analysis of these complex translocations is of great
interest, as they may occur in clusters around particular
chromosomal bands. These rare translocations may provide insight
that may be useful for identifying novel gene rearrangements in
APL. In the present study, a case report of a patient with APL
harboring three-way translocations in a complex karyotype that was
determined by molecular cytogenetic approaches is provided.
Case report
Case presentation
A 37-year-old man was admitted to the Second
Hospital of Jilin University (Changchun, China) in September 2014.
He had an intermittent fever and a sore throat for 6 days, as well
as a nosebleed for 2 days and hematuria for 1 day. A complete blood
examination demonstrated a hemoglobin (Hb) count of 104 g/l (normal
range, 115–150 g/l), a white blood cell (WBC) count of
45.8×109/l (normal range, 3.5–9.5×109/l) with
89% blasts and a platelet count of 10×109/l (normal
range, 125–350×109/l). Coagulation tests identified a
prothrombin time of 16.0 sec (normal, 9.4–12.5 sec), a normal
prothrombin international ratio of 1.35 (normal, 0.8–1.2), a
prothrombin activity of 61% (normal, 80–150%), an activated partial
thromboplastin time of 33.6 sec (normal, 22.0–42.0 sec), a
fibrinogen level of 1.31 g/l (normal, 2.0–4.0 g/l) and a D-dimer of
57.20 µg/ml (normal, 0–1.0 µg/ml). The patient had no
hepatosplenomegaly.
Morphologic analysis
Analysis of the bone marrow (BM) aspirate
demonstrated a markedly hypercellular marrow with the absence of
megakaryocytes and 88% abnormal promyelocytes. Cytochemical
staining of the BM aspirate specimen was performed as described
previously (3), at room temperature.
The film preparation was covered with sufficient 0.3% benzidine
solution (Baso Biotech Co., Ltd., Wuhan, China) for 1 min. An equal
amount of 0.03% H2O2 solution (Baso Biotech
Co., Ltd.) was added. After 4–5 min, the stained smears were washed
with tap water. The smears were counterstained with Wright-Giemsa
solution (Baso Biotech Co., Ltd.) for 10 min. The results
demonstrated that the abnormal promyelocytes were strongly positive
for myeloperoxidase. However, analyses of esterases were not
performed. The core biopsy, which consisted of sheets of abnormal
promyelocytes, of the bone marrow demonstrated a cellularity of
>95%.
Flow cytometry
A total of 50 µl bone marrow (1×106
cells) and antibodies (listed below) were added to four test tubes
at room temperature for 15 min in the dark. Optilyse C lysing
solution (cat. no. 349202; BD Biosciences, San Jose, CA, USA) was
added to the tubes for 10 min at room temperature in the dark.
Then, 1 ml PBS was added to three of the tubes, which were
centrifuged at 300 × g for 5 min; the supernatant was removed and
then the cells were washed with PBS. PBS (500 µl) was added to the
three tubes and the samples were incubated with the following
antibodies: Fluorescein isothiocyanate (FITC)-conjugated anti-CD15
(cat. no. 332778), R-phycoerythrin (PE)-conjugated anti-CD117 (cat.
no. 340529), PerCP-Cy™ 5.5-conjugated anti-CD34 (cat.
no. 347203), PE-conjugated anti-CD13 (cat. no. 347837),
PerCP-Cy5.5-conjugated anti-CD19 (cat. no. 340951; all 1:25),
allophycocyanin (APC)-H7-conjugated anti-HLA-DR (cat. no. 641393),
BD Horizon™ V450-conjugated anti-CD38 (cat. no. 646851),
V450-conjugated anti-CD11b (cat. no. 560480), APC-conjugated
anti-CD4 (cat. no. 340443), APC-H7-conjugated anti-CD14 (cat. no.
641394), V500-conjugated anti-CD45 (cat. no. 560777; all 1:100),
PE-Cy7-conjugated anti-CD33 (cat. no. 333946; 1:50; all BD
Biosciences), PE-Cy7-conjugated anti-CD123 (cat. no. 306010),
APC-conjugated anti-CD7 (cat. no. 343108; both Biolegend, Inc., San
Diego, CA, USA), APC-conjugated anti-CD56 (cat. no. IM2474; all
1:50), FITC-conjugated anti-CD64 (cat. no. IM1604U),
FITC-conjugated anti-CD36 (cat. no. IM0766U), PE-conjugated
anti-CD10 (cat. no. A07760; all 1:25), PE-Cy7-conjugated anti-CD20
(cat. no. IM3629U; 1:100; Beckman Coulter, Inc., Brea, CA, USA).
The first test tube was contained anti-CD15, anti-CD117, anti-CD34,
anti-HLA-DR, anti-CD38, anti-CD45, anti-CD33 and anti-CD7
antibodies. The second test tube was contained anti-CD64,
anti-CD56, anti-CD13, anti-CD34, anti-HLA-DR, anti-CD45, anti-CD11b
and anti-CD123 antibodies. The third test tube was contained
anti-CD36, anti-CD10, anti-CD20, anti-CD19, anti-CD4, anti-CD14,
anti-CD38 and anti-CD45 antibodies.
The sample in the fourth test tube was incubated
with 100 µl intraprep permeabilization reagent A (cat. no. 641776;
BD Biosciences) for 5 min at room temperature. Following the
centrifugation of the fourth test tube at 300 × g for 5 min at room
temperature, the supernatant was removed. A total of 50 µl
intraprep permeabilization reagent B and 20 µl cytoplasmic
antibodies (listed below) were added to the tube and agitated for
15 min. PBS (1 ml) was added to the tube, which was centrifuged at
300 × g for 5 min at room temperature, then the supernatant was
removed. PBS (500 µl) was added to the fourth test tubes and the
samples were incubated with the following antibodies:
FITC-conjugated TDT (cat. no. F7139), PE-conjugated MPO (cat. no.
R7209; both Dako; Agilent Technologies, Inc., Santa Clara, CA,
USA), PE-Cy7-conjugated anti-CD2 (cat. no. 335786; all 1:50),
PerCP-Cy5.5-conjugated anti-CD5 (cat. no. 341109),
PerCP-Cy5.5-conjugated anti-CD9 (cat. no. 341649),
APC-H7-conjugated anti-CD3 (cat. no. 641397), V450-conjugated
anti-cCD3 (cat. no. 558117), V500-conjugated anti-CD45 (cat. no.
560777; all 1:100; BD Biosciences), APC-conjugated anti-CD79a (cat.
no. 333506; 1:50; Biolegend, Inc.). All four samples were then
examined using a BD FACSCanto II flow cytometer and the data were
analyzed using BD FACSDIVA™ 6.1.3 software (both BD
Biosciences). The results demonstrated that the samples were
positive for CD13, CD33 and myeloperoxidase (MPO), and some cells
expressed CD38, CD64 and CD117.
Molecular analysis
The PML-RARA rearrangement was confirmed by reverse
transcription-qualitative polymerase chain reaction (RT-qPCR).
Total RNA was extracted from bone marrow samples using TRIzol
reagent (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Complementary DNA was synthesized using the total RNA, random
hexamer primers (cat. no. 3801), dNTP mixture, 5X M-MLV buffer and
reverse transcriptase M-MLV (cat. no. 2641Q; both Takara Bio, Inc.,
Otsu, Japan). The conditions for reverse transcription reaction
consisted of 10 min at 30°C, 60 min at 42°C, 15 min at 70°C, and
4°C indefinitely. Subsequently, a qPCR analysis was performed using
Premix Ex Taq™ (cat. no. RR390A; Takara Bio, Inc.), and
the following primers: PML-RARA forward,
5′-CCGTCATAGGAAGTGAGGTCT-3′ and reverse,
5′-GGCTGGGCACTATCTCTTCA-3′; and GAPDH forward,
5′-AATGGAAATCCCATCACCATCT-3′ and reverse,
5′-CATCGCCCCACTTGATTTTG-3′. The thermocycling conditions were as
follows: An initial denaturation at 95°C for 10 min, followed by 40
cycles at 95°C for 15 sec and 58°C for 40 sec on the Applied
Biosystems 7500 Fast PCR System (Thermo Fisher Scientific, Inc.).
The values were normalized to GAPDH transcript levels and expressed
as a ratio of PML-RARA to GAPDH by absolute quantification method
(Table I). The results demonstrated
that only the short-type chimeric transcript was expressed, while
RARA-PML was not expressed. The patient was positive for FMS
related tyrosine kinase 3 internal tandem duplication (FLT3-ITD)
gene mutation, which was detected as previously described (4).
 | Table I.Reverse transcription-qualitative
polymerase chain reaction analysis results. |
Table I.
Reverse transcription-qualitative
polymerase chain reaction analysis results.
| Test | Results |
|---|
| PML-RARA
(Long-type) | Negative |
| PML-RARA
(Variant-type) | Negative |
| PML-RARA
(Short-type) | Positive |
| PML-RARA (copy
numbers) | 186,700 |
| GAPDH (copy
numbers) | 183,500 |
| PML-RARA/GAPDH | 101.74% |
Treatment
Based on these results, a diagnosis of APL with
PML-RARA was made. The patient started induction therapy with oral
ATRA [40 mg/(m2day)], intravenous ATO [10
mg/(m2day)] and cytarabine [200 mg/(m2day)]
treatment. On the 3rd day after therapy initiation, the WBC count
reached 61.8×109/l with double lower limb edema and bone
pain syndrome, which are symptoms of APL differentiation (5). Subsequently, the oral ATRA therapy was
stopped and the patient received treatment with dexamethasone (10
mg for 3 days) and daunorubicin (20 mg for 3 days); the APL
differentiation symptoms gradually disappeared. The patient
repeatedly received ATRA induction therapy; the WBC count gradually
decreased to a normal count. The patient received intrathecal
dexamethasone (10 mg) and cytarabine (50 mg) treatment for
prevention. The patient demonstrated hematological recovery after
39 days. A complete hematological remission was achieved and BM
aspiration demonstrated a regenerating marrow without morphologic
evidence of the malignant disease. The patient was negative for
PML-RARA fusion gene. The patient in complete remission (CR)
received consolidation courses every month, with no recurrence for
4 years to date.
Conventional cytogenetic analysis and
fluorescence in situ hybridization (FISH)
The BM sample of the patient was analyzed for
metaphase karyotyping by G-banding after 48 h of unstimulated
culture. Of the 20 metaphase cells examined using CytoVision system
(Leica Microsystems, San Jose, CA, USA), an apparent complex
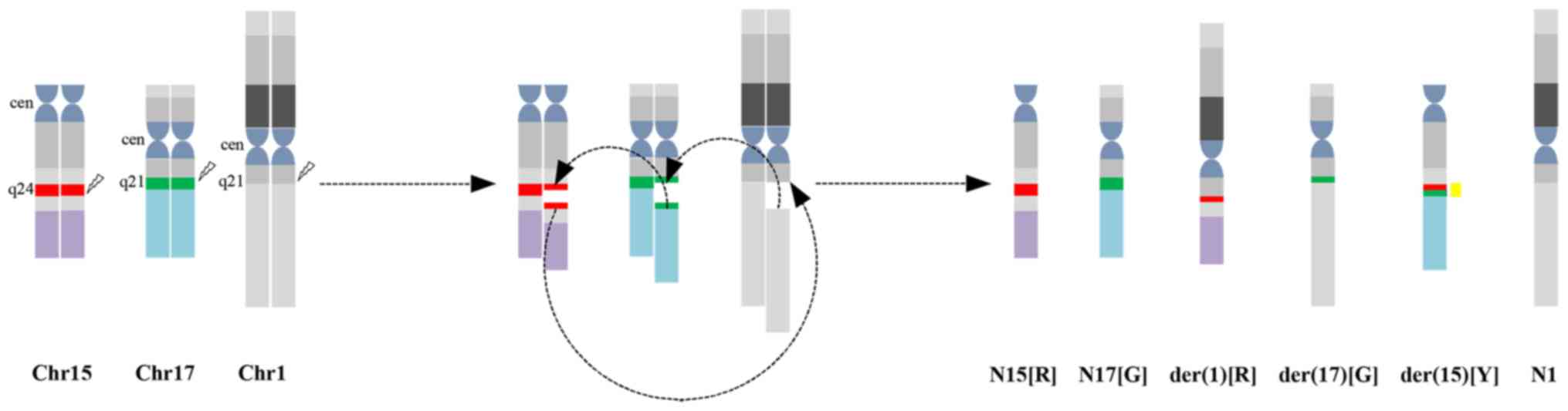
translocation was identified, which involved chromosome 1 in
addition to chromosomes 15 and 17 with breakpoints at 1q21, 15q24
and 17q21. No other consistent structural or numerical
abnormalities were detected (Fig.
1). To confirm this complex chromosomal rearrangement and
determine the diagnosis, FISH was performed using the Vysis
dual-color, dual-fusion probe (Abbott Laboratories, Abbott Park,
IL, USA) according to the manufacturer's protocol. The cells were
counterstained with DAPI II in the dark at −20°C for 30 min prior
to observation. Fluorescent signals were visualized using a
fluorescence microscope at a magnification of ×1,000. Typically, in
APL with PML-RARA, two yellow signals, one red and one green signal
are visible, which indicate the classical t(15;17). In the patient,
392 out of 400 cells (98%) exhibited two red signals, two green
signals and one yellow fusion signal (Fig. 2), which were the result of the
complex translocation. Taken together, the cytogenomic findings can
be described as: 46,XY, t(1;17;15)(q21;q21;q24)[20].nuc ish(PML,
RARA)×3(PML con RARAx1)[400].
Discussion
To date, to the best of our knowledge, 46 cases with
complex translocation have been previously reported (6–8). Among
these, only four cases of three-way translocations involved
chromosome 1, and the breakpoint occurred at 1p31, 1p32, 1p36 and
1q23 (Table II) (2,9–11). The present study identified the fifth
case of APL harboring a three-way translocation involving
chromosome 1;17;15. The clinical characterization of the five cases
of APL with t(1;17;15) is summarized in Table II. All of the patients were male and
the median age at diagnosis was 42 years. No distinct clinical
features were observed in APL with complex translocation regarding
the three-way t(1;17;15) translocation.
 | Table II.Clinical characterization of
previously reported acute promyelocytic leukemia cases harboring a
three-way translocation involving chromosome 1;17;15. |
Table II.
Clinical characterization of
previously reported acute promyelocytic leukemia cases harboring a
three-way translocation involving chromosome 1;17;15.
| Author (year) | Sex | Age (years) | WBC (×109/l) | Hb (g/l) | DIC | FAB | Karyotype | PML-RARA fusion
gene | Treatment | Relapse | Survival
(months) | Breakpoints on
chromosome 1 | (Refs.) |
|---|
| Present study | M | 37 | 45.8 | 109 | No | M3 | 46, XY, der(1)t(1;17)
(q21;q21)t(15;17) (q24;q21),der(15)t(15;17)t(1;17),
der(17)t(15;17)t(1;17)[20] | Short type | ATRA+ATO+Ara-C | No | >48 | 1q21 | NA |
| Grimwade (2000) | M | NA | NA | NA | NA | M3 |
46,XY,t(1;17;15)(p32;q21;q22)/46, idem,
add(21)(p13)/46, XY | Short type | NA | NA | NA | 1p32 | (2) |
| Osella (1991) | M | 46 | 0.7 | NA | Yes | M3 | 46,XY/46,XY,
t(1;15;17)(p36; q22;q21.1)/46,XY, t(1;15;17)
(p36;q22;q21.1),+8 | NA |
Ara-C+daunorubicin | Yes | >11 | 1p36 | (9) |
| Park and Fairweather
(1996) | M | 46 | 4.9 | 94 | Yes | M3 | 46,XY/46, XY,
del(1)(p22), del(3)(p25),der(17)t(1;15;17) (17pter→17q21::15q21→
15q22::1p36→1p31::15q21→ 15q22::17q21→17pter) | NA |
ATRA+Ara-C+daunorubicin | Yes | 9 | 1p31 | (10) |
| Galieni (1996) | M | 41 | NA | NA | NA | M3 | 46,XY, t(1;17;15)
(q23;q23;q22)/47,idem, +10 | NA | ATRA+C | No | >8 | 1q23 | (11) |
According to karyotyping and FISH analysis, the
complex translocation in the present study may be described as
follows: 15q24 translocated to 1q21; a small piece of chromosome 17
(17q21) translocated to 15q24; and 1q21 connected to 17q21 that was
located in der(15)(q24). As a result of the genomic rearrangement,
a part of the PML gene (labeled in red) is present on the
derivative of chromosome 1 and a part of the RARA gene (labeled in
green) is present on the derivative of chromosome 17. In addition,
one yellow signal, one red and one green signal indicates the
PML-RARA fusion on the derivative of chromosome 15, and the normal
chromosomes 15 and 17, respectively (Fig. 3). Correspondingly, FISH identified
two red signals, two green signals and one yellow signal.
The association between complex translocations and
classical t(15;17) remains elusive. Previous studies hypothesized
that the complex translocation possibly evolves from the classical
t(15;17) (7,12). However, as a cell with t(15;17) alone
was not identified, the results of the present suggested that all
of the events happened at the same time and not in two steps. A
recent study proposed a non-homologous chromosome recombination
model as one of the mechanisms that results in chromosome
translocations in leukemia (13). It
is noteworthy that the breakpoint in the present case occurred in a
novel area of the long arm of chromosome 1, 1q21. The breakpoint
1q21 has been identified in AML, predominantly involving two
different chromosome translocations. The first was described in a
patient with AML with t(1;11)(q21;q23), which leads to the fusion
gene MLL-AF1q (14,15), and the second was described in a
1-year-old patient with AML with t(1;21)(q21;q22), which leads to
the fusion gene runt-related transcription factor 1-zinc finger
protein (ZNF)687 (16). Whether the
AF1q or ZNF687 gene is located at the breakpoint and forms a fusion
gene in the present case requires clarification.
The incidence of FLT3-ITD mutations in APL is 12–38%
(17). FLT3-ITD mutations are
associated with higher Hb and WBC levels, as well as short-type
PML-RARA fusion transcripts at diagnosis. Hb>9.6 g/dl and
WBC≥20×109/l are important factors for predicting the
presence of presence FLT3-ITD according to a multivariate analysis
(18,19). The results of the present study are
consistent with those of the aforementioned study (18), supporting the hypothesis that
carriers of FLT3-ITD constitute a biologically distinct group of
patients with APL. Previous studies have demonstrated that FLT3-ITD
in APL has an adverse prognostic value (17,19). Of
note, two previous independent studies demonstrated that the
addition of ATO to frontline therapy overcomes the impact of
previously described adverse prognostic factors, including FLT3-ITD
mutations (20,21). The patient of the present study
received treatment with ATO during induction and consolidation
courses, with no recurrence for 3 years to date. The outcome of
this case is consistent with the results of the aforementioned
studies (20,21).
The majority of previous studies observed no
difference in clinical outcome between APL with typical t(15;17)
and APL with complex translocations (22,23). The
results of the present study are consistent with those of previous
studies, supporting the hypothesis that the presence of the
PML-RARA fusion gene is crucial for achieving the best response to
ATRA (24–26). In previous studies, APL with complex
translocations, including four cases of three-way translocations
involving chromosome 1, appears to be associated with poor
prognosis, which may be due to the absence of targeted therapies
(27–29). Additional genomic alterations besides
the PML-RARA fusion may be implicated in the heterogenicity of
therapy outcomes in APL.
In conclusion, the present study was the first, to
the best of our knowledge, to identify a novel breakpoint in
chromosome 1 in an adult patient with APL using FISH. Furthermore,
at the 3-year follow-up, a good response to the combined treatment
with ATRA and ATO was revealed, as is observed in typical APL. The
significance of complex translocations in APL with PML-RARA
requires further investigation.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
LL and LY analyzed the data, performed all of the
examinations of the patients and drafted the manuscript. TM, LL and
HC contributed to data acquisition and examination of the patient.
All authors agree to the submission and have full responsibility
for all primary data. No part of this paper has been published or
submitted elsewhere.
Ethics approval and consent to
participate
The patient provided written informed consent prior
to the study according to the Declaration of Helsinki and the study
was approved by the Second Hospital of Jilin University.
Patient consent for publication
Written informed consent was obtained from the
patient for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and Vardiman JW:
The 2016 revision to the World Health Organization classification
of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grimwade D, Biondi A, Mozziconacci MJ,
Hagemeijer A, Berger R, Neat M, Howe K, Dastugue N, Jansen J,
Radford-Weiss I, et al: Characterization of acute promyelocytic
leukemia cases lacking the classic t(15;17): Results of the
european working party. Groupe français de cytogénétique
hématologique, groupe de français d'hematologie cellulaire, UK
cancer cytogenetics group and BIOMED 1 european community-concerted
action ‘molecular cytogenetic diagnosis in haematological
malignancies’. Blood. 96:1297–1308. 2000.PubMed/NCBI
|
|
3
|
Chen WX, Zhu HL, Xue M, Zhou H, Zhao F,
Yan N and Chen Y: Quick staining technique for myeloperoxidase
using potassium iodide and oxidized pyronine B. Acta Histochem.
116:292–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gale RE, Hills R, Pizzey AR, Kottaridis
PD, Swirsky D, Gilkes AF, Nugent E, Mills KI, Wheatley K, Solomon
E, et al: Relationship between FLT3 mutation status, biologic
characteristics, and response to targeted therapy in acute
promyelocytic leukemia. Blood. 106:3768–3776. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Montesinos P and Sanz MA: The
differentiation syndrome in patients with acute promyelocytic
leukemia: Experience of the pethema group and review of the
literature. Mediterr J Hematol Infect Dis. 3:e20110592011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Ma J, Liu X, Liu R, Xu L, Wang L,
Cen J and Chu X: A complex translocation (3;17;15) in acute
promyelocytic leukemia confirmed by fluorescence in situ
hybridization. Oncol Lett. 12:4717–4719. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang R, Kim YM, Wang X, Li Y, Pang H, Lee
JY and Li S: Coexistence of t(15;17) and t(15;16;17) detected by
fluorescence in situ hybridization in a patient with acute
promyelocytic leukemia: A case report and literature review. Oncol
Lett. 8:1001–1008. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bennour A, Tabka I, Youssef YB, Zaier M,
Hizem S, Khelif A, Saad A and Sennana H: A PML/RARA chimeric gene
on chromosome 12 in a patient with acute promyelocytic leukemia
(M4) associated with a new variant translocation:
t(12;15;17)(q24;q24;q11). Med Oncol. 30:4092013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osella P, Wyandt H, Vosburgh E and
Milunsky A: Report of a variant t(1;15;17)(p36;q22;q21.1) in a
patient with acute promyelocytic leukemia. Cancer Genet Cytogenet.
57:201–207. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park JP and Fairweather RB: Complex
t(1;15;17) in acute promyelocytic leukemia with duplication of RAR
alpha and PML sequences. Cancer Genet Cytogenet. 89:52–56. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Galieni P, Marotta G, Vessichelli F,
Diverio D, Minoletti F, Bucalossi A, Lo Coco F and Lauria F:
Variant t(1;15;17)(q23;q22;q23) in a case of acute promyelocytic
leukemia. Leukemia. 10:1658–1661. 1996.PubMed/NCBI
|
|
12
|
Miyazaki K, Kikukawa M, Kiuchi A, Shin K,
Iwamoto T and Ohyashiki K: Complex translocations derived stepwise
from standard t(15;17) in a patient with variant acute
promyelocytic leukemia. Cancer Genet Cytogenet. 176:127–130. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y and Rowley JD: Chromatin
structural elements and chromosomal translocations in leukemia. DNA
Repair (Amst). 5:1282–1297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tse W, Zhu W, Chen HS and Cohen A: A novel
gene, AF1q, fused to MLL in t(1;11) (q21;q23), is specifically
expressed in leukemic and immature hematopoietic cells. Blood.
85:650–656. 1995.PubMed/NCBI
|
|
15
|
Busson-Le Coniat M, Salomon-Nguyen F,
Hillion J, Bernard OA and Berger R: MLL-AF1q fusion resulting from
t(1;11) in acute leukemia. Leukemia. 13:302–306. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nguyen TT, Ma LN, Slovak ML, Bangs CD,
Cherry AM and Arber DA: Identification of novel Runx1 (AML1)
translocation partner genes SH3D19, YTHDf2, and ZNF687 in acute
myeloid leukemia. Genes Chromosomes Cancer. 45:918–932. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Beitinjaneh A, Jang S, Roukoz H and
Majhail NS: Prognostic significance of FLT3 internal tandem
duplication and tyrosine kinase domain mutations in acute
promyelocytic leukemia: A systematic review. Leuk Res. 34:831–836.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Souza Melo CP, Campos CB, Dutra ÁP, Neto
JC, Fenelon AJ, Neto AH, Carbone EK, Pianovski MA, Ferreira AC and
Assumpcão JG: Correlation between FLT3-ITD status and clinical,
cellular and molecular profiles in promyelocytic acute leukemias.
Leuk Res. 39:131–137. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lucena-Araujo AR, Kim HT, Jacomo RH, Melo
RA, Bittencourt R, Pasquini R, Pagnano K, Fagundes EM, Chauffaille
Mde L, Chiattone CS, et al: Internal tandem duplication of the FLT3
gene confers poor overall survival in patients with acute
promyelocytic leukemia treated with all-trans retinoic acid and
anthracycline-based chemotherapy: An International Consortium on
Acute Promyelocytic Leukemia study. Ann Hematol. 93:2001–2010.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Poiré X, Moser BK, Gallagher RE, Laumann
K, Bloomfield CD, Powell BL, Koval G, Gulati K, Holowka N, Larson
RA, et al: Arsenic trioxide in front-line therapy of acute
promyelocytic leukemia (C9710): Prognostic significance of FLT3
mutations and complex karyotype. Leuk Lymphoma. 55:1523–1532. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cicconi L, Divona M, Ciardi C, Ottone T,
Ferrantini A, Lavorgna S, Alfonso V, Paoloni F, Piciocchi A,
Avvisati G, et al: PML-RARα kinetics and impact of FLT3-ITD
mutations in newly diagnosed acute promyelocytic leukaemia treated
with ATRA and ATO or ATRA and chemotherapy. Leukemia. 30:1987–1992.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wiernik PH, Sun Z, Gundacker H, Dewald G,
Slovak ML, Paietta E, Kim HT, Appelbaum FR, Cassileth PA and
Tallman MS: Prognostic implications of additional chromosome
abnormalities among patients with de novo acute promyelocytic
leukemia with t(15;17). Med Oncol. 29:2095–2101. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fujishima M, Takahashi N, Miura I,
Kobayashi Y, Kume M, Nishinari T and Miura AB: A PML/RARA chimeric
gene on chromosome 2 in a patient with acute promyelocytic leukemia
(M3) associated with a new variant translocation:
t(2;15;17)(q21;q22;q21). Cancer Genet Cytogenet. 120:80–82. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hernández JM, Martín G, Gutiérrez NC,
Cervera J, Ferro MT, Calasanz MJ, Martínez-Climent JA, Luño E,
Tormo M, Rayón C, et al: Additional cytogenetic changes do not
influence the outcome of patients with newly diagnosed acute
promyelocytic leukemia treated with an ATRA plus anthracyclin based
protocol. A report of the Spanish group PETHEMA. Haematologica.
86:807–813. 2001.PubMed/NCBI
|
|
25
|
De Botton S, Chevret S, Sanz M, Dombret H,
Thomas X, Guerci A, Fey M, Rayon C, Huguet F, Sotto JJ, et al:
Additional chromosomal abnormalities in patients with acute
promyelocytic leukaemia (APL) do not confer poor prognosis: Results
of APL 93 trial. Br J Haematol. 111:801–806. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Botton S, Coiteux V, Chevret S, Rayon
C, Vilmer E, Sanz M, de La Serna J, Philippe N, Baruchel A,
Leverger G, et al: Outcome of childhood acute promyelocytic
leukemia with all-trans-retinoic acid and chemotherapy. J Clin
Oncol. 22:1404–1412. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Freeman CE, Mercer DD, Ye Y, Van Brunt J
III and Li MM: Cytogenetic and molecular characterization of
complex three-way translocations in acute promyelocytic leukemia.
Beijing Da Xue Xue Bao Yi Xue Ban. 41:477–479. 2009.PubMed/NCBI
|
|
28
|
McKinney CD, Golden WL, Gemma NW, Swerdlow
SH and Williams ME: RARA and PML gene rearrangements in acute
promyelocytic leukemia with complex translocations and atypical
features. Genes Chromosomes Cancer. 9:49–56. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Calabrese G, Min T, Stuppia L, Powles R,
Swansbury JG, Morizio E, Peila R, Donti E, Fioritoni G and Palka G:
Complex chromosome translocations of standard t(8;21) and t(15;17)
arise from a two-step mechanism as evidenced by fluorescence in
situ hybridization analysis. Cancer Genet Cytogenet. 91:40–45.
1996. View Article : Google Scholar : PubMed/NCBI
|