Introduction
Melanoma is the most prevalent malignant skin
cancer. Melanoma ranks as the fifth most prevalent cancer in males
and the seventh most prevalent cancer in females, causing ~80% skin
cancer-associated mortalities worldwide (1,2).
Incidence rates of malignant melanoma have increased significantly
since 1992 with an overall 45% increase and estimated 3.1% annual
percent change (1,2). Therefore, exploring the molecular
mechanism during melanoma progression is urgently required as it
may benefit the development of effective strategies for melanoma
treatment.
Long non-coding RNAs (lncRNAs) are a newly
discovered group of small non-coding RNAs >200 nucleotides in
length, comprising 80% of non-coding RNAs (3–5). In
recent years, a large number of lncRNAs have been demonstrated to
participate in various cellular biological processes, such as cell
proliferation, differentiation, apoptosis, cell cycle, motility,
and tumourigenesis (6,7). A number of lncRNAs have been identified
as oncogenes or tumor suppressors in human cancers, including
melanoma (8,9). For instance, the lncRNA growth
arrest-specific 5 serves an inhibitory role in melanoma via
regulating gelatinase A and B both in vitro and in
vivo (10). The lncRNA
BRAF-activated non protein-coding RNA promotes melanoma cell
proliferation by regulating mitogen-activated protein kinase
pathway activity (11).
Among cancer-related lncRNAs, urothelial
cancer-associated 1 (UCA1) generally serves a tumor-promoting role
(12,13). Tian et al (14) previously reported that UCA1 was
significantly upregulated in melanoma tissues compared with its
expression in paired adjacent non-tumor tissues, and melanomas at
advanced stages exhibited higher UCA1 expression than tumors at
early stages. Furthermore, previous studies have demonstrated that
UCA1 functions as an oncogene in certain common human cancers
through directly interacting with its target microRNAs (miRNAs or
miRs) and further affecting the protein expression of the
downstream target genes (15,16). For
instance, UCA1 promotes the proliferation and migration of
pancreatic cancer cells through regulating the miR-96/forkhead box
protein (FOX)O3 axis (17). In
addition, UCA1 promotes the migration and epithelial-mesenchymal
transition of bladder cancer cells by regulating the miR-143/high
mobility group box 1 pathway (12).
In melanoma, UCA1 promotes cancer cell proliferation, cell cycle
progression and migration via modulation of the miR-507-FOXM1 axis
(18). However, whether other miRNAs
and downstream proteins are also associated with UCA1-mediated
melanoma cells remains unclear.
miR-28-5p has been demonstrated to serve different
roles in different cancer types (19,20). For
instance, miR-28-5p promotes ovarian cancer progression through
inhibition of NEDD4 binding protein 1 (20). In contrast, miR-28-5p is
downregulated in colorectal cancer, and overexpression of miR-28-5p
exhibits suppressive effects on colorectal cancer cell
proliferation, migration and invasion in vitro, as well as
tumor growth in vivo (19).
furthermore, homeobox (HOX)B3is hypothesized to be a direct target
gene of miR-28-5p, and the expression of HOXB3 is regulated by
miR-28-5p in colorectal cancer cells (19). However, the detailed role of
miR-28-5p and HOXB3 in melanoma remains unclear. Therefore, the
present study aimed to explore the molecular mechanism of UCA1
underlying melanoma cell proliferation and migration.
Materials and methods
Tissue samples
The present study was approved by the Research
Ethics Committee of Third Xiangya Hospital (Changsha, China). A
total of 22 melanoma tumors and matched adjacent non-tumor tissues
were collected from primary melanoma patients at the Department of
Burn and Plastic Surgery, Third Xiangya Hospital of Central South
University (Changsha, China) between April 2014 and May 2017. These
patients included 10 males and 12 females from 34–60 years old with
a mean age of 48.3 years. In total, 12 I–II stage cases and 10
III–IV stage cases were included. No patient recruited for the
present study had received adjuvant treatment prior to surgical
resection. Written informed consent was obtained from all
patients.
Cell culture
Normal human epidermal melanocyte HEMa-LP cells and
human melanoma cell lines, including A375, SK-MEL-2 and SK-MEL-28,
were obtained from the Cell Bank of the Chinese Academy of Science
(Shanghai, China). Cells were cultured in Dulbeccos modified Eagles
medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10%
fetal bovine serum (Thermo Fisher Scientific, Inc.) and incubated
at 37°C in a humidified atmosphere with 5% CO2.
Cell transfection
SK-MEL-28 cells were seeded (1×105 cells
per well) into a 6-well plate and were transiently transfected with
50 nM UCA1 small interfering (si)RNA (siUCA1; cat. no. NR_015379),
negative control siRNA (siNC; cat. no. SIC001), miR-28-5p mimic
(cat. no. HMI0425), NC miR mimic (miR-NC; cat. no. HMC0002; all
Sigma-Aldrich; Merck KGaA, Darmstradt, Germany), or co-transfected
with UCA1 siRNA and the HOBX3 expression plasmid or the blank
pcDNA3.1 vector (Yearthbio Technology, Changsha, China) using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to
the manufacturers instruction. At 48 h following transfection, the
cells were harvested, and the following assays were conducted.
Luciferase reporter gene assay
miRanda software version 1.0 (http://www.micro-RNA.org/) was used to predict
potential UCA1-miR interactions. TargetScan software 7.2
(www.targetscan.org/) was used to predict
the potential target genes of miR-28-5p. UCA1 sequences containing
the wild-type (WT) or mutated (MT) miR-28-5p binding sites and
HOXB3 3′UTR sequences containing the WT or MT miR-28-5p binding
sites were subcloned into the pmiR-GLo luciferase reporter vector
(Promega Corporation, Madison, WI, USA). SK-MEL-28 cells were
co-transfected with WT or MT UCA1 reporter plasmids together with
miR-28-5p mimic or miR-NC, or co-transfected with the WT or MT
HOXB3 reporter plasmids together with miR-28-5p mimic or miR-NC
using Lipofectamine 2000. At 48 h following transfection,
luciferase activity was examined using the Dual Luciferase Reporter
Assay system (Promega Corporation). The activity of firefly
luciferase was normalized to the activity of Renilla
luciferase.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and, HEMa-LP,
A375, SK-MEL-2 and SK-MEL-28 cells using TRIzol reagent (Thermo
Fisher Scientific, Inc.,) and then reverse transcribed into cDNA
using an OneStep RT-PCR kit (Qiagen, Inc., Valencia, CA, USA)
according to the manufacturers protocol. SYBR® Premix Ex
Taq™ (Takara Biotechnology Co., Ltd., Dalian, China) was
used to perform qPCR to determine UCA1, miR-28-5p and HOXB3
expression with an ABI 7300 Plus Fast Sequence Detection system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction
was performed under the following conditions: 95°C for 3 min
followed by 35 cycles at 95°C for 30 sec and 60°C for 30 sec. The
relative expression was calculated and normalized via the
2−ΔΔCq method (21).
GAPDH and U6 were used as internal controls. The following primer
sequences were utilized in these studies: miR-28-5p,
5′-AAGGAGCUCACAGUCUAUUGAG-3′ and reverse
5′-CUCAAUAGACUGUGAGCUCCUU-3′; UCA1, forward
5′-CTCTCCATTGGGTTCACCATTC-3′ and reverse
5′-GCGGCAGGTCTTAAGAGATGAG-3′; HOXB3, forward
5′-CCAGTGCCACTAGCAACAG-3′ and reverse 5′-CGTTTGCCTCGACTCTTTCATC-3′.
GAPDH, forward 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse
5′-ATGGCATGGACTGTGGTCAT-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and
reverse 5′-AACGCTTCACGAATTTGCGT-3′.
Cell proliferation analysis
Cell Counting Kit (CCK)-8 assays (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) were conducted to study cell
proliferation following the manufacturers protocol. Following
transfection for 48 h, SK-MEL-28 cells (5,000 cells per well) were
seeded onto 96-well plates, and cell proliferation was measured at
0, 24, 48 and 72 h by using the CCK-8. Absorbance was detected at
optical density of 450 nm using a spectrophotometer.
Cell migration analysis
Transfected SK-MEL-28 cells were seeded in 6-well
plates and incubated at 37°C until 90% confluence. Following serum
starvation at 37°C for 24 h, wounds were created using a 100 µl
pipette tip. Wound healing was observed and photographed at 0 and
24 h.
Western blotting
Total proteins were extracted from SK-MEL-28 cells
using RIPA lysis buffer (Beyotime Institute of Biotechnology,
Beijing, China). The protein concentration was measured using a
bicinchoninic acid protein assay kit (Thermo Fisher Scientific,
Inc.). Proteins (50 µg per lane) were separated by SDS-PAGE on 10%
gels and transferred onto polyvinylidene difluoride membranes. Then
membranes were incubated with the following antibodies: HOXB3
(1:500; cat. no. ab83404), MMP-9 (1:500; cat. no. ab73734) and
GAPDH (1:500; cat. no. ab9485) for 3 h at room temperature, and
horseradish peroxidase-conjugated secondary antibody (1:5,000; cat.
no. ab6721; all Abcam, Cambridge, UK) for 1 h at room temperature.
GAPDH was used as the internal control. The protein bands were
detected using an Enhanced Chemiluminescence Western Blotting kit
(Pierce; Thermo Fisher Scientific, Inc.) and quantified using Image
Lab analysis software version 3.1 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Statistical analysis
Data are presented as the mean ± standard deviation.
SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was used to perform
statistical analysis. Students t-test and one-way analysis of
variance with Tukeys post hoc test were used. Pearson correlation
analysis was performed to examine the correlation between UCA1 and
miR-28-5p expression in melanoma tissues. P<0.05 was considered
to indicate a statistically significant difference.
Results
UCA1 is upregulated in melanoma
tissues and cell lines
In the present study, RT-qPCR was initially
performed to examine UCA1 expression in a total of 22 melanoma
issues and their matched adjacent non-tumor tissues. As indicated
in Fig. 1A, UCA1 expression was
significantly higher in melanoma tissues than in adjacent non-tumor
tissues. Furthermore, UCA1 expression was significantly higher in
melanoma tissues at stage III–IV than in tissues at stage I–II
(Fig. 1B). Consistently, UCA1 was
also significantly upregulated in human melanoma cell lines (A375,
SK-MEL-2 and SK-MEL-28) compared with its expression in normal
human epidermal melanocyte HEMa-LP cells (Fig. 1C). These findings suggest that UCA1
is upregulated in melanoma.
Knockdown of UCA1 expression
suppresses SK-MEL-28 cell proliferation and migration
The role of UCA1 in the regulation of melanoma cell
proliferation and migration was then investigated. Given that
SK-MEL-28 cells exhibited the highest expression of UCA1, these
cells were transfected with UCA1 siRNA to knockdown its expression.
Following transfection, UCA1 expression levels were significantly
reduced in the siUCA1 group compared with those in the siNC group
(Fig. 2A). CCK-8 and wound healing
assays were conducted to evaluate the effects of UCA1
downregulation on SK-MEL-28 cell proliferation and migration,
respectively. CCK-8 assay data revealed that inhibition of UCA1
expression caused a significant reduction in SK-MEL-28 cell
proliferation (Fig. 2B). Similarly,
knockdown of UCA1 expression significantly inhibited SK-MEL-28 cell
migration (Fig. 2C). Thus,
downregulation of UCA1 effectively inhibited SK-MEL-28 cell
proliferation and migration.
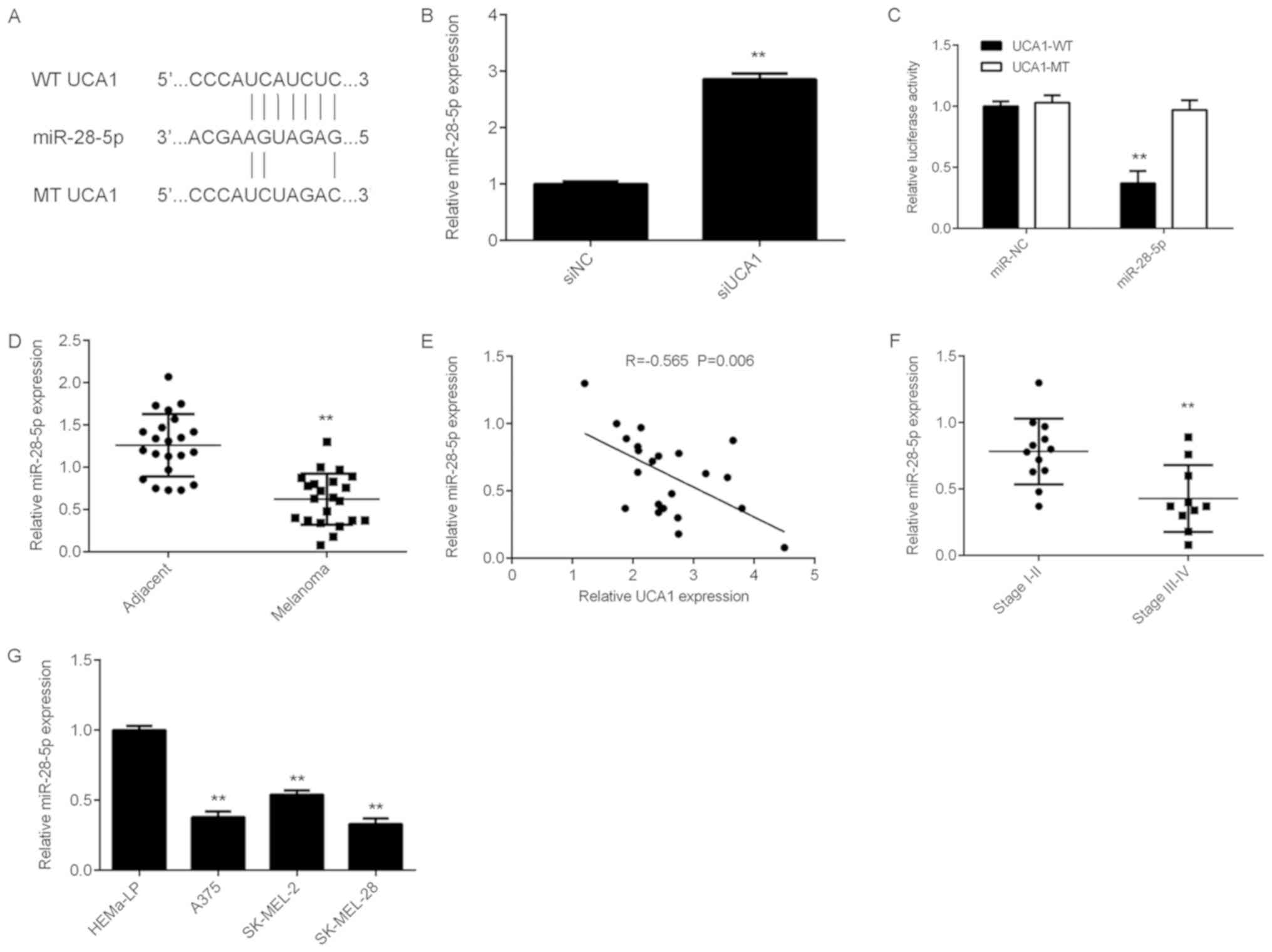
miR-28-5p is a target of UCA1 in
melanoma cells
LncRNAs act as sinks for pools of miRNAs to regulate
the downstream target genes (15,16).
Therefore, miRanda software was used to predict potential UCA1-miR
interactions. The data demonstrated that miR-28-5p harbors a
potential binding site for UCA1 (Fig.
3A). It was then demonstrated that UCA1 knockdown led to
significant upregulation of miR-28-5p expression in SK-MEL-28 cells
(Fig. 3B). To further confirm their
targeting relationship, luciferase reporter gene plasmids were
constructed containing UCA1-WT and UCA1-MT binding sites with
miR-28-5p (Fig. 3A). SK-MEL-28 cells
were then co-transfected with miR-NC or miR-28-5p and UCA1-WT or
UCA1-MT luciferase reporter gene plasmids. Dual luciferase assay
results revealed that miR-28-5p significantly reduced the
luciferase activity of the UCA1-WT luciferase reporter gene plasmid
but did not affect the luciferase activity of the UCA1-MT
luciferase reporter gene plasmid (Fig.
3C). Furthermore, miR-28-5p was significantly downregulated in
melanoma tissues compared with that in adjacent non-tumor tissues
(Fig. 3D), and high expression of
UCA1 exhibited a negative correlation with low miR-28-5p expression
in melanoma tissues (Fig. 3E). In
addition, miR-28-5p expression was significantly lower in melanoma
stage III–IV tissues than in stage I–II tissues (Fig. 3F). Finally, miR-28-5p was also
significantly downregulated in melanoma cell lines compared with
the expression in normal human epidermal melanocyte HEMa-LP cells
(Fig. 3G).
miR-28-5p is associated with
UCA1-mediated melanoma cell proliferation and migration
It was investigated whether miR-28-5p is associated
with the UCA1-mediated proliferation and migration of melanoma
cells. As it was demonstrated that miR-28-5p was upregulated
following knockdown of UCA1 expression, siUCA1-transfected
SK-MEL-28 cells were then transfected with miR-28-5p inhibitor to
reduce its expression. Following transfection, miR-28-5p expression
levels were significantly decreased in the siUCA1+miR-28-5p
inhibitor group compared with those in the siUCA1+NC inhibitor
group (Fig. 4A). Furthermore, CCK-8
assay and wound healing assay data demonstrated that the
proliferation and migration capacities of cells were significantly
increased in the siUCA1+miR-28-5p inhibitor group compared with
those in the siUCA1+NC inhibitor group (Fig. 4B and C). These results suggest that
knockdown of UCA1 expression inhibits SK-MEL-28 cell proliferation
and migration via upregulation of miR-28-5p.
HOXB3 is a target gene of miR-28-5p in
SK-MEL-28 cells
The downstream effector of miR-28-5p in SK-MEL-28
cells was then investigated, and Targetscan software predicted that
HOXB3 was a target gene of miR-28-5p (Fig. 5A). To confirm the prediction,
WT-HOXB3-3′UTR and MT-HOXB3-3′UTR luciferase reporter plasmids were
generated (Fig. 5A). Luciferase
reporter gene assay data indicated that miR-28-5p significantly
reduced the luciferase activity of the HOXB3-WT luciferase reporter
gene plasmid but did not affect the luciferase activity of the
HOXB3-MT luciferase reporter gene plasmid (Fig. 5B), indicating that miR-28-5p directly
binds to the 3′UTR of HOXB3 mRNA in SK-MEL-28 cells. Furthermore,
it was demonstrated that UCA1 knockdown significantly reduced HOXB3
expression, which was abolished by inhibition of miR-28-5p
expression in SK-MEL-28 cells (Fig. 5C
and D). These above findings suggest that UCA1 affects HOXB3
expression via miR-28-5p. In addition, it was demonstrated that
HOXB3 was upregulated in melanoma tissues compared with its
expression in adjacent normal tissues, and HOXB3 protein expression
was high in tumor tissues at advanced stages compared with that in
tissues at earlier stages (Fig. 5E and
F).
HOXB3 acts as a downstream effector of
UCA1 in SK-MEL-28 cells
It was then investigated whether HOXB3 acts as a
downstream effector of UCA1 in SK-MEL-28 cells. As HOXB3 was
downregulated following knockdown of UCA1 expression,
siUCA1-transfected SK-MEL-28 cells were then transfected with the
HOXB3 expression plasmid to upregulate its expression. Following
transfection, HOXB3 mRNA and protein expression levels were
significantly increased in the siUCA1+HOXB3 group compared with
those in the siUCA1+blank group (Fig. 6A
and B). Furthermore, CCK-8 assay and wound healing assay data
revealed that the proliferation and migration capacities of cells
were significantly increased in the siUCA1+HOXB3 group compared
with those in the siUCA1+blank group (Fig. 6C and D). These findings indicate that
HOXB3 overexpression significantly rescued the suppressive effects
of UCA1 downregulation on the proliferation and migration of
SK-MEL-28 cells. Taken together, these findings suggest that
inhibition of UCA1 expression suppresses SK-MEL-28 cell
proliferation and migration through modulating the miR-28-5p/HOXB3
axis.
Discussion
UCA1 serves a promoting role in different human
cancers (15,22), but the detailed regulatory mechanism
of UCA1 underlying melanoma cell proliferation and migration
remains largely unknown. In the present study it was demonstrated
that UCA1 expression was increased in melanoma tissues and cell
lines. Inhibition of UCA1 expression markedly reduced melanoma cell
migration and proliferation. Further investigation revealed that
UCA1 functioned in melanoma cells through directly binding to
miR-28-5p. The expression of miR-28-5p was significantly reduced in
melanoma tissues and exhibited an inverse correlation with UCA1
expression. Furthermore, HOXB3 was identified as a target gene of
miR-28-5p in melanoma cells, and HOXB3 overexpression reversed the
suppressive effects of UCA1 downregulation on melanoma cell
proliferation and migration.
In recent years, an increasing number of lncRNAs
have been demonstrated to be deregulated in human cancers, and some
of these lncRNAs have tumor promoting or suppressive roles in
melanoma (10,23). For instance, the lncRNA H19 promotes
glucose metabolism and cell growth in malignant melanoma via
modulating the expression of miR-106a-5p and its target gene E2F3
(24). In contrast, the upregulation
of lncRNA cancer susceptibility 2 has suppressive effects on the
malignant progression of melanoma through regulating the
miR-18a-5p/runt related transcription factor 1 axis (23). However, the molecular mechanism of
lncRNAs in the regulation of melanoma cell proliferation and
migration remains unknown. In the present study, it was
demonstrated that the lncRNA UCA1 was significantly upregulated in
a total of 21 melanoma tissues compared with adjacent non-tumor
tissues. These findings were consistent with a previous report
(14).
miRNAs, another type of non-coding small RNA that
are 22–25 nucleotides in length, regulate the expression of their
target genes through directly binding to the complementary sites in
the 3′-UTR region of target mRNAs (25–27).
Similar to lncRNAs, miRs are also associated with various cancers
(28–32). For instance, a previous study
demonstrated that miR-33a-5p increased the radiosensitivity of
melanoma cells by inhibition of glycolysis (33). In addition, it was also reported that
miR-18b inhibited the growth of malignant melanoma via inhibition
of hypoxia inducible factor-1α-mediated glycolysis (34). Through interacting with miRs, lncRNAs
can regulate the expression of miRs and thus the downstream target
genes (35,36). For instance, the lncRNA X inactive
specific transcript acts as a competing endogenous lncRNA to
regulate transforming growth factor-β1 by sponging miR-185 in
gastric cancer (37). The lncRNA H19
promotes glucose metabolism and cell growth in malignant melanoma
via inhibition of miR-106a-5p and thus upregulation of E2F3
(24). In the present study, it was
demonstrated that UCA1 directly targets miR-28-5p, and inhibition
of UCA1 enhanced the miR-28-5p expression. Furthermore, it was
demonstrated that downregulation of miR-28-5p in melanoma tissues
was inversely correlated with the upregulation of UCA1, which
further suggests that increased UCA1 expression contributes to the
decreased expression of miR-28-5p in melanoma. Further
investigation results suggest that the oncogenic role of UCA1 in
melanoma cells occurs through regulation of miR-28-5p.
Given that miRs function through regulating the
expression of their target genes (28,29),
bioinformatics analysis was conducted to predict the potential
target gene of miR-28-5p. We selected HOXB3 for the following
studies. A previous study reported that miR-28-5p regulates HOXB3
expression in colorectal cancer cells (19), but the targeting relationship between
miR-28-5p and HOXB3 has not been confirmed. In the present study,
luciferase reporter gene assay results indicated that HOXB3 was a
direct target gene of miR-28-5p in SK-MEL-28 cells. Furthermore,
UCA1 downregulation reduced HOXB3 expression, which was rescued by
knockdown of miR-28-5p in SK-MEL-28 cells. These findings suggest
that UCA1 affects HOXB3 expression via miR-28-5p. Furthermore, it
was demonstrated that HOXB3 overexpression significantly abolished
the suppressive effects of UCA1 downregulation on SK-MEL-28 cell
migration and proliferation, suggesting that UCA1 regulates
SK-MEL-28 cell proliferation and migration via the miR-28-5p/HOXB3
axis.
In addition to the UCA1/miR-28-5p/HOXB3 axis,
several other lncRNA/miRNA axes have been demonstrated to
participate in the regulation of melanoma cell proliferation and
migration. For instance, Chen et al (38) recently reported that lncRNA ILF3-AS1
promoted melanoma cell proliferation, migration, and invasion via
negatively regulating miR-200b/a/429. Zhao et al (39) demonstrated that hepatocellular
carcinoma upregulated EZH2-associated lncRNA enhanced the
proliferation, migration and invasion of melanoma cells via
inhibition of miR-200b/a/429. However, these studies did not
investigate the downstream proteins of these axes. Therefore, the
findings in the present study expand the understanding of the
function of the lncRNA/miR/protein pathways in melanoma cell
proliferation and migration.
To the best of our knowledge, the present study is
the first to report that the UCA1/miR-28-5p/HOXB3 axis has a key
role in the regulation of melanoma cell proliferation and
migration, which expands the understanding of the molecular
mechanism underlying melanoma progression. The limitation of the
present study is lack of animal experiments. Thus, future studies
should further confirm the function of this signaling pathway in
vivo.
Acknowledgements
Not applicable.
Funding
The present study was supported by New Xiangya
Talent Project of the Third Xiangya Hospital of Central South
University (grant no. JY201607).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors contributions
CH wrote the manuscript. JZ designed the study and
revised the manuscript. FT collected clinical tissues. CH, JC, DX,
XL, YX and SW performed all experiments.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Third Hospital of Central South University (Changsha,
China). Written informed consent was obtained from all patients
involved in the present study.
Patient consent for publication
Written informed consent was obtained from all
patients involved in the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Trotter SC, Sroa N, Winkelmann RR, Olencki
T and Bechtel M: A global review of melanoma follow-up guidelines.
J Clin Aesthet Dermatol. 6:18–26. 2013.PubMed/NCBI
|
|
2
|
Linos E, Swetter SM, Cockburn MG, Colditz
GA and Clarke CA: Increasing burden of melanoma in the United
States. J Invest Dermatol. 129:1666–1674. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gao YL, Zhao ZS, Zhang MY, Han LJ, Dong YJ
and Xu B: Long non-coding RNA PVT1 facilitates cervical cancer
progression via negative regulating of miR-424. Oncol Res.
25:1391–1398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hua F, Li CH, Chen XG and Liu XP: Long
Noncoding RNA CCAT2 knockdown suppresses tumorous progression by
sponging mir-424 in epithelial ovarian vancer. Oncol Res.
26:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li J, Zi Y, Wang W and Li Y: LncRNA MEG3
inhibits cell proliferation and metastasis in chronic myeloid
leukemia via targeting MiR-184. Oncol Res. 26:297–305. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang S, Hui Y, Li X and Jia Q: Silencing
of lncRNA-CCDC26 restrains the growth and migration of glioma cells
in vitro and in vivo via targeting miR-203. Oncol Res.
26:1143–1154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang JJ, Wang DD, Du CX and Wang Y: Long
noncoding RNA ANRIL promotes cervical cancer development By acting
as a sponge of miR-186. Oncol Res. 26:345–352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiong W, Huang C, Deng H, Jian C, Zen C,
Ye K, Zhong Z, Zhao X and Zhu L: Oncogenic non-coding RNA NEAT1
promotes the prostate cancer cell growth through the SRC3/IGF1R/AKT
pathway. Int J Biochem Cell Biol. 94:125–132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao L, Chen J, Ou B, Liu C, Zou Y and Chen
Q: GAS5 knockdown reduces the chemo-sensitivity of non-small cell
lung cancer (NSCLC) cell to cisplatin (DDP) through regulating
miR-21/PTEN axis. Biomed Pharmacother. 93:570–579. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen L, Yang H, Xiao Y, Tang X, Li Y, Han
Q, Fu J, Yang Y and Zhu Y: LncRNA GAS5 is a critical regulator of
metastasis phenotype of melanoma cells and inhibits tumor growth in
vivo. Onco Targets Ther. 9:4075–4087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L
and Sha N: Long non-coding RNA BANCR promotes proliferation in
malignant melanoma by regulating MAPK pathway activation. PLoS One.
9:e1008932014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo J, Chen J, Li H, Yang Y, Yun H, Yang S
and Mao X: LncRNA UCA1 promotes the invasion and EMT of bladder
cancer cells by regulating the miR-143/HMGB1 pathway. Oncol Lett.
14:5556–5562. 2017.PubMed/NCBI
|
|
13
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
malat-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xue M, Pang H, Li X, Li H, Pan J and Chen
W: Long noncoding RNA UCA1 promotes bladder cancer cell migration
and invasion via hsa-miR-145/ZEB1/2/FSCN1 pathway. Cancer Sci.
107:18–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu
Y, Qian H and Dai T: LncRNA UCA1 impacts cell proliferation,
invasion, and migration of pancreatic cancer through regulating
miR-96/FOXO3. IUBMB Life. 70:276–290. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang
Y, Zang W and Zhao G: LncRNA UCA1-miR-507-FOXM1 axis is involved in
cell proliferation, invasion and G0/G1 cell cycle arrest in
melanoma. Med Oncol. 33:882016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Almeida MI, Nicoloso MS, Zeng L, Ivan C,
Spizzo R, Gafà R, Xiao L, Zhang X, Vannini I, Fanini F, et al:
Strand-specific miR-28-5p and miR-28-3p have distinct effects in
colorectal cancer cells. Gastroenterology. 142:886–896 e889. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Jiang N, Shi H, Zhao S, Yao S and
Shen H: miR-28-5p promotes the development and progression of
ovarian cancer through inhibition of N4BP1. Int J Oncol.
50:22362017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li F and Hu CP: Long non-coding RNA
urothelial carcinoma associated 1 (UCA1): Insight into its role in
human diseases. Crit Rev Eukaryot Gene Expr. 25:191–197. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Qian W, Feng F, Cao Q, Li Y, Hou
Y, Zhang L and Fan J: Upregulated lncRNA CASC2 may inhibit
malignant melanoma development through regulating miR-18a-5p/RUNX1.
Oncol Res. doi.org/10.3727/096504018X15178740729367.
|
|
24
|
Luan W, Zhou Z, Ni X, Xia Y, Wang J, Yan Y
and Xu B: Long non-coding RNA H19 promotes glucose metabolism and
cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J
Cancer Res Clin Oncol. 144:531–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Z, Guo J, Ma Y, Lin Z and Zhang L:
Oncogenic role of MicroRNA-30b-5p in glioblastoma through targeting
proline-rich transmembrane protein 2. Oncol Res. 26:219–230. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu H, Huang L, Zhu S, Li X, Li Z, Yu C
and Yu X: Regulation of autophagy by systemic admission of
microRNA-141 to target HMGB1 in l-arginine-induced acute
pancreatitis in vivo. Pancreatology. 16:337–346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zeng F, Xue M, Xiao T, Li Y, Xiao S, Jiang
B and Ren C: MiR-200b promotes the cell proliferation and
metastasis of cervical cancer by inhibiting FOXG1. Biomed
Pharmacother. 79:294–301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Cao KE, He Q, Yin Z and Zhou J:
miR-199a-5p induces cell invasion by suppressing E-cadherin
expression in cutaneous squamous cell carcinoma. Oncol Lett.
12:97–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su Y, Xiong J, Hu J, Wei X, Zhang X and
Rao L: MicroRNA-140-5p targets insulin like growth factor 2 mRNA
binding protein 1 (IGF2BP1) to suppress cervical cancer growth and
metastasis. Oncotarget. 7:68397–68411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou J, Xu D, Xie H, Tang J, Liu R, Li J,
Wang S, Chen X, Su J, Zhou X, et al: miR-33a functions as a tumor
suppressor in melanoma by targeting HIF-1α. Cancer Biol Ther.
16:846–855. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou J, Liu R, Wang Y, Tang J, Tang S,
Chen X, Xia K, Xiong W, Xu D, Wang S, et al: miR-199a-5p regulates
the expression of metastasis-associated genes in B16F10 melanoma
cells. Int J Clin Exp Pathol. 7:7182–7190. 2014.PubMed/NCBI
|
|
32
|
Zhou J, Liu R, Luo C, Zhou X, Xia K, Chen
X, Zhou M, Zou Q, Cao P and Cao K: MiR-20a inhibits cutaneous
squamous cell carcinoma metastasis and proliferation by directly
targeting LIMK1. Cancer Biol Ther. 15:1340–1349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cao K, Li J, Chen J, Qian L, Wang A, Chen
X, Xiong W, Tang J, Tang S, Chen Y, et al: microRNA-33a-5p
increases radiosensitivity by inhibiting glycolysis in melanoma.
Oncotarget. 8:83660–83672. 2017.PubMed/NCBI
|
|
34
|
Chen Y, Zhang Z, Luo C, Chen Z and Zhou J:
MicroRNA-18b inhibits the growth of malignant melanoma via
inhibition of HIF-1α-mediated glycolysis. Oncol Rep. 36:471–479.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu S, Song L, Zeng S and Zhang L:
MALAT1-miR-124-RBG2 axis is involved in growth and invasion of
HR-HPV-positive cervical cancer cells. Tumour Biol. 37:633–640.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie CH, Cao YM, Huang Y, Shi QW, Guo JH,
Fan ZW, Li JG, Chen BW and Wu BY: Long non-coding RNA TUG1
contributes to tumorigenesis of human osteosarcoma by sponging
miR-9-5p and regulating POU2F1 expression. Tumour Biol.
37:15031–15041. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Q, Chen B, Liu P and Yang J: XIST
promotes gastric cancer (GC) progression through TGF-beta1 via
targeting miR-185. J Cell Biochem. 119:2787–2796. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Liu S, Zhao X, Ma X, Gao G, Yu L,
Yan D, Dong H and Sun W: Long noncoding RNA ILF3-AS1 promotes cell
proliferation, migration, and invasion via negatively regulating
miR-200b/a/429 in melanoma. Biosci Rep. 37:BSR201710312017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao H, Xing G, Wang Y, Luo Z, Liu G and
Meng H: Long noncoding RNA HEIH promotes melanoma cell
proliferation, migration and invasion via inhibition of
miR-200b/a/429. Biosci Rep. 37:BSR201706822017. View Article : Google Scholar : PubMed/NCBI
|