Introduction
Breast cancer is a common malignant tumor with an
increasing incidence rate (1,2).
According to the World Cancer Research Fund, 450,000 mortalities
are caused by breast cancer worldwide each year, accounting for
13.7% of cancer-associated mortalities in women (3). Great efforts have been made to improve
the diagnosis and treatment of breast cancer in recent years;
however, the pathogenic mechanism of breast cancer is yet to be
fully elucidated. At present, doxorubicin (DOX) is considered to be
one of the most effective drugs for the treatment of breast cancer
(4). However, following several
treatments, drug resistance may develop, which leads to therapy
failure (5,6). Currently, the mechanism of DOX
resistance is poorly understood.
Long non-coding RNA (lncRNA) are RNA molecules
comprising >200 nucleotides that do not possess any
protein-coding capacity (7,8). However, lncRNAs serve key roles in the
transcriptional, post-transcriptional and epigenetic regulation of
gene expression (9). Numerous
studies have indicated that the abnormal expression of lncRNA is
associated with various tumor functions, including tumor formation,
drug resistance and metastasis (10,11).
Additionally, the abnormal expression of the lncRNA, HOX transcript
antisense RNA (HOTAIR), was identified in breast cancer cell lines
and tissue (12,13). A previous study demonstrated that
lncRNA-HOTAIR increased cisplatin resistance in gastric cancer by
targeting microRNA-126 via the phosphoinositide 3-kinase
(PI3K)/protein kinase B (AKT)/multidrug resistance-associated
protein 1 (MRP1) pathway (14).
However, It is not clear whether LncRNA-HOTAIR is involved in
resistance to DOX in breast cancer cells.
The PI3K/AKT/mechanistic target of rapamycin (mTOR)
signal transduction pathway serves a key role in the cell cycle,
the proliferation of cells, cellular metabolism and in protein
synthesis (15). A previous study
has indicated that the PI3K/AKT/mTOR pathway is frequently
activated in ovarian cancer (16).
Furthermore, the aberrant activation of the signaling pathway is
associated with the development of breast cancer and has therefore
been targeted for the treatment of breast cancer (17).
The present study aimed to assess whether
lncRNA-HOTAIR is associated with the resistance of breast cancer
cells to DOX. The results revealed that the silencing of
lncRNA-HOTAIR decreased the resistance of breast cancer cells to
DOX by suppressing the PI3K/AKT/mTOR pathway. These results may
provide a novel intervention target to reverse DOX-resistance in
breast cancer.
Materials and methods
Cell culture
The human breast cancer cell lines, MCF-7 and SKBR3,
were obtained from the American Type Culture Collection (Manassas,
VA, USA). Cells were cultivated in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and
incubated at 37°C with 5% CO2.
Establishment of DOX-resistant cell
lines
DOX was purchased from the First Affiliated Hospital
of Kunming Medical University (Kunming, China). MCF-7 and SKBR3
cells were cultured in DOX-free Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc.) until cells were in
the logarithmic growth phase, at which point the culture medium was
replaced with DMEM medium containing a low dose of DOX (0.2 µg/ml).
Subsequently, the concentration of DOX was increased from 0.2 to
0.6 µg/ml in 0.04 µg increments every 3 weeks. The DOX-resistant
MCF-7 and SKBR3 cell lines were considered to have been
successfully established when cells were able to survive under high
DOX treatment (0.6 µg/ml). Prior to subsequent experimentation,
cells were cultured in the drug-free DMEM medium containing 5%
CO2 at 37°C for 2 days.
Cell transfection and RNA
interference
LncRNA-HOTAIR small interfering RNAs (siRNAs
or siR; siR-HOTAIR1, 3′-GAACGGGAGUACAGAGAGAUU-5′; siR-HOTAIR1-2,
3′-CCACAUGAACGCCCAGAGAUU-5′) were designed and synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China). Negative control
siRNA (3′-CUACAACAGCCACAACGUCdTdT-5′) was purchased from Sangon
Biotech Co., Ltd. (Shanghai, China). Briefly, 500 ng siR-HOTAIR1
was transfected into MCF-7 cells and DOXR-MCF-7 cells using 1 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
cells were incubated at 37°C in an atmosphere containing 5%
CO2 for 6 h. Subsequently, transfected cells were
incubated at 37°C for 72 h. Cells were then divided into the
following experimental groups: Untransfected MCF-7 cells, negative
control siRNA-transfected MCF-7 cells, siR-HOTAIR1 transfected
MCF-7 cells; untransfected DOXR-MCF-7 cells, negative control
siRNA-transfected DOXR-MCF-7 cells and siR-HOTAIR1 transfected
DOXR-MCF-7 cells.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from MCF-7 and DOXR-MCF-7
cells using TRIzol reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Total RNA was then
reverse-transcribed into cDNA using an RT kit (Takara Biotechnology
Co., Ltd., Dalian, China) following the manufacturer's protocol.
qPCR was performed using the SYBR Premix Ex Taq (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with the following
thermocycling conditions: Initial denaturation at 95°C for 10 min,
followed by 40 cycles of denaturation for 30 sec at 95°C, annealing
at 60°C for 30 sec and extension at 72°C for 30 sec. Relative
expression levels were calculated using the 2−ΔΔCq
method (18). GAPDH was used as the
reference gene. The following primers were used for amplification:
GAPDH forward, 5′-ATTGATGGATGCTAFGAGTATT-3′ and reverse,
5′-AGTCTTCTGGGTGGCAGTGAT-3′; lncRNA-HOTAIR forward,
5′-CGGAGTGAGTTTATCGCAG-3′ and reverse,
5′-GGCGACCGGAGCTCATCTTACC-3′.
MTT assay
Following 72 h incubation, cell proliferation
analysis was performed using an MTT assay kit (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). MCF-7 and DOXR-MCF-7 cells
(5×105 cells/well) were seeded into 96-well plates.
Subsequently, cell proliferation was detected at different time
points (24, 48 and 72 h). MTT (5 mg/Ml, 20 µl) was added to cells
and incubated for 4 h at 37°C. The precipitate was dissolved in 100
µl dimethyl sulfoxide and absorbance at 490 nm was detected using a
microplate spectrophotometer.
Colony formation assay
A colony formation assay was performed following
transfection. Cells (500 cells/well) were seeded into 6-well plates
and incubated for 10 days in 5% CO2 at 37°C to form
colonies. Cells were fixed in 95% methanol for 15 min at 4°C and
stained with 1% crystal violet solution for 30 min at room
temperature. Images were captured under a light microscope
(magnification, ×100). Colony number was quantified using
Alpha-View Software 3.1 (FluorChem Q; ProteinSimple, San Jose, CA,
USA).
Flow cytometric assay
Cell apoptosis was assessed using a fluorescein
isothiocyanate (FITC) apoptosis detection kit (Oncogene Research
Products, La Jolla, CA, USA) in accordance with the manufacturer's
protocol. Cells were harvested, washed twice with pre-cooled PBS
and stained successively with propidium iodide (10 µl) and
Annexin-V-FITC (10 µl; Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China). Following 15 min of incubation at room temperature, a flow
cytometer (FACSVantage SE; BD Biosciences, San Jose, CA, USA) was
used for sample analysis using the FlowJo software package (version
10.0.7; Tree Star, Inc., Ashland, OR, USA). The number of cells in
each classification was presented.
Western blot analysis
Western blotting was performed to assess the
expression of proteins of interest. Proteins were isolated using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) and protein concentration was
measured using a bicinchoninic acid protein assay kit (Beyotime
Institute of Biotechnology). Proteins (20 µg/per lane) were
subsequently separated via SDS-PAGE on 10% gels using
polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica,
MA, USA) for protein transfer. Membranes were then blocked with 5%
nonfat milk for 2 h at room temperature and incubated with the
following primary antibodies obtained from Cell Signaling
Technology, Inc. (Danvers, MA, USA): Anti-Bcl-2-associated X
protein (Bax; 1:1,000; cat. no. 2774), anti-B-cell lymphoma 2
(Bcl2; 1:1,000; cat. no. 2872), anti-cleaved caspase3 (1:500; cat.
no. 9664), anti-PI3K (1:1,000; cat. no. 4255), anti-AKT (1:500;
cat. no. 9272), anti-mTOR (1:500; cat. no. 2972), anti-phospho
(p)-mTOR (ser2448; 1;1,000; cat. no. 5536), anti-p-PI3K (tyr458;
1:1000; cat. no. 4228), anti-p-AKT (ser473; 1:500; cat. no. 4060),
anti-MRP1 (1:500; cat. no. 14685), anti-multidrug resistance
protein 1 (MDR1; 1:500; cat. no. 13978), anti-ATP binding cassette
subfamily B member 1 (ABCB1; 1:1,000; cat. no. 12683) and
anti-GAPDH (1:2,000; cat. no. 5174). The membrane was then
incubated with goat anti-rabbit IgG secondary antibodies (1:3,000;
cat. no. ab6721; Abcam, Cambridge, UK) for 1 h at room temperature.
An enhanced chemiluminescence kit (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used for visualization and the images were
analyzed using ImageJ v1.8.0 (National Institutes of Health,
Bethesda, MD, USA). The relative quantification of protein
expression was analyzed using Image-Pro Plus 6.0 (Media
Cybernetics, Inc., Rockville, MD, USA).
Statistical analysis
Statistical analysis was performed using SPSS
version 20.0 software (IBM Corp, Armonk, NY, USA). Each experiment
was repeated in triplicate and all data are presented as the mean ±
standard deviation. Differences among multiple groups were detected
using one-way analysis of variance followed by the Dunnett's post
hoc-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Detection of lncRNA-HOTAIR
interference efficiency
The results of RT-qPCR revealed that HOTAIR was
upregulated in the drug resistant breast cancer cell line,
DOXR-MCF-7, compared with the other cell lines. HOTAIR expression
also significantly differed between MCF-7 and MCF-7-DOX cell lines
(Fig. 1A). Therefore, MCF-7 and
MCF-7-DOX cell lines were used for the subsequent experimentation.
The interference efficiency of HOTAIR siRNA was determined via
RT-qPCR. The results revealed that the expression of lncRNA-HOTAIR1
was significantly reduced in the siR-HOTAIR1 and siR-HOTAIR1-2
groups compared with the negative control (Fig. 1B). The siR-HOTAIR1 plasmid, which
exhibited the greatest interference effect, was selected for
subsequent experimentation.
Effect of siR-HOTAIR on the
proliferation of MCF-7 cells
An MTT assay was performed to assess the
proliferation of siR-HOTAIR1-transfected MCF-7 cells using optical
density values (Fig. 1C). The
results indicated that proliferation was markedly decreased in the
siR-HOTAIR1 group compared with the control and NC groups.
Subsequently, a colony formation assay was performed to further
assess the proliferation rate of MCF-7 cells (Fig. 1D). The results demonstrated that cell
colony formation ability was significantly decreased in the
siR-HOTAIR1 group compared with the control and NC groups.
Furthermore, the results of flow cytometry revealed that the rate
of apoptosis was increased in the siR-HOTAIR1 group compared with
the control and NC groups (Fig. 2).
These results indicate that the knockdown of lncRNA-HOTAIR may
suppress the proliferation of MCF-7 cells.
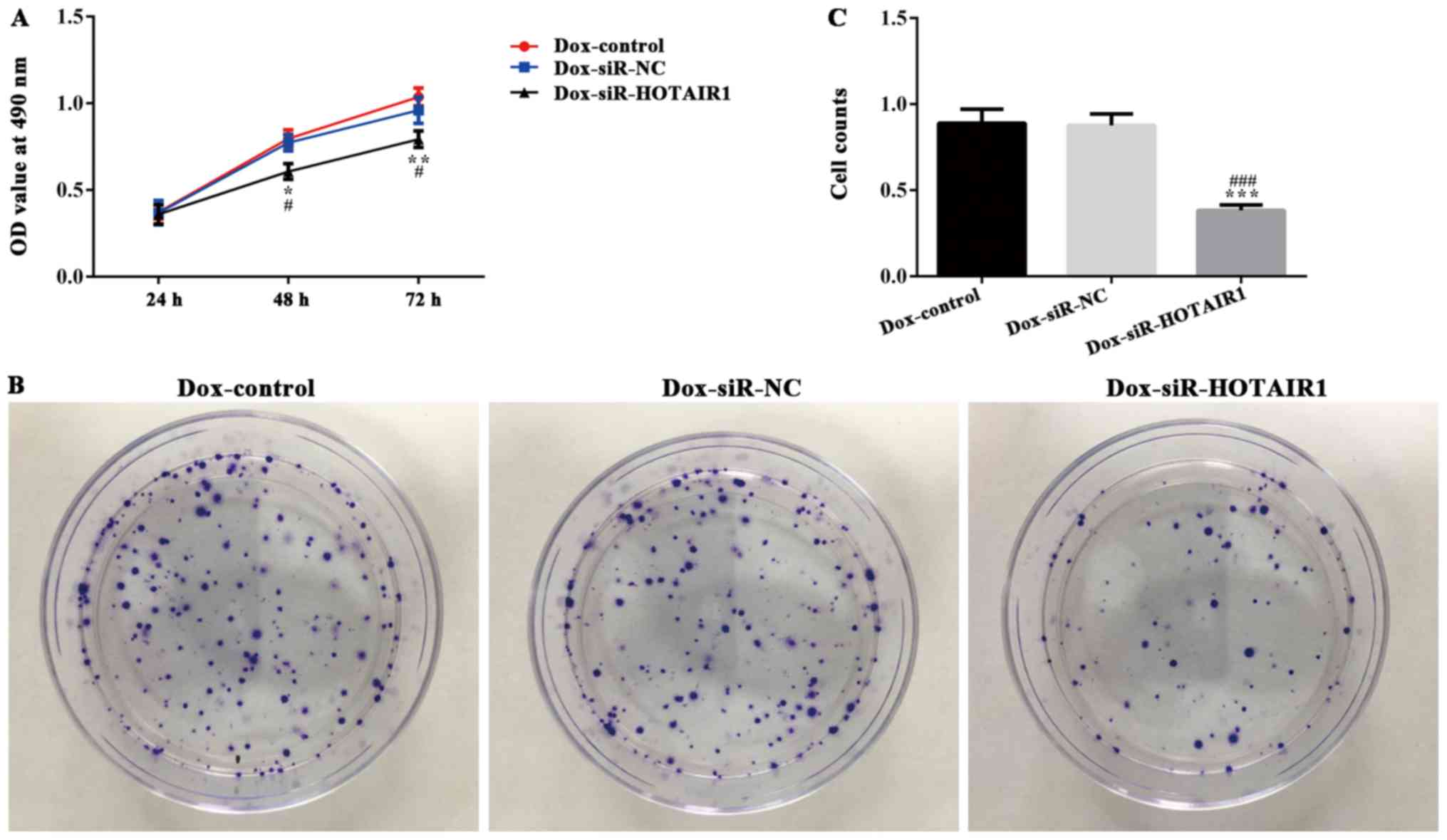
HOTAIR silencing reduces the
sensitivity of drug resistance in drug-resistant MCF-7 cells
DOXR-MCF-7 cells were transfected with siR-HOTAIR1
and an MTT assay revealed that proliferation was significantly
decreased in the siR-HOTAIR1 group compared with the control and NC
groups (Fig. 3A). Additionally, cell
colony formation was significantly decreased in the Dox-siR-HOTAIR1
group (Fig. 3B and C). Furthermore,
the apoptosis rate of Dox-siR-HOTAIR1 cells was significantly
increased compared with the control group (Fig. 4). The results indicate that
lncRNA-HOTAIR may serve a key role in the resistance to DOX in
MCF-7 cells.
HOTAIR silencing inhibits
proliferation and promotes apoptosis by regulating apoptosis
associated proteins
The activation of programmed cell death machinery is
considered to be associated with the inhibition of cell
proliferation. To assess the effect of siR-HOTAIR1 on cell
apoptosis, the expression of apoptosis-associated proteins was
determined via western blot analysis. Caspase-3, Bcl-2 and Bax have
essential roles in cell apoptosis and tumorigenesis. As
demonstrated in Fig. 5, the protein
levels of caspase-3 and Bax were increased, and the expression of
Bcl-2 was markedly decreased in the DOXR-MCF-7 siR-HOTAIR1 group
compared with control group. The results indicated that HOTAIR
silencing may affect proliferation and promote apoptosis by
regulating the expression of apoptosis-associated proteins.
Effect of HOTAIR silencing on
drug-resistant proteins
Classical multidrug-resistant proteins, MDR1, MRP1
and ABCB1 were detected to determine the effect of lncRNA-HOTAIR on
the drug resistance of DOXR-MCF-7 cells. The protein levels of
MDR1, MRP1 and ABCB1 were significantly decreased in DOXR-MCF-7
siR-HOTAIR1 cells compared with the siR-NC DOXR-MCF-7 cells
(Fig. 6).
HOTAIR affects the PI3K/AKT/mTOR
signaling pathway in drug-resistant MCF-7 cells
The PI3K/AKT/mTOR pathway serves a key role in cell
proliferation, cellular metabolism, protein synthesis and in the
cell cycle (14). It is considered
to be one of the most frequently mutated pathways in cancer
(14). To determine the effect of
HOTAIR silencing on the PI3K/AKT/mTOR pathway, the phosphorylation
of these proteins were analyzed via western blotting. The protein
expression of PI3K, AKT and mTOR remained unchanged, while the
phosphorylation of PI3K, AKT and mTOR was significantly reduced by
HOTAIR silencing in DOXR-MCF-7 cells compared with the control and
NC groups (Fig. 7). This series of
analyses indicate that siR-HOTAIR alters the resistance of MCF-7
cells to DOX.
Discussion
Breast cancer is the most common cause of
cancer-associated mortality among women (1,2) and
chemoresistance is a major cause of treatment failure (19,20).
Previous research has revealed that drug resistance is associated
with DNA repair, aberrant gene expression and epigenetics (21).
LncRNAs have gained increasing attention as they
have been reported to be associated with the progression of several
types of cancer (22). Previous
research has indicated that lncRNAs may be involved in numerous
cellular activities and the roles of lncRNAs in various cancer
types have been discussed. LncRNA-HOTAIR has been identified as a
regulator of trans-silencing (23).
A study by Yu et al (24)
demonstrated that the knockdown of lncRNA-HOTAIR significantly
inhibited MCF-7 cell proliferation and promoted apoptosis (24). The present study revealed that
lncRNA-HOTAIR silencing decreased proliferation and increased
apoptosis in breast cancer cells, which supports these previous
findings.
A previous study has demonstrated that lncRNA-HOTAIR
is also involved in regulating the resistance of lung cancer cells
to cisplatin (25). Furthermore,
previous studies have indicated that the overexpression of HOTAIR
promotes the proliferation and enhances doxorubicin resistance in
GC and TCC cells; however, HOTAIR silencing exhibits the opposite
regulative effects (26,27). In the present study, the expression
of multidrug-resistant proteins was significantly decreased by
lncRNA-HOTAIR silencing, which also inhibited cell viability and
induced apoptosis in DOXR-MCF-7 cells, indicating that the
knockdown of lncRNA-HOTAIR decreased DOX resistance in breast
cancer cells, which is consistent with previous findings.
Furthermore, a previous study demonstrated that lncRNA-HOTAIR
silencing suppresses the proliferation and metastasis of
osteosarcoma cells by inhibiting the PI3K/AKT/mTOR pathway
(28). The results of the present
study revealed that the phosphorylation of PI3K, AKT and mTOR were
significantly altered in DOXR-MCF-7 cells, which indicated that the
knockdown of lncRNA-HOTAIR decreased DOX resistance breast cancer
cells by inhibiting the PI3K/AKT/mTOR pathway. Caspase-3, Bcl-2 and
Bax serve essential roles in cell apoptosis and tumorigenesis. In
the current study, the protein levels of caspase-3, Bcl-2 and Bax
were significantly altered by HOTAIR silencing in DOXR-MCF-7 cells,
which indicated that the knockdown of lncRNA-HOTAIR promoted
apoptosis in DOXR-MCF-7 cells.
In summary, the results of the present study
indicated that lncRNA-HOTAIR silencing inhibits cell proliferation
and promotes apoptosis in MCF-7 and DOXR-MCF-7 cell lines.
Furthermore, the knockdown of lncRNA-HOTAIR reduces DOX resistance
in breast cancer cells via the PI3K/AKT/mTOR signaling pathway,
indicating that lncRNA-HOTAIR has the potential to become a
therapeutic target for the treatment of breast cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the current study. JQ and CZ performed
the experiments. JQ and JL analyzed the data. ZL drafted the
manuscript and analyzed data. ZL and JQ interpreted data and
revised the final manuscript. ZL wrote the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All experimental protocols were performed in
accordance with the principles of the Declaration of Helsinki and
were approved by the Clinical Research Ethics Committee of bengbu
medical college (Anhui, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Howell A, Anderson AS, Clarke RB, Duffy
SW, Evans DG, Garcia-Closas M, Gescher AJ, Key TJ, Saxton JM and
Harvie MN: Risk determination and prevention of breast cancer.
Breast Cancer Res. 16:4462014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bargostavan MH, Eslami G, Esfandiari N and
Shams Shahemabadi A: MMP9 promoter polymorphism (−1562C/T) does not
affect the serum levels of soluble MICB and MICA in breast cancer.
Iran J Immunol. 13:45–53. 2016.PubMed/NCBI
|
|
3
|
Kakarala M, Rozek L, Cote M, Liyanage S
and Brenner DE: Brenner Breast cancer histology and receptor status
characterization in Asian Indian and Pakistani women in the U. S.
-a SEER analysis. BMC Cancer. 10:1912010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H, Yu Y, Jiang Z, Cao WM, Wang Z, Dou
J, Zhao Y, Cui Y and Zhang H: Next-generation proteasome inhibitor
MLN9708 sensitizes breast cancer cells to doxorubicin-induced
apoptosis. Sci Rep. 6:264562016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Palmieri C, Krell J, James CR,
Harper-Wynne C, Misra V, Cleator S and Miles D: Rechallenging with
anthracyclines and taxanes in metastatic breast cancer. Nat Rev
Clin Oncol. 7:561–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu X, Fu Y, Wang Y, Wan S and Zhang J:
Gaining insight into crizotinib resistance mechanisms caused by
L2026M and G2032R mutations in ROS1 via molecular dynamics
simulations and free-energy calculations. J Mol Model. 23:1412017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen LL and Zhao JC: Functional analysis
of long noncoding RNAs in development and disease. Adv Exp Med
Biol. 825:129–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu YR, Tang RX, Huang WT, Ren FH, He RQ,
Yang LH, Luo DZ, Dang YW and Chen G: Long noncoding RNAs in
hepatocellular carcinoma: Novel insights into their mechanism.
World J Hepatol. 7:2781–2791. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang T, Alvarez A, Hu B and Cheng SY:
Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer.
32:582–593. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng G and Sui G: Noncoding RNA in
oncogenesis: A new era of identifying key players. Int J Mol Sci.
14:18319–18349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gökmen-Polar Y, Vladislav IT, Neelamraju
Y, Janga SC and Badve S: Prognostic impact of HOTAIR expression is
restricted to ER-negative breast cancers. Sci Rep. 5:87652015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhuang Y, Nguyen HT, Burow ME, Zhuo Y,
El-Dahr SS, Yao X, Cao S, Flemington EK, Nephew KP, Fang F, et al:
Elevated expression of long intergenic non-coding RNA. HOTAIR in a
basal-like variant of MCF-7 breast cancer cell. Mol Carcinog.
54:1656–1667. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan J, Dang Y, Liu S, Zhang Y and Zhang G:
LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by
targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol.
21:235–256. 2015.
|
|
15
|
Austreid E, Lonning PE and Eikesdal HP:
The emergence of targeted drugs in breast cancer to prevent
resistance to endocrine treatment and chemotherapy. Expert Opin
Pharmacother. 15:681–700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mabuchi S, Kuroda H, Takahashi R and
Sasano T: The PI3K/AKT/mTOR pathway as a therapeutic target in
ovarian cancer. Gynecol Oncol. 137:173–179. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JJ, Loh K and Yap YS: PI3K/Akt/mTOR
inhibitors in breast cancer. Cancer Biol Med. 12:342–354.
2015.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
Relative Gene Expression Data Using Real-Time Quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Amiri-Kordestani L, Basseville A, Kurdziel
K, Fojo AT and Bates SE: Targeting MDR in breast and lung cancer:
Discriminating its potential importance from the failure of drug
resistance reversal studies. Drug Resist Updat. 15:50–61. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jia H, Truica CI, Wang B, Wang Y, Ren X,
Harvey HA, Song J and Yang JM: Immunotherapy for triple-negative
breast cancer: Existing challenges and exciting prospects. Drug
Resist Updat. 32:1–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jang JE, Eom JI, Jeung HK, Cheong JW, Lee
JY, Kim JS and Min YH: Targeting AMPK-ULK1-mediated autophagy for
combating BET inhibitor resistance in acute myeloid leukemia stem
cells. Autophagy. 13:761–762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Woo CJ and Kingston RE: HOTAIR lifts
noncoding RNAs to new levels. Cell. 129:1257–1259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu Y, Lv F, Liang D, Yang Q, Zhang B, Lin
H, Wang X, Qian G, Xu J and You W: HOTAIR may regulate
proliferation, apoptosis, migration and invasion of MCF-7 cells
through regulating the P53/Akt/JNK signaling pathway. Biomed
Pharmacother. 90:555–561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W,
De W, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21(WAF1/CIP1) expression. PLoS One.
8:e772932013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang H, Qin R, Guan A, Yao Y, Huang Y, Jia
H, Huang W and Gao J: HOTAIR enhanced paclitaxel and doxorubicin
resistance in gastric cancer cells partly through inhibiting
miR-217 expression. J Cell Biochem. 119:7226–7234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shang C, Guo Y, Zhang H and Xue YX: Long
noncoding RNA HOTAIR is a prognostic biomarker and inhibits
chemosensitivity to doxorubicin in bladder transitional cell
carcinoma. Cancer Chemother Pharmacol. 77:507–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li E, Zhao Z, Ma B and Zhang J: Long
noncoding RNA HOTAIR promotes the proliferation and metastasis of
osteosarcoma cells through the AKT/mTOR signaling pathway. Exp Ther
Med. 14:5321–5328. 2017.PubMed/NCBI
|