Introduction
Laryngeal carcinoma is a common head and neck
malignancy with high incidence, accounting for approximately 2.4%
of new malignancies worldwide each year (1,2).
Despite extensive application of different treatment modalities,
invasion and metastasis remain the main causes of mortality for
laryngeal squamous cell carcinoma (LSCC) patients. Current
treatments, including surgical intervention, radiation therapy and
chemotherapy, are moderately effective in the early-stage cases,
but are less effective in more advanced cases (3). The mechanisms behind the occurrence
and development of LSCC remain unclear. Therefore, a novel
therapeutic strategy for the treatment of LSCC is urgently
required.
Research over the past years clearly implicates
multiple genetic alterations in the development and progression of
laryngeal carcinoma (4). In
laryngeal carcinoma, alterations in the expression profiles of
several genes have been reported, including those that have
important functions in cell adhesion, signal transduction, cell
differentiation, metastasis, DNA repair and glycosylation (5).
B-cell-specific MLV integration site-1 (Bmi-1) has
been detected in a variety of human carcinoma specimens, from the
pre-cancerous to the advanced stages. In particular, Bmi-1 is
overexpressed in a number of malignancies, including mantle cell
lymphoma (6), B-cell
non-Hodgkin’s lymphoma (7),
myeloid leukemia (8), non-small
cell lung cancer (9), colorectal
cancer (10), as well as breast
and prostate cancers (11,12).
Recently, our group reported that Bmi-1 is also overexpressed in
head and neck cancers (13),
particularly in nasopharyngeal and oral cancers (14,15). Gene chip analysis also showed that
Bmi-l can be used to predict the rapid recurrence of a variety of
cancers following treatment, as well as to predict metastasis
(16), by cooperating with H-RAS
in promoting breast cancer cell proliferation and invasion,
resisting apoptosis, and enhancing transfer capabilities, including
brain metastasis (17). Bmi-1 is
a key protein responsible for cell cycle arrest in response to DNA
damage and plays a role in genetic stability and cancer
susceptibility. It mediates cell cycle arrest at the G1/S, S and
G2/M phase to prevent the processing of damaged DNA (18).
RNA interference (RNAi) is a new technique for the
effective suppression of gene expression (19). RNAi mediated by small interfering
RNA (siRNA) has become an excellent tool for exploring gene
function in basic research and may become a promising strategy for
cancer gene therapy due to its high efficiency and specificity in
blocking target gene expression (20–22). However, to our knowledge, siRNA
targeting Bmi-1 for the treatment of laryngeal carcinoma has not
been reported thus far. In this study, we designed and constructed
siRNA against Bmi-1 and investigated its efficacy in suppressing
Bmi-1 expression in the laryngeal carcinoma cell line, HEp-2, which
expresses high levels of Bmi-1, to develop an antitumor reagent. We
show that the specific downregulation of Bmi-1 by siRNA is
sufficient to inhibit the growth of HEp-2 cells in vitro and
in vivo. Furthermore, we explored the mechanisms through
which Bmi-1 induces oncogenesis and tumor progression.
Materials and methods
Construction of lentiviral vector
We selected four different target sequences specific
to Bmi-1 (Table I) with the
online siRNA tool provided by Invitrogen (http://www.invitrogen.com/rnai), using the Bmi-1
reference sequence (GenBank accession no. NM 005180.5).
Double-stranded DNA containing the interference sequences was
synthesized according to the structure of a pGCSIL-GFP viral vector
(Gikai Gene Company, Shanghai, China) (Fig. 1), and then inserted into a
linearized vector. All the constructs were cloned and sequenced to
confirm their structure. The positive clones were identified as
lentiviral vectors that expressed human Bmi-1 short hairpin RNA
(shRNA), hereinafter designated as pGCSIL/Bmi-1-A, pGCSIL/Bmi-1-B,
pGCSIL/Bmi-1-C and pGCSIL/Bmi-1-D. The four lentiviral vectors were
transfected into HEp-2 cells separately to evaluate their RNAi
effects. We then co-transfected the pGCSIL/Bmi-1-D vector and the
viral packaging system (containing an optimized mixture of two
packaging plasmids, pHelper 1.0 and pHelper 2.0 vector; Gikai Gene
Company) into HEp-2 cells to replicate competent lentivirus. Viral
supernatant was harvested at 48 h after transfection, filtered
through a 0.45-mm cellulose acetate filter, and then frozen at
−70°C. The lentivirus containing the human Bmi-1 shRNA expressing
cassette (pGCSIL/Bmi-1-D) was used as the positive control for
lentivirus production, denoted as Bmi-1-RNAi-LV. The pGCSIL/U6 mock
vector was also packaged and used as the negative control, denoted
as NC-GFP-LV. Viral concentrations were determined by serial
dilution of the concentrated vector stocks in HEp-2 cells in
96-well plates. The number of green fluorescent protein
(GFP)-positive cells was measured at four days post-transduction
under a microscope. The titers averaged 1×108 TU/ml.
 | Table ICore target recognition sequences of
siRNAs from human Bmi-1 cDNA. |
Table I
Core target recognition sequences of
siRNAs from human Bmi-1 cDNA.
| Marker | Species | Target
sequence |
|---|
| Bmi-1-A | Human |
GGAGGAACCTTTAAAGGAT |
| Bmi-1-B | Human |
CCAGAGAGATGGACTGACA |
| Bmi-1-C | Human |
ACAGGAAACAGTATTGTAT |
| Bmi-1-D | Human |
GGAGGAGGTGAATGATAAA |
Cell culture and lentiviral transduction
of HEp-2
For lentiviral transduction, the HEp-2 cells were
plated into six-well plates at a density of 1×106
cells/well. When the cells reached 30% confluence (typically on day
three after subculturing), the medium was replaced with 1 ml of
fresh medium containing lentivirus at a multiplicity of infection
(MOI) of 5 and 6 μg/ml polybrene (Gikai Gene Company). The medium
was replaced with fresh medium on the following day.
Real-time reverse-transcription PCR
(real-time RT-PCR)
The expression of Bmi-1 mRNA in first passage
untransfected or transfected HEp-2 cells was detected, with actin
as the normalizing control. The specific PCR primer sequences of
these genes designed by Beacon Designer 2 software were as follows:
Bmi-1 forward, 5′-TGGCTCGCATTCATTTTCTG-3′ and reverse,
5′-AGTAGTGGTCTGGTCTT GTG-3′; actin forward, 5′-GTG
GACATCCGCAAAGAC-3′ and reverse, 5′-AAAGGGTGT AACGCAACTA-3′.
Transduced HEp-2 cells were trypsinized and harvested at five days
after transduction. Total RNA was isolated using TRIzol reagent
(Invitrogen-Gibco, Grand Island, NY) and cDNA was acquired
according to the M-MLV reverse transcription procedures (Promega,
Madison, WI) with 2 μg of total RNA. Two-step real-time RT-PCR
reactions were carried out using the TP800 Real-Time PCR System
(Takara), which included cycle 1 (1X) at 95°C for 15 sec and cycle
2 (45X) at 95°C for 5 sec, and 60°C for 30 sec. Absorbance data
were collected at the end of every extension (60°C) and graphed
using GraphPad Prism 4.0 software. The real-time PCR data were
analyzed using the 2−ΔΔCt method (Table II).
 | Table IIQuantitative results of Bmi-1 mRNA
expression by real-time PCR. |
Table II
Quantitative results of Bmi-1 mRNA
expression by real-time PCR.
| Vector | ΔΔCt |
2−ΔΔCt |
|---|
| Control | 0.053±0.05 | 0.972±0.124 |
| NC-GFP-Bmi-1 | −0.003±0.15 | 0.999±0.106 |
| Bmi-1-RNAi-LV | −2.663±0.32 | 0.16±0.036 |
Western blot analysis
For western blot analysis, cells were washed three
times with PBS, homogenized in cell lysis buffer (50 mmol/l Tris,
pH 7.8, 150 mmol/l NaCl, 1% nonidet P-40) containing 10 μl/ml
protease inhibitor (Sigma, St. Louis, MO), incubated on ice for 30
min, and then centrifuged for 30 min at 10,000 rpm. The aqueous
supernatant was collected and quantified using Bradford Reagent
(Sigma). Individual samples, each containing 30 μg of protein, were
separated on a precast 12.5% SDS polyacrylamide gel in a Tris-HCl
buffer (pH 7.4) and blotted onto a polyvinylidene fluoride membrane
(Bio-Rad, Hercules, CA). The membrane was incubated at room
temperature for 1 h in PBST buffer (PBS, pH 7.4, 0.05% Triton
X-100) containing 1% (w/v) bovine serum albumin to block
non-specific protein binding sites. After blocking, the blots were
probed with a goat anti-Bmi-1 antibody (ab25791, 1:200; Abcam)
overnight at 4°C, followed by five washes with PBST. The blots were
then incubated with an anti-goat IgG (1:5,000, sc-2020; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA) for 1 h at room temperature.
After washing, the immunoreactive bands were detected using a
chemiluminescent substrate (NEN Life Science, Inc.). Subsequently,
the blots were reprobed with a mouse anti-GAPDH control antibody
(1:6,000, sc32233; Santa Cruz Biotechnology, Inc.).
MTT assay
Transfected and control HEp-2 cells
(5×105/well) were plated into a 96-well plate in
octuple. For six consecutive days, 20 μl of MTT (5 mg/ml) were
added daily to each well, and the cells were incubated at 37°C for
an additional 4 h. The reaction was stopped by lysing the cells
with 200 μl of DMSO for 5 min. Quantification was carried out at
570 nm and expressed as a percentage of the control.
Cell cycle analysis by flow
cytometry
Parental HEp-2 cells and control siRNA-transfected
cells were harvested with 0.125% trypsin, washed twice in PBS,
counted, and fixed overnight with 70% (v/v) ethanol. Cells were
then centrifuged at 1,000 × g for 10 min, resuspended at a
concentration of 1×106 well/ml in PBS, followed by RNase
incubation at 37°C for 30 min. The nuclei of cells were then
stained with propidium iodide (PI) for another 30 min. A total of
10,000 nuclei were examined in a FACSCalibur flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA); DNA histograms were analyzed
by ModFit software.
Apoptosis detected by flow cytometry
Each of the cell lines were seeded in 24-well
culture plates and divided into the following groups: Bmi-1-RNAi-LV
group, negative control group and control group. Each group
contained five culture flasks. When the cells reached 70–80%
confluence after 48 h, the cells were harvested and washed in cold
PBS. Annexin V and PI staining were carried out using the Annexin
V-FITC Apoptosis Detection Kit (BD Biosciences) according to the
manufacturer’s instructions. After 20 min of incubation in the dark
at room temperature, the cells were immediately analyzed by an
Epics Elite ESP flow cytometer (Beckman-Coulter, Inc.).
Migration assay
The migration of the cells was assessed by a
modified Boyden chamber method using microchemotaxis chambers
(Transwell; Corning-Costar, Acton, MA) with a polycarbonate
membrane filter (8.0 μm pore size, 0.33 cm2 growth
area). In all the experiments, both sides of the membrane were
pre-coated with laminin (5 μg/ml) at 37°C for 1 h, and then
air-dried. Medium containing 10% fetal bovine serum (FBS) was added
to the lower chambers as a chemoattractant. The subconfluent HEp-2
cells at passage 1, with or without Bmi-1-RNAi-LV treatment, were
trypsinized and resuspended in 2% FBS to a concentration of
7×105 cells/ml. After 24 h of incubation at 37°C, the
upper surface of the membrane filters was scraped with a cotton
swab to remove non-migrated cells, and then fixed with 4%
paraformaldehyde for 15 min at room temperature. Migrating cells
that passed through the Transwell filter pores and attached on the
lower surface of the filters were counted in five non-overlapping
fields following nuclear staining with hematoxylin. The experiments
were carried out in triplicate.
Wound healing assay
Cells were cultured and grown to 100% confluence. A
clear area was then scraped in the monolayer with a 200-μl pipette
tip. After washing with serum-free RPMI-1640 medium, the cells were
incubated in RPMI-1640 medium containing 5% FBS at 37°C. The
migration of cells into the wounded areas was evaluated at the
indicated times using an inverted microscope, and then
photographed. The healing rate was quantified with measurements of
the gap size during culturing. Three different areas in each assay
were selected to measure the distance of the migrating cells to the
origin of the wound.
Colony formation analysis
Minimal concentrations of FBS (2%) were added to the
medium to measure the cell colony formation ability. Briefly, HEp-2
cells, transfected with anti-Bmi-1 siRNA and negative control RNA,
were plated in six-well plates in RPMI-1640 supplemented with 10%
FBS at a density of 1,000 cells/well. After 4 h, the medium was
changed to FBS (2%)-supplemented medium. After ten days of
incubation at 37°C in an incubator under a humidified 5%
CO2 atmosphere, cells were fixed for 2 min with
methanol, dyed for 20 min with 1% crystal violet, and the numbers
of colonies (at least 50 cells) formed in the whole well were
counted.
Determination of active caspase-3, -8 and
-9
The activities of caspase-3, -8 and -9 were measured
using caspase-3, -8 and -9 Colorimetric Assay kits (Keygen BioTech,
Nanjing, China), respectively, to determine the potential role of
the caspase-3, -8 and -9 proteases in the Bmi-1-knockdown-induced
apoptosis. Briefly, 1×106 cells 72 h after transfection
were lysed at 4°C for 30 min. The supernatant were then transferred
to a clean microfuge tube, and protein concentration was assayed.
Subsequently, 50 μl of 2× reaction buffer and 5 μl of caspase-3, -8
and -9 substrates were added, and the cells were incubated at 37°C
for 4 h in the dark. The absorbance was measured at 405 nm on a
multidetection microplate reader (Synergy™ HT; Bio-Tek, Winooski,
VT). The activities of caspase-3, -8 and -9 were determined by
calculating the ratio of OD 405 nm of the transfected cells to OD
405 nm of the parental cells following the instructions of the
manufacturer.
In vivo tumor model
Six-week-old female athymic nude mice were
subcutaneously injected with 5×106 cells in 0.2 ml of
PBS into the right scapular region. Three groups (five mice each)
of mice were examined. Group 1 was injected with HEp-2 cells alone;
group 2 was injected with HEp-2 cells transfected with scrambled
nucleotide control siRNA (NC-GFP-LV); and group 3 was injected with
HEp-2 cells transfected with Bmi-1 siRNA (Bmi-1-RNAi-LV). Tumor
size was measured every two days using calipers. Tumor volume was
calculated with the formula (L × W2)/2, where L is the
length and W is the width. The mice were sacrificed after three
weeks and the weight of the tumors was measured. The investigation
conforms with the Guide for the Care and Use of Laboratory Animals
published by the US National Institutes of Health (NIH Publication
No. 85-23, revised 1996).
Statistical analysis
SPSS 10.0 was used for statistical analysis. One-way
analysis of variance (ANOVA) was used to analyze the significance
between groups. The LSD method of multiple comparisons with
parental and control vector groups was used when the probability
for ANOVA was statistically significant. A P-value of 0.05 was
considered to indicate a statistically significant difference.
Results
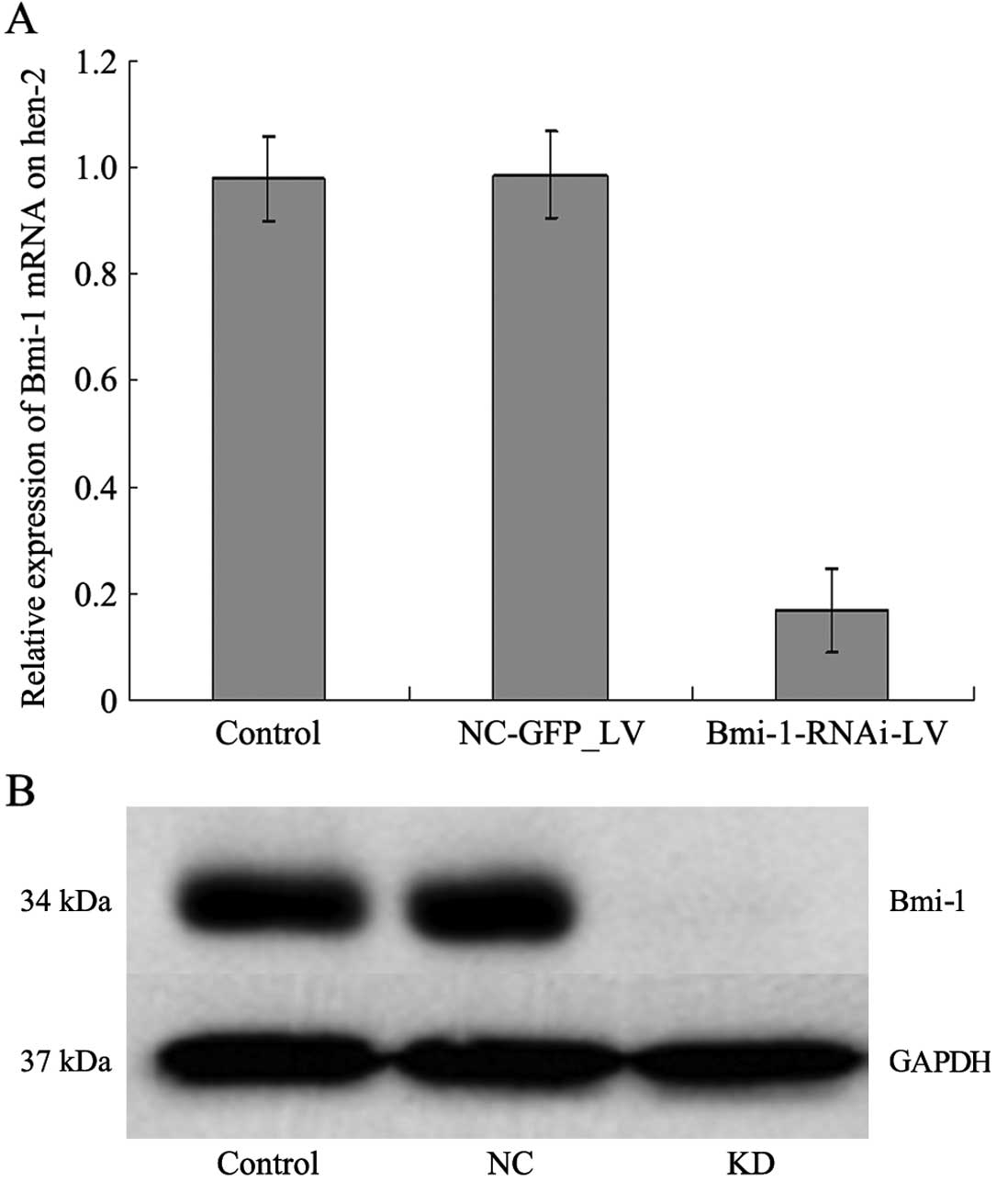
Bmi-1 mRNA and protein expression
Among the four candidate target sequences screened,
the lentiviral vector containing the human Bmi-1 shRNA-expressing
cassette (sequence, 5′-GGAGGAGGTGAATGATAAA-3′) achieved the
greatest efficacy in silencing Bmi-1 expression. This construct was
therefore denoted as Bmi-1-RNAi-LV; the negative control containing
pGCSIL/U6 mock vector only was denoted as NC-GFP-LV. After the
Bmi-1-RNAi-LV construct was transfected into HEp-2 cells, Bmi-1
mRNA expression levels in the transfected cells were compared with
those in the untransfected and control-transfected (NC-GFP-LV)
HEp-2 cells by quantitative RT-PCR. As a result, cells transfected
with Bmi-1-RNAi-LV showed an 80% reduction in the level of Bmi-1
mRNA expression (Fig. 2A). Bmi-1
protein expression was determined by western blot analysis to
further confirm the specificity of Bmi-1-RNAi-LV-mediated Bmi-1
silencing. Bmi-1 protein expression in the cells transfected with
Bmi-1-RNA-LV decreased significantly compared with that of the
control cells (Fig. 2B). These
results indicated that lentivirus-mediated RNAi is an effective
method for modulating Bmi-1 expression in cultured HEp-2 cells.
No significant change (P>0.05) was observed in
the levels of retinoblastoma (Rb) and human telomerase reverse
transcriptase (hTERT) in the cells following transfection with
Bmi-1 siRNA. However, phosphatase and tensin homolog (PTEN) levels
increased significantly (P<0.05) and Bcl-2, p14, cyclin D1 and
homeobox A9 (HOXA9) levels decreased significantly (P<0.05) in
the cells treated with Bmi-1 siRNA (Fig. 3).
Proliferation of transfected HEp-2
cells
The siRNA-transfected cells proliferated at a
significantly lower rate compared to the control and parental
cells, as measured by MTT assay (Fig.
4). Cell growth assay was performed, demonstrating a
significant decrease in cell viability in the Bmi-1-RNAi cells
compared with the control cells in a time-dependent manner, and the
highest inhibitory rate was 51.25±2.86% for the HEp-2 cells on day
six (P<0.05). However, no significant difference in cell
proliferation was observed between the negative control and the
control group in each cell line (P>0.05).
Inhibition of Bmi-1 expression increases
cell apoptosis
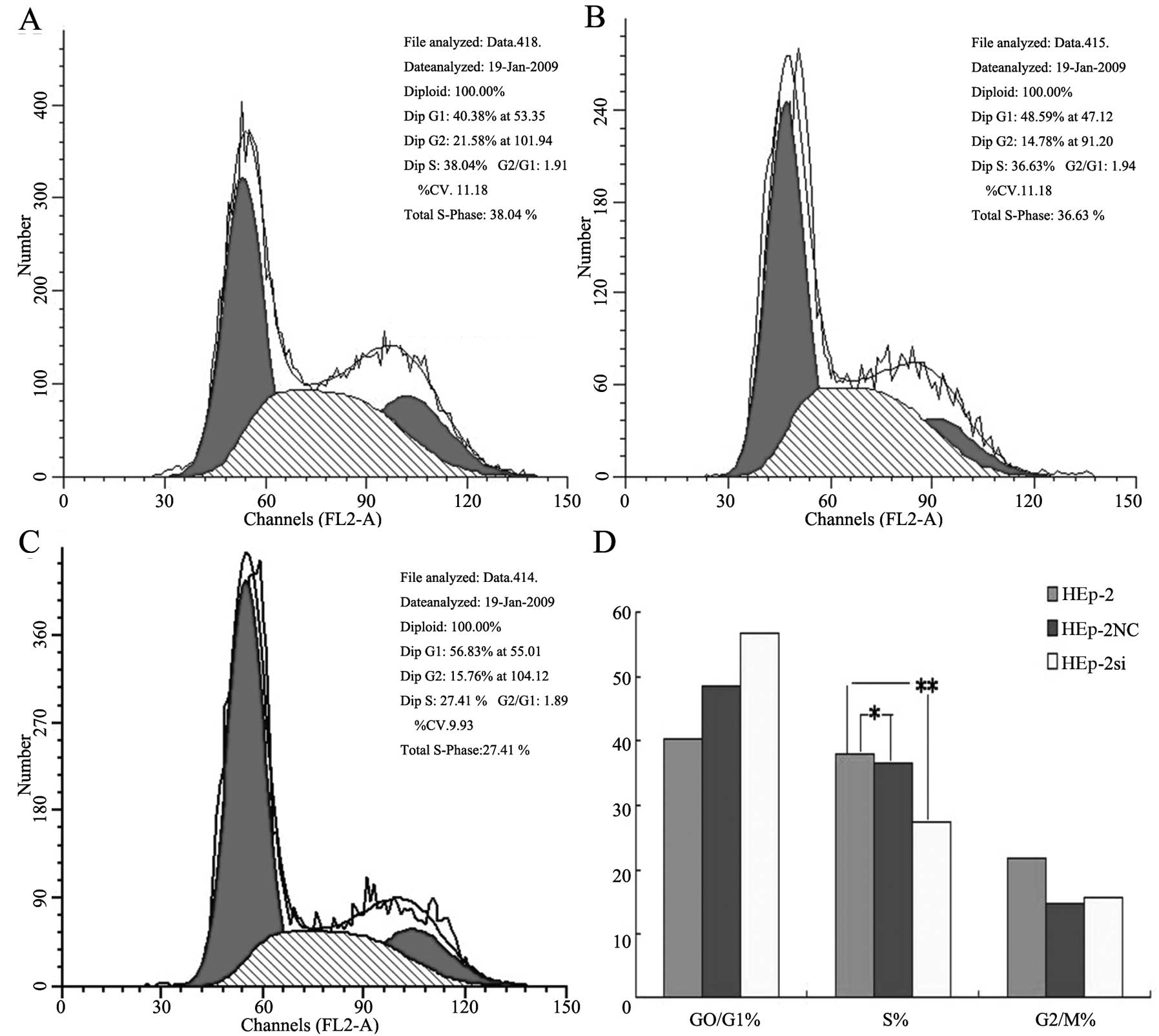
Cell cycle distribution analysis (Fig. 5) indicated that significant
changes were observed in the Bmi-1-RNAi cells, compared with the
control group cells; several cells were blocked in the G0/G1 phase
by 87.5±2.5% (P<0.05) and reduced in the G2/M phase by 3.5±1.3%
(P<0.05), whereas no significant difference was observed in the
cell cycle distribution between the negative control and control
group (P>0.05).
The cells were stained with Annexin V and PI to
further evaluate the induction of apoptosis. The proportion of
Annexin V stained cells to the total Bmi-1 siRNA-transfected cells
was increased (Fig. 6). A small
amount of necrotic cells stained with PI, but not Annexin V, was
also observed. The apoptotic rates in the experimental group after
the cells were treated with RNAi were 21.83, 8.59 and 6.38% in the
Bmi-1-RNAi-LV, NC-GFP-LV group and control group, respectively. The
apoptotic rate of HEp-2 cells increased to 21.83% (P<0.05)
following treatment with Bmi-1-RNAi-LV, whereas no obvious cell
apoptosis was observed in the negatuve conrol and control group
(P>0.05). These results indicated that anti-Bmi-1 siRNA induced
HEp-2 cells apoptosis.
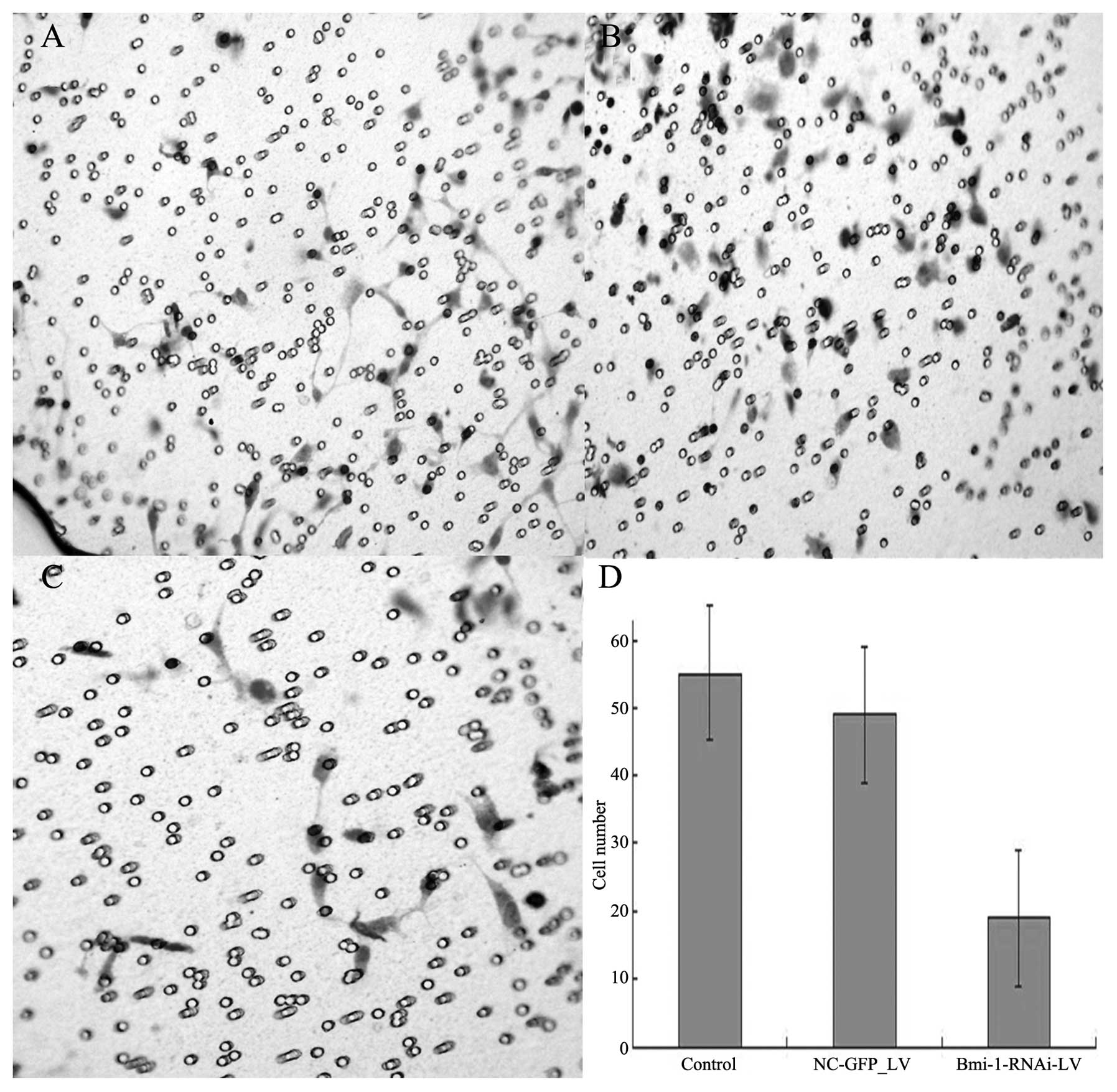
Boyden chamber assay and wound healing
assay
After the cultivation of the cells for 48 h,
invasions of 18.5±3.8, 55.6±11.3, and 49±13.5 cells/field of view
through the porous Transwell membranes were observed for the
Bmi-1-RNAi-LV, control and NC-GFP-LV group, respectively (t=12.62,
P<0.01) (Fig. 7).
HEp-2 cells transfected with Bmi-1 siRNA had a
slower wound-healing rate compared with the cells transfected with
scrambled nucleotide control siRNA (NC-GFP-LV) (0.38±0.046:1,
P<0.05) (Fig. 8).
Activation of caspase-3, -8 and -9 via
inhibition of Bmi-1 expression
The activities of caspase-3, -8 and -9 in the HEp-2
cells transfected with Bmi-1 siRNA or with negative RNA were
examined (Fig. 9) by colorimetric
assay. In addition to executioner caspase-3, initiator caspase-8
and -9 are also important for apoptosis. The ratio of OD 405 nm of
the transfected cells to OD 405 nm of the scrambled nucleotide
control siRNA cells was calculated. The OD value of caspase-3 and
-9 was approximately 4 and that of caspase-8 was approximately
2.
Downregulation of Bmi-1 inhibits the
tumorigenicity of HEp-2 cells in vivo
Tumor volume was measured every two days until the
mice were sacrificed on day 21. The mice in group 3 had
substantially smaller tumors compared to the mice in the other two
groups (Fig. 10). At the time of
sacrifice, the tumor volume for the mice injected with
Bmi-1-RNAi-LV was 274.68±154.79 mm3 compared with
664.31±246.30 mm3 for the mice injected with HEp-2 cells
(P=0.003) or 643.67±270.12 mm3 for the mice in the
negative control group (P=0.008). In addition, the mean tumor
weight at the end of the experiment was significantly lower in the
Bmi-1-RNAi-LV group (0.58±0.12 g) compared to that in the negative
control (1.15±0.19 g, P=0.005) or the control group (0.98±0.20 g,
P=0.007).
TUNEL assay
TUNEL assay showed that groups 1 and 2 had scattered
apoptotic cells, whereas group 3 showed a large number of apoptotic
cells (x400) compared with the HEp-2 and negative control group
(P<0.05) (Fig. 11).
Western blot analysis
The western blot analysis results showed the
significant inhibitory effect of Bmi-1 lentiviral-mediated RNAi on
Bmi-1, cyclin D1 and p14 protein expression. Bmi-1, cyclin D1 and
p14 protein expression in the HEp-2 cells transfected with Bmi-1
lentiviral-mediated RNAi was significantly decreased (Fig. 12).
Discussion
Laryngeal carcinoma accounts for 1 to 5% of all
tumors, and is the second most common cancer of the respiratory
system (next to lung cancer). It is also the second most common
epithelial malignancy in the head and neck region (next to
nasopharyngeal cancer), and accounts for 7.9 to 35% of all ear,
nose and throat (ENT) malignancies (23). If treated early, the prognosis of
laryngeal carcinoma is good, with a five-year survival rate of 90%
and the possibility of retaining laryngeal functions (24). However, the prognosis of
late-stage laryngeal carcinoma is poor due to tumor invasion and
metastasis. The five-year survival rate is below 60%, and patients
usually succumb to the diseased due to local recurrence, neck lymph
node recurrence, or distant metastasis (25,26). The molecular mechanisms behind the
development and progression of laryngeal carcinoma are poorly
understood.
Bmi-1 plays a critical role in the immortalization
of normal epithelial cells and early malignant transformation, as
well as in the maintenance of the self-renewal of stem cells.
Polycomb group (PcG) proteins play an important role
as epigenetic gene silencers during development (27). In addition to their role in
development, these proteins have been reported to be overexpressed
in a variety of human cancers, such as malignant lymphomas and
various solid tumors (28). In
particular, Bmi-1 is overexpressed in a number of malignancies,
such as mantle cell lymphoma (6),
B-cell non-Hodgkin’s lymphoma (9), myeloid leukemia (8), non-small cell lung cancer (29), colorectal cancer (10), as well as breast and prostate
cancers (16,12). In addition, we recently reported
that Bmi-1 is also overexpressed in laryngeal carcinoma (30). Our present study demonstrates that
a significant difference in Bmi-1 expression at both the protein
and mRNA level exists between laryngeal carcinoma cells and the
adjacent normal laryngeal tissue.
In the present study, we designed and constructed a
siRNA recombinant expression vector targeting Bmi-1 to investigate
its effect on laryngeal carcinoma cell proliferation. Cell
proliferation was significantly inhibited after the cells were
treated with RNAi. We demonstrate that lentiviruses can efficiently
deliver Bmi-1 siRNAs into HEp-2 cells and that the transfection of
Bmi-1 shRNA virtually eliminates Bmi-1 protein expression,
indicating that siRNA sequences targeting Bmi-1 may be a potential
therapeutic strategy for the gene therapy of LSCC. Previous studies
have reported that the downregulation of Bmi-1 by the
adenovirus-mediated delivery of Bmi-1 siRNA reduces the invasion,
angiogenesis and metastasis of lung cancer cells (31). Moreover, the adenovirus-mediated
expression of Bmi-1 siRNA in human lung xenografts in vivo
has been shown to result in increased immunostaining of Fas, Fas-L,
cleaved Bid and TIMP-3, which in turn promotes apoptosis in lung
cancer (32).
The initiation and progression of laryngeal
carcinoma involve a series of genetic events, including the
activation of oncogenes and the inactivation of tumor suppressors
(33).
The regulatory mechanism of the PcG proteins relies
upon epigenetic modifications of specific histone tails that are
inherited through cell division (34,35). Bmi-1 is a member of the PcG
family, thus, Bmi-1 overexpression can repress the p16Ink4a
(CDKN2A) and p19Arf targets (10,15). In the absence of p16Ink4a, the
cyclin D/Cdk4/6 complex can phosphorylate pRb, allowing the
E2F-dependent transcription, which leads to cell cycle progression
and DNA synthesis. Bmi-1-deficient mouse embryonic fibroblasts
overexpress the INK4a/ARF locus-encoded genes, CDKN2A and p19ARF
(mouse homologue of human p14ARF), and undergo premature senescence
in culture (36).
Previous studies using in vivo mouse and
in vitro cell culture models have shown that Bmi-1 regulates
the expression of the INK4A/ARF locus, which encodes two
important tumor suppressors, namely, p16INK4A and p19ARF (p14ARF in
humans) (36,37). Bmi-1 can potentially regulate the
p16-pRb and p53-p21 pathways of senescence by downregulating
p16INK4A and p19ARF (38).
PTEN regulates multiple signal transduction
pathways, involved in the inhibition of proliferation and
migration. PTEN exerts a variety of tumor suppressive effects, on
various downstream signaling pathways, such as the pI3K/AKT
pathway, and regulates gene transcription and protein translation,
inducing apoptosis and promoting cell cycle arrest.
Bmi-1 expression is regulated by the cell cycle and
the highest expression is observed in the G2/M phase (39). Our data showed that suppressing
the expression of Bmi-1 in HEp-2 cells significantly inhibits cell
proliferation and induces spontaneous cell apoptosis and G0/G1 cell
cycle arrest. These results are in accordance with thos form
previous studies on esophageal carcinoma cells (40) and cervical carcinoma cells. These
data suggest that Bmi-1 plays an important role in the development
of laryngeal carcinoma.
The suppression of Bmi-1 expression by
lentiviral-mediated RNAi, induces cell cycle arrest at the G1 phase
of the cell cycle and promotes apoptosis. Thus, this method of
suppressing Bmi-1 expression which leads to cell growth inhibition
and the induction of apoptosis, in turn reduces the number of
proliferating cells, abnormal proliferation and suppresses the
growth of laryngeal carcinoma cells; thus it has a significant
therapeutic effect.
In the present study, BALB/c nude mice were injected
with HEp-2 laryngeal carcinoma cells transfected with Bmi-1 siRNA
(Bmi-1-RNAi-LV). The results showed that both the average tumor
weight and volume were significantly lower in the
Bmi-1-RNAi-LV-treated group than those in the control group. The
study by Abdouh showed that downregulating the expression of Bmi-1
by lentiviral shRNA led to the decline of colony formation and
glioma growth both in vitro and in vivo (41), which is consistent with our
findings.
Our results demonstrated that Bmi-1 gene silencing
cannot eliminate the tumor completely, suggesting that Bmi-1 is not
the only protease involved in the invasion and growth of LSCC. The
functions of Bmi-1 associated with stem cells in head and neck
cancer have not yet been fully elucidated. It is also important to
investigate the associated genes and signal transduction pathways
in order to effectively contribute to the gene targeted therapy of
head and neck tumors, thus increasing the possibility of developing
a more potent therapy for head and neck tumors.
We directly approached the important role of Bmi-1
in the invasion and growth of LSCC by analyzing the effects of
stable Bmi-1 silencing by recombinant lentivirus-mediated shRNA. To
our knowledge, we demonstrate for the first time the inhibition of
Bmi-1 expression by lentivirus-mediated shRNA, suggesting that
Bmi-1 may be a candidate gene for the gene targeted therapy of
human LSCC.
Acknowledgements
The present study was supported by a grant from the
Science and Technology Project of Shanxi Province (no.
2010D40).
References
|
1
|
Marioni G, Marchese-Ragona R, Cartei G,
Marchese F and Staffieri A: Current opinion in diagnosis and
treatment of laryngeal carcinoma. Cancer Treat Rev. 32:504–515.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Papadas TA, Alexopoulos EC, Mallis A,
Jelastopulu E, Mastronikolis NS and Goumas P: Survival after
laryngectomy: a review of 133 patients with laryngeal carcinoma.
Eur Arch Otorhinolaryngol. 267:1095–1101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun YN, Liu M, Yang B, Li B and Lu J: Role
of siRNA silencing of MMP-2 gene on invasion and growth of
laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol.
265:1385–1391. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao JX and Xie XL: Regulation of gene
expression in laryngeal carcinama by microRNAs. Int J Pathol Clin
Med. 32:222–225. 2012.
|
|
5
|
Srivastava R: Apoptosis, Cell Signaling,
and Human Diseases Molecular Mechanisms. 1. Humana Press; Totowa:
2007
|
|
6
|
Bea S, Tort F, Pinyol M, et al: BMI-1 gene
amplification and overexpression in hematological malignancies
occur mainly in mantle cell lymphomas. Cancer Res. 61:2409–2412.
2001.
|
|
7
|
Bhattacharyya J, Mihara K, Ohtsubo M, et
al: Overexpression of BMI-1 correlates with drug resistance in
B-cell lymphoma cells through the stabilization of survivin
expression. Cancer Sci. 103:34–41. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sawa M, Yamamoto K, Yokozawa T, et al:
BMI-1 is highly expressed in M0-subtype acute myeloid leukemia. Int
J Hematol. 82:42–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kimura M, Takenobu H, Akita N, et al: Bmi1
regulates cell fate via tumor suppressor WWOX repression in
small-cell lung cancer cells. Cancer Sci. 102:983–990. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim JH, Yoon SY, Kim CN, et al: The Bmi-1
oncoprotein is overexpressed in human colorectal cancer and
correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett.
203:217–224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Glinsky GV, Berezovska O and Glinskii AB:
Microarray analysis identifies a death-from-cancer signature
predicting therapy failure in patients with multiple types of
cancer. J Clin Invest. 115:1503–1521. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song W, Tao K, Li H, et al: Bmi1 is
related to proliferation, survival and poor prognosis in pancreatic
cancer. Cancer Sci. 101:1754–1760. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yao X, Wang X, Zhang S and Zhu H: Effects
of Bmi-1 RNAi gene on laryngeal carcinoma Hep-2 cells. Lin Chung Er
Bi Yan Hou Tou Jing Wai Ke Za Zhi. 26:550–557. 2012.(In
Chinese).
|
|
14
|
Kang MK, Kim RH, Kim SJ, et al: Elevated
Bmi-1 expression is associated with dysplastic cell transformation
during oral carcinogenesis and is required for cancer cell
replication and survival. Br J Cancer. 96:126–133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song LB, Zeng MS, Liao WT, et al: Bmi-1 is
a novel molecular marker of nasopharyngeal carcinoma progression
and immortalizes primary human nasopharyngeal epithelial cells.
Cancer Res. 66:6225–6232. 2006. View Article : Google Scholar
|
|
16
|
Glinsky GV: Death-from-cancer signatures
and stem cell contribution to metastatic cancer. Cell Cycle.
4:1171–1175. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hoenerhoff MJ, Chu I, Barkan D, et al:
BMI1 cooperates with H-RAS to induce an aggressive breast cancer
phenotype with brain metastases. Oncogene. 28:3022–3032. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu CR, Lee SS, Ho C, et al: Bmi1 functions
as an oncogene independent of Ink4A/Arf repression in hepatic
carcinogenesis. Mol Cancer Res. 7:1937–1945. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Elbashir SM, Harborth J, Lendeckel W, et
al: Duplexes of 21-nucleotide RNAs mediate RNA interference in
cultured mammalian cells. Nature. 411:494–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mittal V: Improving the efficiency of RNA
interference in mammals. Nat Rev Genet. 5:355–365. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moffat J and Sabatini DM: Building
mammalian signalling pathways with RNAi screens. Nat Rev Mol Cell
Biol. 7:177–187. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim DH and Rossi JJ: Strategies for
silencing human diseaseusing RNA interference. Nat Rev Genet.
8:173–184. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang XZ and Wang JB: Practical
otolaryngology. People’s Medical Publishing House; Beijing: pp.
502–503. 1998
|
|
24
|
Tu Gy, Tang PZ and Jia CY:
Horizontovertical laryngectomy for supraglottic carcinoma.
Otolaryngol Head Neck Surg. 117:280–286. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sessions DG, Lenox J, Spector GJ, et al:
Management of T3N0M0 glottic carcinoma: therapeutic outcomes.
Laryngoscope. 112:1281–1288. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu YZ, Tang PZ, Qi YF and Xu Z: The
management of stomal recurrence after laryngectomy. Zhonghua Er Bi
Yan Hou Ke Za Zhi. 37:380–383. 2002.(In Chinese).
|
|
27
|
Ringrose L and Paro R: Epigenetic
regulation of cellular memory by the Polycomb and Trithorax group
proteins. Annu Rev Genet. 38:413–443. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Raaphorst FM: Deregulated expression of
Polycomb-group oncogenes in human malignant lymphomas and
epithelial tumors. Hum Mol Genet. 14:R93–R100. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kikuchi J, Kinoshita I, Shimizu Y, et al:
Distinctive expression of the polycomb group proteins Bmi1 polycomb
ring finger oncogene and enhancer of zeste homolog 2 in nonsmall
cell lung cancers and their clinical and clinicopathologic
significance. Cancer. 116:3015–3024. 2010. View Article : Google Scholar
|
|
30
|
Yao XB, Wang XX, Zhang SQ, Yan LY and Zhu
HL: Association of Bmi-1 mRNA overexpression with differentiation,
metastasis and prognosis of laryngocarcinoma. Xi’An Jiaotong Daxue
Xuebao. 32:246–249. 2011.(In Chinese).
|
|
31
|
Wang YX, Wei YM, Wang XH and Zhu XY:
Construction of eukaryotic expression vector targeting Bmi-1 and
its influences on proliferation of SW480 cells. Xian Dai Sheng Wu
Yi Xue Jin Zhan. 9:2270–2272. 2009.(In Chinese).
|
|
32
|
Wang ZX, Lu BB, Yang JS, Wang KM and De W:
Adenovirus-mediated siRNA targeting c-Met inhibits proliferation
and invasion of small-cell lung cancer (SCLC) cells. J Surg Res.
171:127–135. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li PH, Wang HJ, Liu W and Xu XG: PRDM1
gene expression in laryngeal carcinoma and its clinical
significance. Head Neck Surg. 24:179–180. 2010.
|
|
34
|
Pirrotta V: Polycombing the genome: PcG,
trxG, and chromatin silencing. Cell. 93:333–336. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Orlando V: Polycomb, epigenomes, and
control of cell identity. Cell. 112:599–606. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jacobs JJ, Kieboom K, Marino S, Depinho RA
and van Lohuizen M: The oncogene and Polycomb-group gene bmi-1
regulates cell proliferation and senescence through the ink4a
locus. Nature. 397:164–168. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Itahana K, Zou Y, Itahana Y, et al:
Control of the replicative life span of human fibroblasts by p16
and the polycomb protein Bmi-1. Mol Cell Biol. 23:389–401. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dimri GP: What has senescence got to do
with cancer? Cancer Cell. 7:505–512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu Z, Liu H, Lv X, Liu Y, Li S and Li H:
Knockdown of the Bmi-1 oncogene inhibits cell proliferation and
induces cell apoptosis and is involved in the decrease of Akt
phosphorylation in the human breast carcinoma cell line MCF-7.
Oncol Rep. 25:409–418. 2011.PubMed/NCBI
|
|
40
|
Liu WL, Guo XZ, Zhang LJ, et al:
Prognostic relevance of Bmi-1 expression and autoantibodies in
esophageal squamous cell carcinoma. BMC Cancer. 10:4672010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Abdouh M, Facchino S, Chatoo W, et al:
BMI1 sustains human glioblastoma multiforme stem cell renewal. J
Neurosci. 29:8884–8896. 2009. View Article : Google Scholar : PubMed/NCBI
|