Introduction
Breast cancer (BC) is the most frequently diagnosed
cancer and the leading cause of cancer mortality in women,
accounting for 23% of all cancer cases and 14% of cancer-related
deaths (1). For 2011, an
estimated 230,480 new cases of invasive BC and 39,520 BC deaths
were expected among US women (2,3).
Reliable markers are still lacking, and the most promising serum
markers, CEA and CA153, are not sensitive or specific enough for
screening asymptomatic women. Currently, the most efficacious
clinical screening tool for BC is mammography (1). This approach is too expensive to be
feasible in most developing countries, where screening rates
continue to be lower for poor women (1,2).
Serum-based screening is easier, less invasive and more affordable
than mammography. Thus, leading to an increased interest in
identifying and validating serum-based biomarkers for the diagnosis
of BC.
microRNAs (miRNAs) are a class of 20–25 nucleotide
nonprotein-coding small RNAs that negatively regulate gene
expressions by cleaving, inhibiting or degrading mRNA translation.
Negative regulation by miRNAs controls crucial physiological
processes, such as cell differentiation, proliferation, apoptosis,
development and cell metabolism (4–8).
Numerous studies have shown that aberrant miRNA expression is
associated with the development and progression of various types of
human cancer (9–13). Although their biological function
remains largely unknown, some miRNAs have functions similar to
those of oncogenes or tumor suppressors (14–16). Bioinformatic data indicate that
miRNAs have the potential to regulate at least 20–30% of all human
genes. A single miRNA can control the expression of hundreds of
miRNA gene targets (17,18). More than 50% of the annotated
human miRNA genes are located in cancer-associated genomic regions
or in fragile sites and may thus play important roles in
tumorigenesis (19).
Recently, miRNAs in serum and plasma have been
investigated as promising novel biomarkers for cancer diagnosis and
prognosis (20). Many studies
have emerged recently showing that miR-181a is expressed at low
levels in a variety of tumors including lung (21), oral (22), hepatocellular (23), ovarian (24) and glioblastoma (25). However, published studies
regarding serum miR-181a in BC do not exist. On the basis of
findings in other types of cancer, we hypothesized that deregulated
miR-181a may also be present in the serum of patients with BC and
might even serve as a diagnostic marker.
In the present study, we first investigated
circulating miR-181a levels in the sera from a small sample of BC
and healthy subjects. Based on promising results in the small
sample, we also analyzed a larger sample and compared the
sensitivity of miR-181a in diagnosing early stage BC to the
sensitivity of CA153 and CEA. Further analysis explored the
relationship of serum miR-181a levels with BC clinical and
pathological features.
Materials and methods
Study subjects
In this study, 152 women were recruited who had a
histologically confirmed diagnosis of primary BC (median age, 50.38
years; range, 25–83 years). Serum samples were collected from 75
age-matched healthy women who underwent medical examinations at the
Beijing Cancer Hospital between March and October 2010 (median age,
48.4 years; range, 22–60 years).
Indicators
The level of CEA and CA153 were determined by
electrochemiluminescence immunoassays using the Modular E170
automated analyzer (Roche, Basel, Switzerland). The normal upper
limits were 5.0 mg/l for CEA and 25 kU/l for CA153. Hormone
receptor (ER, PR) status was determined by routine
immunohistochemical methods. Tumors were staged according to the
AJCC tumor-node-metastasis (TNM) staging system, and
histopathological grading was also performed (lymph node
metastasis/no lymph node metastasis and ER and PR status).
Clinicopathological characteristics of the BC patients/samples are
shown in Table I. This study was
approved by the Clinical Research Ethics Committee of the Peking
University Cancer Hospital.
 | Table I.Clinical characteristics and serum
miR-181a levels in the breast cancer patients. |
Table I.
Clinical characteristics and serum
miR-181a levels in the breast cancer patients.
| Characteristic | n | miR-181a levels
(mean ± SD) | Mann-Whitney
U/Kruskal-Wallis H/t-testa |
|---|
| Group | | | P<0.001 |
| Healthy
group | 75 | 1.0908±0.50821 | |
| Breast cancer
group | 152 | 0.8385±0.69453 | |
| TNM stage | | | P=0.285 |
| DCIS | 4 | 1.0546±0.60466 | |
| I | 40 | 0.9220±0.71448 | |
| II | 79 | 0.8837±0.75118 | |
| III | 13 | 0.5700±0.37882 | |
| IV | 16 |
0.57103±0.47530 | |
| Invasive ductal
carcinoma histological grade | 137 | | P=0.269 |
| I | 5 |
1.20271±0.87914 | |
| II | 104 |
0.79574±0.68236 | |
| III | 19 |
0.94171±0.82403 | |
| Lymph node
metastasis | | | P=0.742 |
| Positive | 72 | 0.8425±0.72189 | |
| Negative | 72 | 0.8201±0.66547 | |
| ER status | | | P=0.213 |
| Positive | 104 | 0.8425±0.74682 | |
| Negative | 45 | 0.8632±0.57218 | |
| PR status | | | P=0.944 |
| Positive | 92 | 0.7723±0.71820 | |
| Negative | 57 | 0.9425±0.64144 | |
Sample processing
We collected 6 ml of venous blood from the
antecubital fossa into an EDTA tube. The blood was centrifuged at
1600 rpm at 4°C (Sorvall Biofuge Stratos, Heraeus, Germany) for 5
min to spin down the blood cells. The supernatant was removed to a
1.5 ml Eppendorf tube followed by a second centrifugation at 12,000
rpm for 15 min at 4°C (Heraeus Fresco 21 microcentrifuge, Thermo
Electron Corporation, Langenselbold, Germany) to completely remove
cell debris. Serum samples were frozen within 4 h of blood
collection and stored at −70°C until analysis.
Total-RNA extraction
Total-RNA was extracted from 500 μl serum using
TRIzol LS reagent (Invitrogen, Carlsbad, CA, USA) in accordance
with the manufacturer’s protocol. Total-RNA was suspended in a
fixed volume of 100 μl diethyl pyrocarbonate (DEPC)-treated water
for use in the reverse transcription (RT) reaction.
Stem-loop primer based RT reaction
Mature miRNA sequences were obtained from the
miRBase database (http://microrna.sanger.ac.uk/). Stem-loop primers,
specific primers and universal primers were designed using Primer
5.0 (Table II). cDNA was
synthesized from total-RNA via RT. First, 10 μl of purified total
RNA plus 3 μl stem-loop RT primer was denatured at 70°C for 5 min
before quenching on ice for 5 min. We then added 0.5 μl dNTP
(Tiangen Biotech Co., Ltd., Beijing, China), 2 μl MDTT, 0.2 μl
RNase inhibitor, 1 μl M-MLV reverse transcriptase (Promega,
Madison, WI, USA) and 3.3 μl 1X M-MLV RT buffer (Promega) for a
final reaction volume of 20 μl (Table
III). Each reverse transcription reaction was performed in a
96-well plate using a Bio-Rad MyCycler™ Thermal Cycler (Bio-Rad,
USA) for 30 min at 16°C, 30 min at 37°C and 10 min at 70°C.
Reactions were held at 4°C.
 | Table II.Sequences of the miR-16 and miR-181a
stem-loop reverse transcriptase (RT) primers and PCR primers. |
Table II.
Sequences of the miR-16 and miR-181a
stem-loop reverse transcriptase (RT) primers and PCR primers.
| Primer | Sequence |
|---|
| RT primers |
| miR-16 |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGCCAAT-3′ |
| miR-181a |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTCACCGA-3′ |
| Specific
primers |
| miR-16 |
5′-GGCGTAGCAGCACGTAAATAT-3′ |
| miR-181a |
5′-GAACATTCAACGCTGTCGGTG-3′ |
| Universal
primer |
5′-ATCCAGTGCAGGGTCCGAGGTA-3′ |
 | Table III.Comparison of miR-181a and the
conventional tumor markers CA153 and CEA for identifying breast
cancer patients. |
Table III.
Comparison of miR-181a and the
conventional tumor markers CA153 and CEA for identifying breast
cancer patients.
| TNM | miR-181a positive
(<0.7486), % | CA153 positive
(>25.0), % | CEA positive
(>5.0), % |
|---|
| DCIS | 50.00 (2/4) | 0.00 (0/4) | 0.00 (0/4) |
| I | 57.50 (23/40) | 2.50 (1/40) | 2.50 (1/40) |
| II | 54.43 (43/79) | 11.39 (9/79) | 10.13 (8/79) |
| III | 69.23 (9/13) | 7.69 (1/13) | 0.00 (0/13) |
| IV | 75.00 (12/16) | 31.25 (5/16) | 31.25 (5/16) |
| Early stage | 55.28 (68/123) | 8.13 (10/123) | 7.32 (9/123) |
| Later stage | 72.41 (21/29) | 20.69 (6/29) | 17.24 (5/29) |
| Sum | 58.55 (89/152) | 10.53 (16/152) | 9.21 (14/152) |
Real-time PCR
The SYBR-Green qRT-PCR assay was used for miRNA
quantification and was performed on an ABI 7500 Real-Time PCR
instrument (Applied Biosystems). Each reaction was performed in a
final volume of 20 μl that contained 1 μl of cDNA, 0.5 mmol/l of
each primer and 1X SYBR-Green PCR Master Mix (Roche). The
amplification profile was as follows: denaturing at 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec.
Melting curve analysis was performed to validate the specificity of
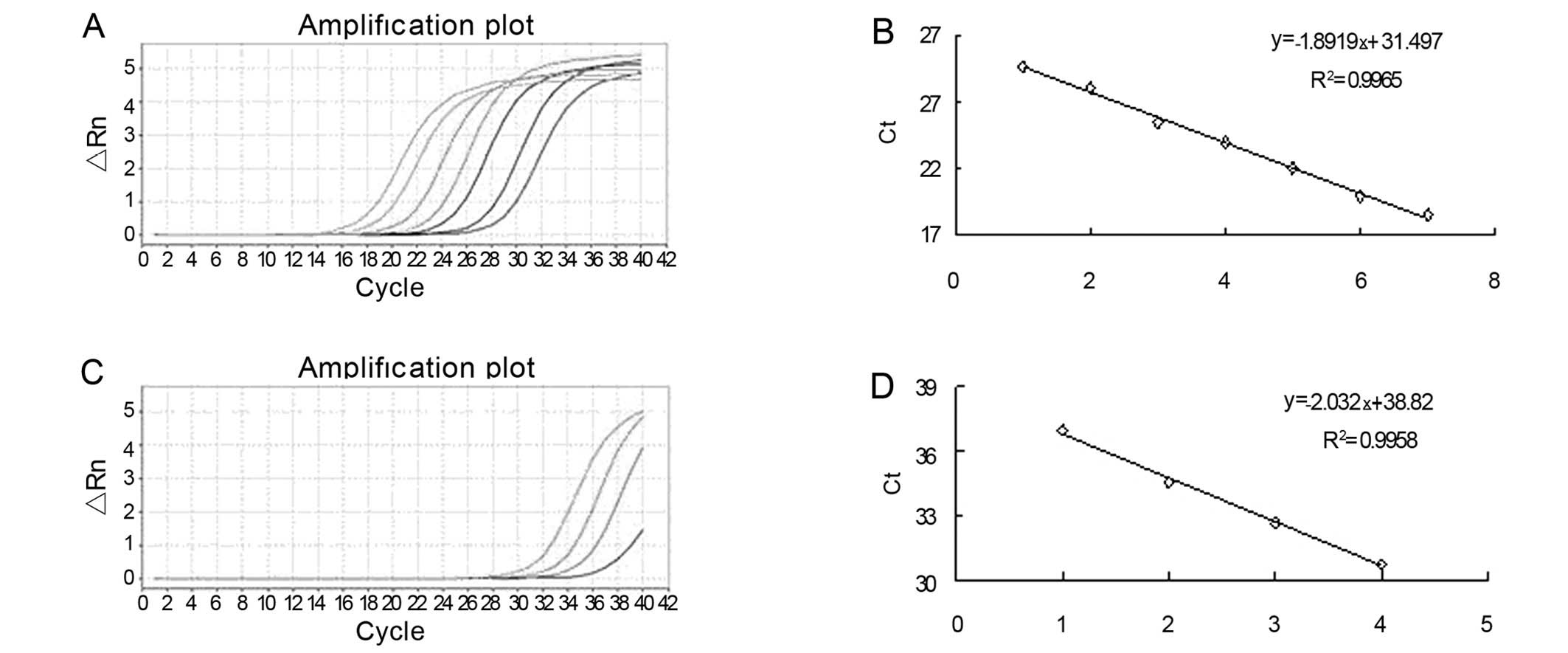
the expected PCR product (Fig.
1). All samples were run in duplicate, including blank controls
without cDNA. The cycle threshold (Ct) is defined as the number of
cycles required for the fluorescent signal to cross the threshold
in qPCR. Serial dilution of cDNA was used to generate the standard
curves. There was excellent linearity between the log of the miRNA
concentration and the cycle threshold (Ct) value (Fig. 2). miR-16: y = −1.8919x + 31.497
(R2=0.9965); miR-181a: y = −2.032x + 38.82
(R2=0.9958).
Statistical analysis
The significance of serum miR-181a levels was
determined by the Mann-Whitney U test, t-test or Kruskal-Wallis H
test as appropriate. Data were expressed as mean values ± standard
deviations (SD), and the relative expression of miR-181a and the
fold-change of expression was determined by the 2−ΔΔCT
method: RQ = 2−ΔΔCT, ΔΔCT = (CT miR-181a -CT miR-16)
tumor - (CT miR-181a - CT miR-16) mean normal (17,18). Receiver operating characteristic
(ROC) curves were established for discriminating between patients
with or without BC. All P-values are two-sided, and values <0.05
were considered statistically significant. All statistical
calculations were performed using SPSS software (version 13.0).
Results
Expression of miR-181a in the serum of BC
and healthy subjects
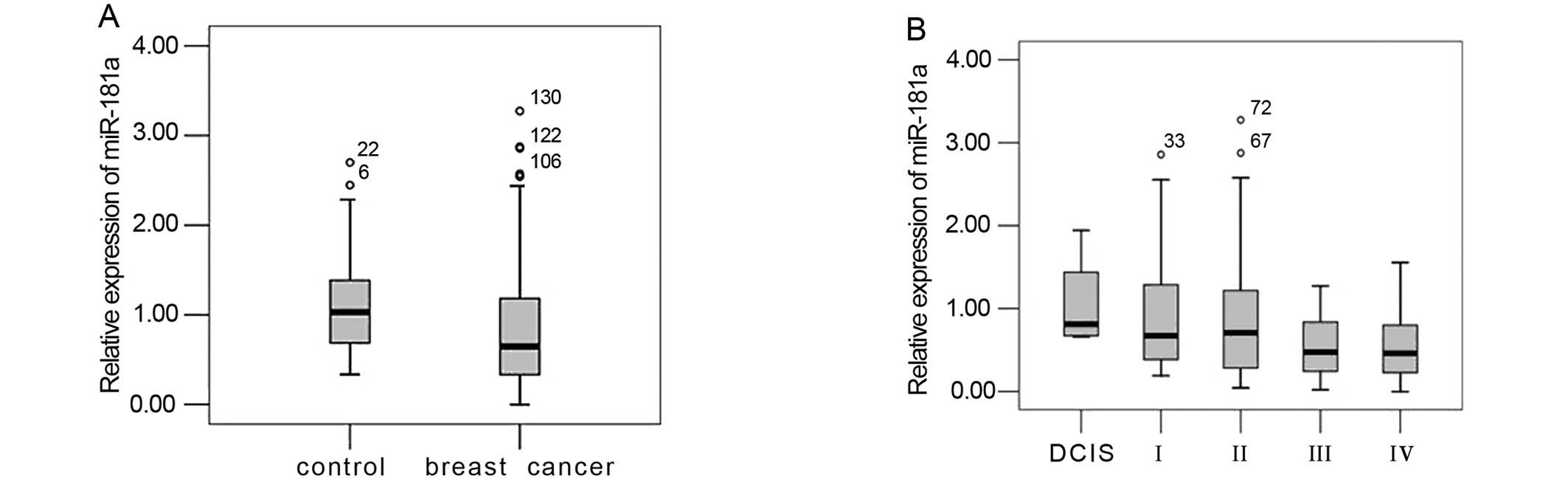
We first analyzed the profiles of serum miR-181a in
a small sample of 10 healthy controls and 10 BC patients. The
median level of miR-181a was significantly lower in patients with
BC than in normal controls (NC): 0.502592±0.44508 vs.
1.412649±0.465748, respectively (P=0.001). The median fold-change
of miR-181a for BC/NC was 0.35578. A similar trend was obtained in
a larger sample of 75 healthy controls and 152 BC patients:
1.0908±0.50821 vs. 0.8385±0.69453 (P=0.001), with the levels of
serum miR-181 significantly downregulated in patients vs. controls
(Fig. 3A). A ROC curve analysis
showed AUC =0.671±0.031. At the cut-off value of 0.7486 for
miR-181a, the sensitivity and specificity for this marker were 70.7
and 59.9%, respectively (Fig.
4).
Comparison of miR-181a with the
conventional BC markers CEA and CA153
To investigate whether miR-181a was a potential
biomarker for the diagnosis of BC, we compared the sensitivity of
miR-181a, CA153 and CEA in each BC stage (in situ, I, II,
III and IV). As shown in Table I,
miR-181a sensitivity for the diagnosis of BC was 58.55%, while the
sensitivity of CA153 and CEA were only 10.53 and 9.21%,
respectively. Serum miR-181a also showed higher sensitivity
(55.28%) for the diagnosis of early stage BC (in situ, I,
II) compared with CA153 (8.13%) and CEA (7.32%) (Table III).
Association of miR-181a expression with
clinical and pathological features
We compared the expression of miR-181a in patients
with different clinical TNM classification stages (Fig. 3B), histological grades and hormone
receptor status. Statistical analysis revealed no significant
correlations between miR-181a and ER or PR status in BC
(P>0.05). Study subject characteristics are summarized in
Table I.
Discussion
CEA and/or CA153 are currently used as serum markers
for BC. Unfortunately, these markers have low sensitivity and
specificity for BC screening. miRNAs are small non-coding RNA
molecules 20–25 nucleotides in length that help regulate a variety
of biological signaling pathways. Some miRNAs are closely related
to the development, invasion, metastasis and other characteristics
of tumors. Calin et al (19) showed that half of the known miRNAs
are in cancer-associated genomic regions/fragile sites, suggesting
they may be involved in the initiation and progression of human
malignancy. miRNAs are thus potential markers for diagnosing and
monitoring patients with BC and may be utilized in novel
therapeutic strategies.
Studies have emerged showing that miRNAs derived
from epithelial tumors are rapidly released into the blood stream
(26,27). Previous reports have shown that
miR-181a deregulation is one of the most commonly reported
miRNA-related events associated with different cancers. To our
knowledge, there are no reports concerning the relationship between
serum miR-181a and BC diagnosis.
Since qRT-PCR is the gold standard for gene
expression quantification and miR-16 is a commonly used internal
reference, we measured serum levels of miR-181a by qRT-PCR and
normalized the levels to those of miR-16. In initial experiments in
a small set of serum samples (n=20), we found that miR-181a
expression was reduced in early BC subjects (0.502592±0.44508)
compared with healthy subjects (1.412649±0.465748) (P=0.001). The
median fold-change of miR-181a for BC/NC was 0.35578, showing that
miR-181a was significantly decreased more than 2-fold. We validated
this finding in a larger sample of 75 healthy controls
(1.0908±0.50821) and 152 BC patients (0.8385±0.69453), showing that
serum miR-181a was significantly downregulated in BC patients
compared to controls (P=0.001). ROC curve analysis showed
AUC=0.671±0.031. At the cut-off value of 0.7486 for miR-181a, the
sensitivity and specificity for this marker was 70.7 and 59.9%,
respectively (Fig. 4). Thus,
serum levels of miR-181a are potential markers for discriminating
BC patients from healthy controls, which is consistent with
previous studies (21–25). Gao et al (21) also reported that miR-181a
(P=0.000) was downregulated in lung carcinoma tissues and showed
potential as a novel diagnostic or prognostic biomarker for non
small cell lung cancer. Our study indicated that miR-181a was a
potential marker for BC. However, previous studies suggest that
miR-181a may be a universal serum marker for several cancers and
not a specific marker for BC. One dissenting report by Zhao et
al (28) found that miR-181a
levels were upregulated in BC patients compared with healthy
controls. The discrepancy might be due to ethnic group, cancer
subtype or individual differences. In our study, all of the
subjects were Han Chinese, and 80.26% of the BCs were stage I and
II, 90.13% were invasive ductal cancer, 69.79% were ER positive and
61.745% were PR positive. It is possible that differences in
clinical characteristics, together with the small sample size in
the present study, might contribute to differences in the findings.
To investigate whether miR-181a could be used as a biomarker in the
diagnosis of BC, we compared the sensitivity of miR-181a, CA153and
CEA in each BC stage (in situ, I, II, III and IV). As shown
in Table III, the sensitivity of
miR-181a for the diagnosis of BC was 58.55%, while the sensitivity
of CA153 and CEA were only 10.53% and 9.21%. We also found that
serum miR-181a had higher sensitivity (55.28%) for the diagnosis of
early stage (in situ, I, II) BC compared with CA153 (8.13%)
and CEA (7.32%).
These findings imply that serum miR-181a may be
useful in detecting certain carcinomas at early stages,
particularly when the individuals are clinically asymptomatic for
extended periods. We speculate that miR-181a may even play a role
in the early events of multistep breast carcinogenesis.
We analyzed miRNA expression in groups of tumors
classified according to TNM staging and other specific
biopathological features, such as hormone receptor expression,
grade and disease stage. Our experimental data did not show a
significant relationship between miR-181a expression, TNM staging
and clinicopathological parameters. These results are consistent
with previous studies which reported that circulating miRNAs did
not show a correlation to different stages, histological subtypes,
or grades of cancer (29–32). These studies also suggested
circulating miRNAs may be potential biomarkers for cancer
detection.
Although extensive research has been conducted on
miRNAs and miRNA-based therapy, there is a limited understanding of
miRNA target genes, of the mechanisms of miRNA-mediated gene
regulation and of exactly how regulation of target genes relates to
tumor progression. The precise targets of miR-181a in BC remain
elusive, and it is unclear how miR-181a may be involved in tumor
formation or how the levels relate to prognosis. We screened for
target genes of miR-181a using the Target Scan program (http://www.targetscan.org) and PicTar (http://pictar). Hundreds of genes were predicted as
targets, many of which were involved in cancers, including K-ras,
OPN, Hoxb5, E2f5, CARD11 and others). Some of the genes had been
validated in previous experiments. In particular, one study
demonstrated that miR-181a had a tumor suppressive effect by
downregulating K-ras in oral squamous cell carcinoma cells and that
miR-181a decreased K-ras protein levels and the luciferase activity
of reporter vectors of the K-ras gene (22). It seems clear that KRAS activation
occurs in the early stage of lung cancer carcinogenesis (33). In our study, the miR-181a
expression level was lower in early stage BC, leading our
speculation that the link between K-ras and miR-181a may play a
role in the etiology of this malignancy. Bhattacharya et al
(34) reported that miR-181a
regulates OPN-dependent metastatic function in hepatocellular
cancer cell lines. Another study (35) showed that miR-181a functioned as a
tumor suppressor by inducing apoptosis, triggering growth
inhibition and inhibiting invasion in glioma cells. If this is also
the case in BC, it raises the possibility that miR-181a could be
exploited as a therapeutic intervention for BC. Some studies
evaluate the usefulness of miRNAs as both targets and tools in
anticancer therapy (36,37). In addition to being used as
targeted therapies and chemotherapy, miRNAs could also alter cancer
cell sensitivity to radiotherapy, as reported by Weidhaas et
al (38).
Although our results are promising, the present
study has several limitations. First, the sample size is small.
Larger samples are needed to validate the feasibility of using
serum miR-181a as a non-invasive diagnostic test for BC. Second,
miR-181a expression has not been observed in benign tumors or in
many different subtypes of BC, so we cannot generalize the findings
to all types of BC. Third, the precise mechanisms by which miR-181a
and its target miRNAs regulate BC progression remain unclear.
Further in vitro and in vivo studies are needed to
determine how miR-181a contributes to breast tumorigenesis.
In conclusion, our data indicate that serum miR-181a
is more sensitive for BC detection than either CEA or CA153 and
thus shows potential for use as a novel diagnostic biomarker for
BC. This study examined the relationship between serum miR-181a
levels and target genes in vivo, which may be exploited as a
minimally invasive treatment for BC. These findings merit further
verification in more extensive studies.
References
|
1.
|
A JemalF BrayMM CenterJ FerlayE WardD
FormanGlobal cancer statisticsCA Cancer J
Clin616990201110.3322/caac.20107
|
|
2.
|
C DeSantisR SiegelP BandiA JemalBreast
cancer statisticsCA Cancer J Clin61409418201110.3322/caac.20134
|
|
3.
|
R SiegelE WardO BrawleyA JemalCancer
statistics: the impact of eliminating socioeconomic and racial
disparities on premature cancer deathsCA Cancer J
Clin61212236201110.3322/caac.2012121685461
|
|
4.
|
S YektaIH ShihDP BartelMicroRNA-directed
cleavage of HOXB8
mRNAScience304594596200410.1126/science.109743415105502
|
|
5.
|
DP BartelMicroRNAs: genomics, biogenesis,
mechanism, and
functionCell116281297200410.1016/S0092-8674(04)00045-514744438
|
|
6.
|
S BaggaJ BrachtS HunterRegulation by let-7
and lin-4 microRNAs results in target mRNA
degradationCell122553563200510.1016/j.cell.2005.07.03116122423
|
|
7.
|
BJ ReinhartFJ SlackM BassonThe
21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis
elegansNature403901906200010.1038/3500260710706289
|
|
8.
|
PH OlsenV AmbrosThe lin-4 regulatory RNA
controls developmental timing in Caenorhabditis elegans by
blocking lin-14 protein synthesis after the initiation of
translationDev Biol216671680199910642801
|
|
9.
|
HH ZhangXJ WangGX LiE YangNM YangDetection
of let-7a microRNA by real-time PCR in gastric carcinomaWorld J
Gastroenterol1328832888200717569129
|
|
10.
|
W WuM SunGM ZouJ ChenMicroRNA and cancer:
Current status and prospectiveInt J
Cancer120953960200710.1002/ijc.2245417163415
|
|
11.
|
R VisoneF PetroccaCM CroceMicro-RNAs in
gastrointestinal and liver
diseaseGastroenterology13518661869200810.1053/j.gastro.2008.10.07419013167
|
|
12.
|
MV IorioP CasaliniC PiovanMicroRNA-205
regulates HER3 in human breast cancerCancer
Res6921952200200910.1158/0008-5472.CAN-08-292019276373
|
|
13.
|
R GarzonCE HeaphyV HavelangeMicroRNA 29b
functions in acute myeloid
leukemiaBlood11453315341200910.1182/blood-2009-03-21193819850741
|
|
14.
|
A CimminoGA CalinM FabbrimiR-15 and miR-16
induce apoptosis by targeting BCL2Proc Natl Acad Sci
USA1021394413949200510.1073/pnas.050665410216166262
|
|
15.
|
KA O’DonnellEA WentzelKI ZellerCV DangJT
Mendellc-Myc-regulated microRNAs modulate E2F1
expressionNature435839843200515944709
|
|
16.
|
CD JohnsonA Esquela-KerscherG StefaniThe
let-7 microRNA represses cell proliferation pathways in human
cellsCancer
Res6777137722200710.1158/0008-5472.CAN-07-108317699775
|
|
17.
|
J BrenneckeA StarkRB RussellSM
CohenPrinciples of microRNA-target recognitionPLoS
Biol3e85200510.1371/journal.pbio.003008515723116
|
|
18.
|
BP LewisCB BurgeDP BartelConserved seed
pairing, often flanked by adenosines, indicates that thousands of
human genes are microRNA
targetsCell1201520200510.1016/j.cell.2004.12.03515652477
|
|
19.
|
GA CalinC SevignaniCD DumitruHuman
microRNA genes are frequently located at fragile sites and genomic
regions involved in cancersProc Natl Acad Sci
USA10129993004200410.1073/pnas.030732310114973191
|
|
20.
|
MA CortezGA CalinMicroRNA identification
in plasma and serum: a new tool to diagnose and monitor
diseasesExpert Opin Biol
Ther9703711200910.1517/1471259090293288919426115
|
|
21.
|
W GaoY YuH CaoDeregulated expression of
miR-21, miR-143 and miR-181a in non small cell lung cancer is
related to clinicopathologic characteristics or patient
prognosisBiomed
Pharmacother64399408201010.1016/j.biopha.2010.01.01820363096
|
|
22.
|
KH ShinSD BaeHS HongRH KimMK KangNH
ParkmiR-181a shows tumor suppressive effect against oral squamous
cell carcinoma cells by downregulating K-rasBiochem Biophys Res
Comm404896902201110.1016/j.bbrc.2010.12.05521167132
|
|
23.
|
SD BhattacharyaJ GarrisonMicro-RNA-181a
regulates osteopontin-dependent metastatic function in
hepatocellular cancer cell
linesSurgery148292297201010.1016/j.surg.2010.05.00720576283
|
|
24.
|
SFM HauslerA KellerPA ChandranWhole
blood-derived miRNA profiles as potential new tools for ovarian
cancer screeningBr J
Cancer103693700201010.1038/sj.bjc.660583320683447
|
|
25.
|
AJ LoweryN MillerRE McNeillMJ
KerinMicroRNAs as prognostic indicators and therapeutic targets:
potential effect on breast cancer managementClin Cancer
Res14360365200810.1158/1078-0432.CCR-07-099218223209
|
|
26.
|
PS MitchellRK ParkinEM KrohCirculating
microRNAs as stable blood-based markers for cancer detectionProc
Natl Acad Sci
USA1051051310518200810.1073/pnas.080454910518663219
|
|
27.
|
Z HuangD HuangS NiZ PengW ShengX DuPlasma
microRNAs are promising novel biomarkers for early detection of
colorectal cancerInt J
Cancer127118126201010.1002/ijc.2500719876917
|
|
28.
|
H ZhaoJ ShenL MedicoA pilot study of
circulating miRNAs as potential biomarkers of early stage breast
cancerPLoS One5e13735201010.1371/journal.pone.001373521060830
|
|
29.
|
EK NgWW ChongH JinDifferential expression
of microRNAs in plasma of patients with colorectal cancer: a
potential marker for colorectal cancer
screeningGut5813751381200910.1136/gut.2008.16781719201770
|
|
30.
|
M TanakaK OikawaM TakanashiDown-regulation
of miR-92 in human plasma is a novel marker for acute leukemia
patientsPLoS One4e5532200910.1371/journal.pone.000553219440243
|
|
31.
|
J WangJ ChenP ChangMicroRNAs in plasma of
pancreatic ductal adenocarcinoma patients as novel blood-based
biomarkers of diseaseCancer Prev
Res2807813200910.1158/1940-6207.CAPR-09-009419723895
|
|
32.
|
KE ResnickH AlderJP HaganDL RichardsonCM
CroceDE CohnThe detection of differentially expressed microRNAs
from the serum of ovarian cancer patients using a novel real-time
PCR platformGynecol
Oncol1125559200910.1016/j.ygyno.2008.08.03618954897
|
|
33.
|
M SagawaY SaitoS FujimuraRI LinnoilaK-ras
point mutation occurs in the early stage of carcinogenesis in lung
cancerBr J Cancer77720723199810.1038/bjc.1998.1189514049
|
|
34.
|
SD BhattacharyaJ GarrisonH
GuoMicro-RNA-181a regulates osteopontin-dependent metastatic
function in hepatocellular cancer cell
linesSurgery148291297201010.1016/j.surg.2010.05.00720576283
|
|
35.
|
L ShiZ ChengJ Zhanghsa-mir-181a and
hsa-mir-181b function as tumor suppressors in human glioma
cellsBrain
Res1236185193200810.1016/j.brainres.2008.07.08518710654
|
|
36.
|
F MengR HensonH Wehbe-JanekK GhoshalST
JacobT PatelMicroRNA-21 regulates expression of the PTEN tumor
suppressor gene in human hepatocellular
cancerGastroenterology133647658200710.1053/j.gastro.2007.05.02217681183
|
|
37.
|
FY MengR HensonM LangInvolvement of human
microRNA in growth and response to chemotherapy in human
cholangiocarcinoma cell
linesGastroenterology13021132129200610.1053/j.gastro.2006.02.05716762633
|
|
38.
|
JB WeidhaasI BabarSM NallurMicroRNAs as
potential agents to alter resistance to cytotoxic anticancer
therapyCancer
Res671111111116200710.1158/0008-5472.CAN-07-285818056433
|