Introduction
Ovarian cancer is the leading cause of mortality
from gynecological malignancies in the Western world, and the high
case-fatality ratio associated with this malignancy may be
attributed in part to its vague and non-specific symptoms (1). Although ovarian cancer confined to
the ovaries has a 5-year survival of 92%, the majority of women
with this type of cancer are diagnosed at an advanced stage (67%)
and have a 5-year survival of only 30% (2). Due to the high mortality rate
associated with ovarian cancer, a number of studies have been
carried out in an attemtp to discover novel therapeutic approaches
(3). Several biomarkers have been
identified, such as human epididymis protein 4 (HE4) (4), transthyretin (5) and cancer antigen 125 (CA125)
(6). However, these biomarkers
are not sufficient for the early diagnosis of ovarian cancer.
Therefore, there is an urgent need to discover novel therapeutic
targets and to explore their molecular mechanisms of action in
ovarian cancer.
Ras-association domain family 1, isoform A (RASSF1A)
is a tumor suppressor gene which is usually inactivated in human
cancers (7). Donninger et
al (8) discovered that
RASSF1A is pro-apoptotic and may serve to integrate pro-growth and
pro-death signaling pathways. Kassler et al (9) observed a strong correlation between
RASSF1A expression and the development of resistance to Taxol in
ovarian cancer. In addition, RASSF1A has been shown to be closely
associated with the progression of prostate (10), thyroid (11), blood-based breast (12) and primary non-small cell lung
cancer (13). Furthermore,
RASSF1A has been shown to suppress melanoma development by encoding
a microtubule-associated protein which may regulate cell
proliferation, migration and apoptosis (14). Besides, RASSF1A interacts with
effectors of apoptotic pathways, including macrophage stimulating
protein (MST)1/2 and modulator of apoptosis 1 (MOAP1), resulting in
the induction of apoptosis (15).
It has been demonstrated that RASSF1A expression inhibits the
tumorigenic potential of A375 cells in nude mice, which correlates
with decreased cell proliferation and increased apoptosis (14). It has also been reported that
RASSF1A suppresses melanoma development by modulating apoptosis and
cell cycle progression (14);
however, the underlying mechanisms have not yet been elucidated.
Thus, RASSF1A possesses great potential as a therapeutic target in
human cancer.
Furthermore, the development of computational tools
and resources for genetic analysis has accelerated rapidly over the
past decades (16). A number of
researchers have utilized bioinformatics approaches to explore the
molecular mechanisms of diseases and have discovered novel
biomarkers (17,18). In this study, the expression of
RASSF1A in 4 ovarian cancer cell lines and in differently treated
SKOV-3 cells was detected. The effects of RASSF1A on cell
morphology, structure, apoptosis and proliferation were also
examined. Moreover, the mechanisms responsible for these effects
were explored using a bioinformatics approach. Our data suggest
that RASSF1A is a novel biomarker for ovarian cancer and may aid in
the early detection, prevention and treatment of the disease.
Materials and methods
Ovarian tumor tissuses and cell line
collection
A total of 47 patients with malignant ovarian
epithelial tumors (43 serous cystadenocarcinoma cases and 4
borderline cases), who underwent surgery (one-time treatment) at
the hospital of China Medical University Graduate School from
September 2005 to January 2008, were recruited for sample
collection. Among these 47 cases, 5 cases were at clinical stage I,
4 cases at stage II, 37 cases at stage III and 1 case at stage IV.
Ovarian samples from 10 patients who underwent an ovarian
anatomical examination or prophylactic ovariotomy were collected as
control group. All the ovarian samples were collected under sterile
conditions and pathologically validated. After being excised from
the human body, the ovarian samples were quickly frozen in liquid
nitrogen and transferred to −80°C freezer. The ovarian epithelial
cancer cells (HO8910, HO8910PM, SKOV-3 and OVCAR-3) were purchased
from the Cell Bank of the Chinese Academy of Sciences.
All patients provided written informed consent, and
the Ethics Committee of China Medical University Graduate School
(Shenyang, China) approved all aspects of this study.
Quantitative reverse
transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the samples using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the protocol
was performed according to the manual provided by the manufacturer.
cDNA was synthesized from 2 μg of total RNA in a 10-μl reaction
system containing 2 μl MgCl2, 1 μl 10X RT buffer, 3.75
μl RNase-Free H2O, 1 μl dNTP mixture (10 mM), RNase
inhibitor 0.25 μl, AMV reverse transcriptase 0.5 μl, random 9 mers
0.5 μl and RNA 1 μl.
The primers for RASSF1A and β-actin were designed as
follows and synthetized by Saibaisheng Biological Engineering Co.,
Ltd., Beijing, China: RASSF1A forward, 5′-CTTCATCTGGGGCGTCGTG-3′
and reverse, 5′-GCATCCTTGGGCAGGTAAAA-3′. The target fragment length
for RASSF1A was 420 bp. The primers for β-actin were as follows:
forward, 5′-TGCGTGACATTAAGGAGAAGC-3′ and reverse,
5′-GAAGGTGGACAGCGAGGC-3′. The target fragment length for β-actin
was 431 bp.
PCR was carried out according using the Takara RNA
PCR kit version 3.00 (Takara Bio, Dalian, China) in a 20-μl
reaction volume (cDNA template 4 μl, 5X PCR buffer 4 μl, distilled
water 7.9 μl, Ex Taq Hot Start (HS) 0.1 μl, sense primer 2 μl and
reverse primer 2 μl) with the following protocol: 94°C for 30 sec,
57°C for 30 sec and 72°C for 40 sec for 30 cycles. All the PCR
reactions were conducted using a Biometra PCR system (Biometra,
Göttingen, Germany). PCR products were separated on 1.5%
Tris-borate EDTA agarose gels with a100-bp DNA Ladder Marker
(Dalian Bao Biological Engineering Co., Dalian, China), hybridized
and visualized using an electrophoresis gel imaging system
(ChemiImager 5500; Alpha Innotech, San Leandro, CA, USA).
Transfection in vitro
The SKOV-3 cells were cultured in medium which
contained 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal
bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA) in a
humidified atmosphere of 5% CO2. The exponential cells
were then collected and seeded on a 6-well cell culture cluster at
a concentration of 5×105 cells/well and allowed to grow
overnight. When grown to 85–90%, the SKOV-3 cells were transfected
with green fluorescent protein (GFP)-labeled adenovirus with or
without RASSF1A (Baisai Biological Co., Beijing, China).
Untransfected SKOV-3 cells were used as controls.
Quantification of RASSF1A expression in
SKOV-3 cells
The SKOV-3 cells were collected after 24–48 h of
transfection, and the differential expression of RASSF1A was
detected by qRT-PCR. The protocol of RT-PCR was similar to the one
described above.
Analysis of cell morphology, structure
and apoptosis
A fluorescence microscope (BX5O; Olympus, Tokyo,
Japan) was used to analyze the morphology of the SKOV-3 cells
following transfection and observe the location of fluorescence. A
transmission electron microscope (TEM; JEM 1010, Jeol, Tokyo,
Japan) was then used to observe the structure of the transfected
and untransfected SKOV-3 cells. Furthermore, a flow cytometer (TOA
Medical Electronics Co., Ltd., Kobe, Japan) was utilized to analyze
the cycle and apoptosis of SKOV-3 cells treated with adenovirus
with or without RASSF1A. The protocol involved digestion with 0.25%
pancreatin, washing with phosphate-buffered saline (PBS) buffer,
fixation with 70% ethanol at 20°C for 10 h, and dying with
propidium iodide for 30 min. The cells, including SKOV-3 cells
transfected with adenovirus with or without RASSF1A and
untransfected SKOV-3 cells were analyzed by flow cytometry using an
Apoptosis detection kit (KGI Biotechnology Development Co., Ltd.,
Nanjing, China). Each group had 10 duplications and the results
were analyzed using multifunctional software systems.
Preparation and quantification of protein
expression
The SKOV-3 cells, including untreated cells, and
those transfected with adenovirus with or without RASSF1A, were
cultured in 6-well plates and each type of cell was allocated 2
wells. The cells were washed with 1X PBS buffer (pH 7.4) 3 times
and digested by 0.25% pancreatin. Pancreatin was removed by
centrifugation at 2,000 rpm for 5 min (high-speed refrigerated
centrifuge 31K5C; Sigma-Aldrich) followed by the addition of 100 μl
RIPA buffer which contained 1X PBS buffer, 1% NP-40, 0.5% sodium
deoxycholate, 0.1% sodium dodecyl sulfate (SDS) and protease
inhibitor, such as phenylmethylsulfonyl fluoride (PMSF; Takara
Bio). Following ultrasonic dispersion for 30 sec, the samples were
incubated on ice for 30 min. The supernatant was transferred to a
new tube after centrifugation at 10,000 × g for 10 min at 4°C
(high-speed refrigerated centrifuge 31K5C; Sigma-Aldrich) and then
the centrifugation was repeated once. In addition, the proteins
were quantified using the Bradford method, as previously described
(19).
Western blot analysis
The protein samples were separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by
transfer onto polyvinylidene fluoride (PVDF) membranes (Millipore,
Billerica, MA, USA). The PVDF membranes were then blocked overnight
with 0.1% Tween-20, 1X PBS and 5% non-fat milk. The membranes were
then washed with buffer containing 0.1% Tween-20 and 1X PBS for 5
min. Following incubation with rabbit anti-human Livin antibody
(1:400; Imgenex Corp, San Diego, CA, USA) or antibodies against
cyclin D1, survivin, p27, caspase-3 and GAPDH (Wuhan Boshide
Bioengineering Co., Ltd., Wuhan, China) for 2 h at room
temperature, the membranes were hybridized with goat anti-rabbit
antibodies (Zhongshan Golden Bridge Biotechnology Co., Beijing,
China) for 1 h and washed with 0.1% Tween-20 and 1X PBS 3 times for
5 min each. The membranes were subsequently treated with alkaline
phosphatase for 5–30 min and the results were analyzed using Scion
Image software (Scion Corp., Frederick, MD, USA) by calculating the
target protein/GAPDH ratio, as previously described (20).
Statistical analysis
Statistical analysis was carried out using SPSS 12.0
software. P-values <0.05 and <0.01 were considered to idicate
statistically significant and highly statistically significant
differences, respectively.
Derivation of genetic data
The gene expression profile data of GSE14407
(21) was downloaded from a
public functional genomics data repository, the Gene Expression
Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. A total of
24 specimens, including 12 healthy ovarian surface epithelial
samples (OSE) and 12 laser capture microdissected serous ovarian
cancer epithelial samples (CEPI), were available based on the
Affymetrix 3′ expression array.
Detection of the expression of
RASSF1A
The derived genetic data were analyzed using
Spotfire DecisionSite software (http://spotfire.tibco.com; TIBCO Software Inc., Palo
Alto, CA, USA) (22). The
differentially expressed genes (DEGs) were identified with a fold
change value >3 and a value of P<0.05 (Student’s t-test).
Furthermore, we detected whether RASSF1A was in the list of
DEGs.
Construction of protein-protein
interaction (PPI) network
The online database resource Search Tool for the
Retrieval of Interacting Genes (STRING) provides uniquely
comprehensive coverage and ease of access to both experimental and
predicted interaction information (23). In the present study, the
interactions between RASSF1A and other genes were derived based on
STRING and the associations with a correlation coefficient >0.4
were identified as PPIs. The PPI network was constructed and
visualized using Cytoscape software, as previously described
(24). Cytoscape is an open
source software project for integrating biomolecular interaction
networks with high-throughput expression data and other molecular
states into a unified conceptual framework.
Pathway enrichment analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID) contains an integrated biological
knowledge base and analytic tools, aiming at systematically
extracting biological meaning from large gene/protein lists
(25). The Kyoto Encyclopedia of
Genes and Genomes (KEGG) is a knowledge base for the systematic
analysis of gene functions, linking genomic information with higher
order functional information (26). In this study, pathway enrichment
analysis was performed for the PPI network by DAVID and the
significantly enriched pathways were identified with a value of
P<0.05.
Results
Expression of RASSF1A
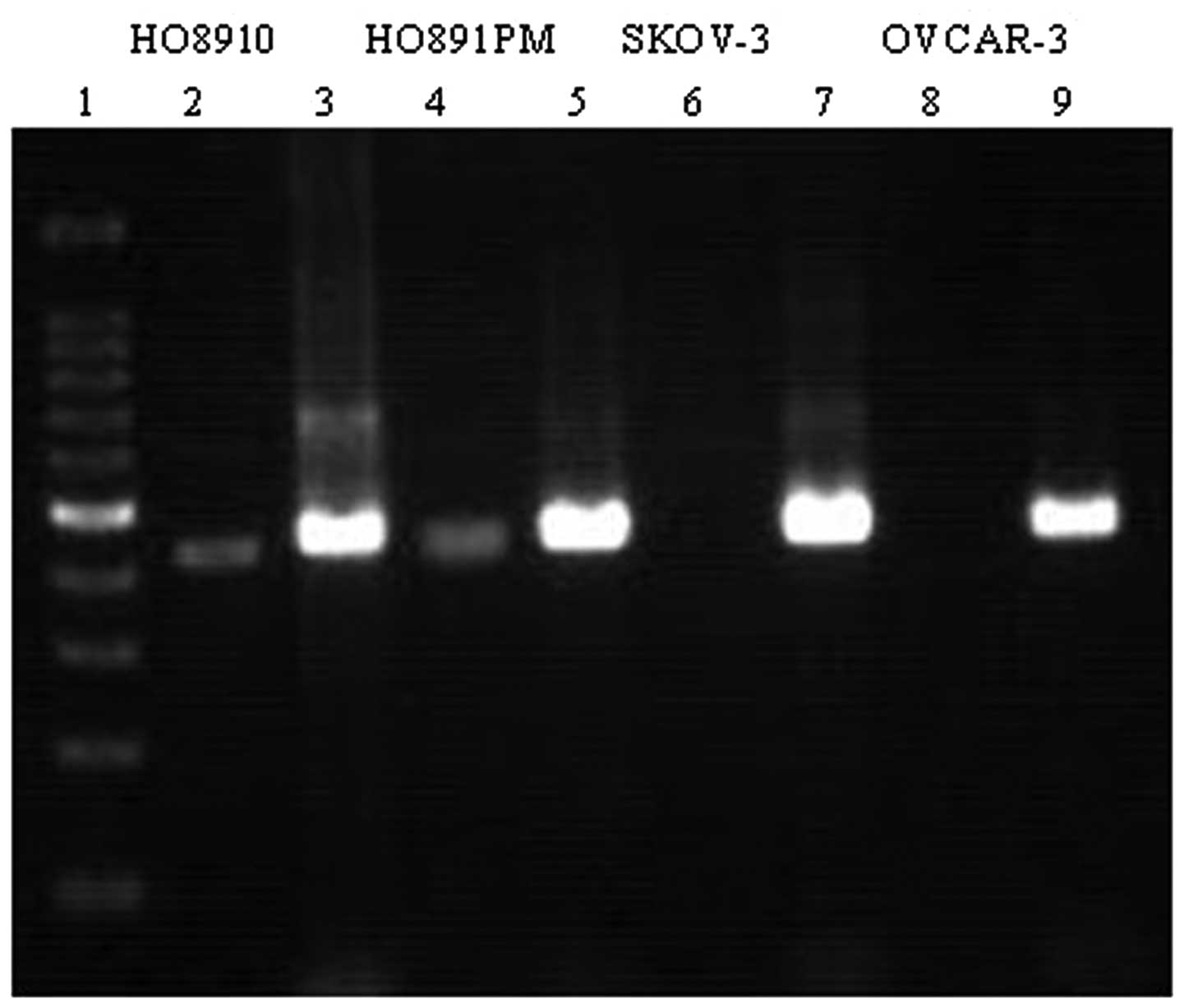
RASSF1A mRNA was expressed in the HO8910 and
HO8910PM cells and was absent in the SKOV-3 and OVCAR-3 cells
(Fig. 1). RASSF1A mRNA expression
was detected in all 10 normal ovarian tissues (100%), while RASSF1A
mRNA was detected in 2 cases among the 47 ovarian tumor samples
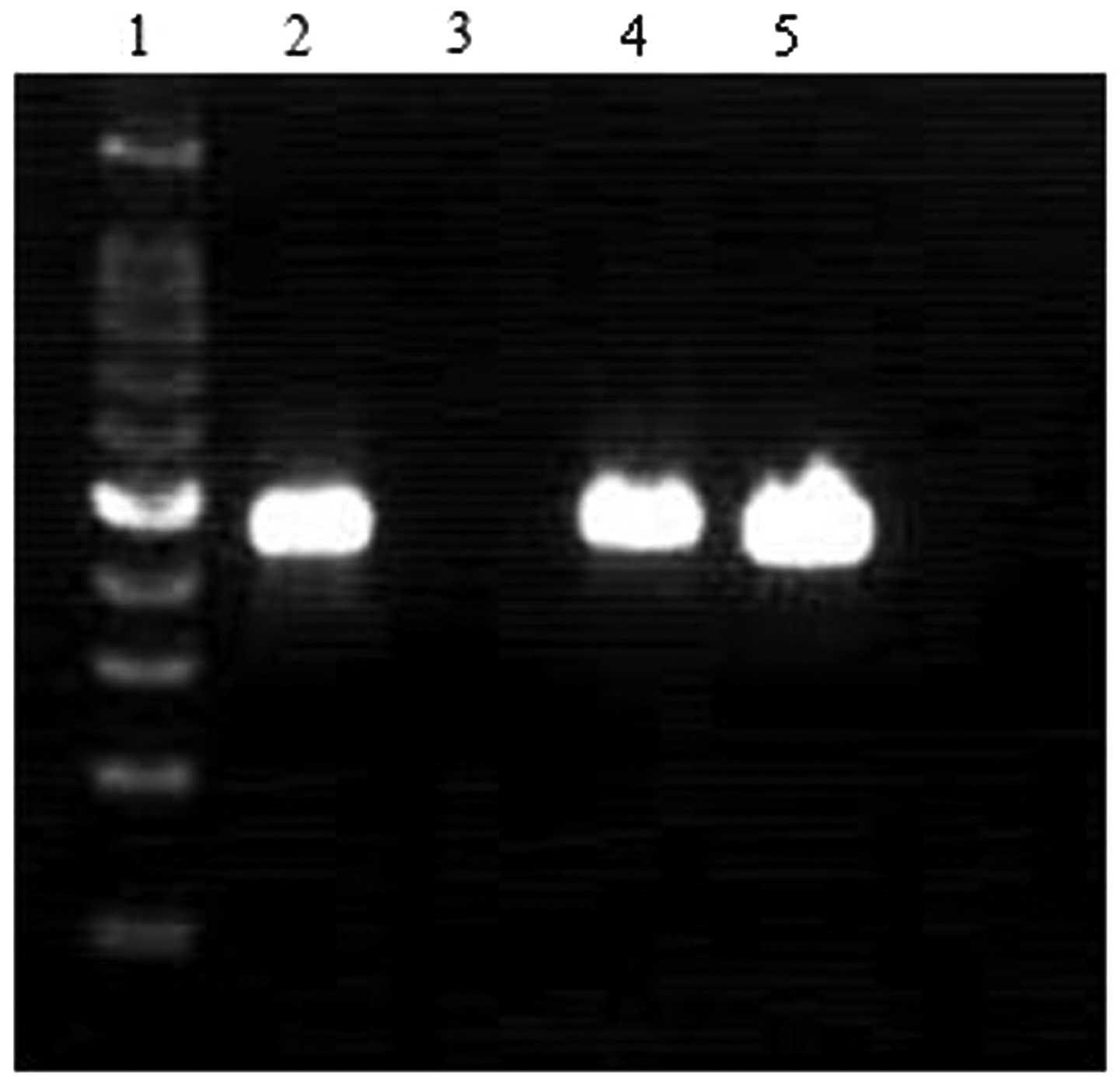
(4.3%). The expression of RASSF1A in the differently treated SKOV-3
cells was detected by qRT-PCR; no RASSF1A expressoin was detected
in the SKOV-3 cells transfected with adenovirus without RASSF1A
(Fig. 2). However, high
expression levels of the RASSF1A gene were found in the SKOV-3
cells transfected with adenovirus carrying RASSF1A.
Morphology and structure of SKOV-3
cells
The morphology of the transfected and untransfected
SKOV-3 cells was examined under fluorescence microscope after 24 h
of transfection. The untreated SKOV-3 cells showed the stripping
away of the cell wall and had projections on their surface
(Fig. 3A). However, the
transfected cells were round in shape and the volumes were
increased; the nucleus was vacuolated (Fig. 3B). In addition, the fluorescence
location in the transfected SKOV-3 cells was observed after 48 h of
treatment. Fluorescent aggregates were observed in the cytoplasm
and were mainly distributed in the cytoskeleton (Fig. 3C). Moreover, a TEM was utilized to
study the structure of the SKOV-3 cells. The untransfected cells
showed a regular growth (Fig.
3D), whereas the nuclear chromatin of the transfected cells
displayed obvious shrinkage, condensation and apoptotic bodies had
formed (Fig. 3E).
Analysis of apoptosis
With the use of a flow cytometer and an apoptosis
detection kit, the cycle and apoptosis of SKOV-3 cells (untreated
cells and cells transfected with adenovirus with or without
RASSF1A) were analyzed (Fig. 4).
In the S phase of the cell cycle, a significantly greater number of
transfected SKOV-3 cells (cells transfected with adenovirus with or
without RASSF1A) was observed, compared with the untreated cells
(P<0.05). Furthermore, the rate of apoptosis of the SKOV-3 cells
transfected with RASSF1A was significantly greater than that of the
other cells (untreated cells and those transfected with adenovirus
without RASSF1A) (P<0.01).
Results of western blot analysis
The expression of cyclin D1 and survivin in the
SKOV-3 cells transfected with adenovirus carrying RASSF1A was
decreased significantly compared with the untreated cells and those
transfected with adenovirus without RASSF1A (P<0.05). However,
the expression of p27 and caspase-3 in the SKOV-3 cells transfected
with RASSF1A was increased highly significantly compared with the
other SKOV-3 cells (untreated cells and those transfected with
adenovirus without RASSF1A) (P<0.01) (Fig. 5).
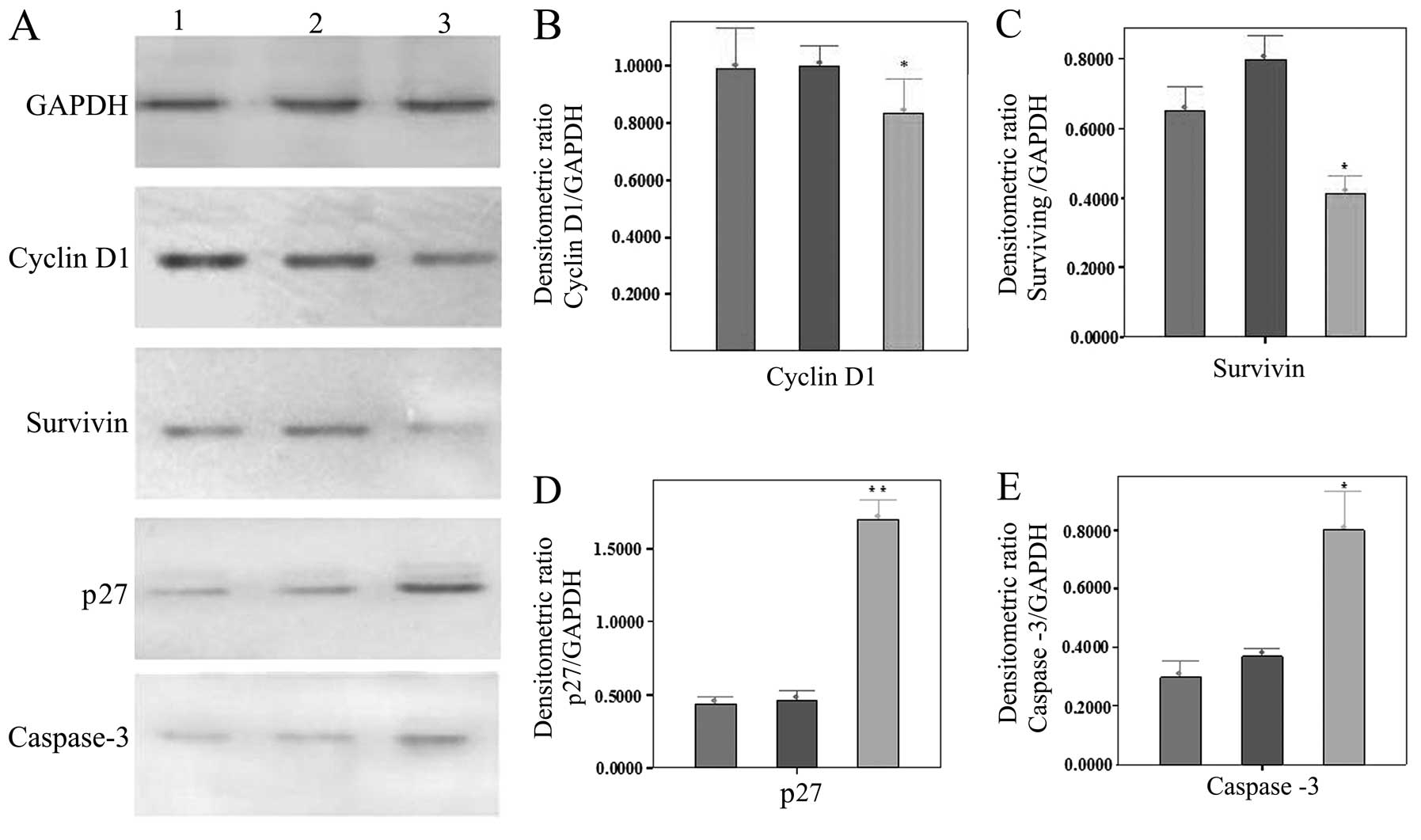 | Figure 5Expression of 4 types of protein in
the differently treated SKOV-3 cells. (A) Expression of GAPDH,
cyclin D1, survivin, p27 and caspase-3. Lanes 1–3 represent
untransfected SKOV-3 cells, those transfected without and those
transfected with adenovirus carrying Ras-association domain family
1, isoform A (RASSF1A), respectively. (B-E) Quantification of the
results of cyclin D1, survivin, p27 and caspase-3, respectively.
*P<0.05 and **P<0.01, statistically
significant differences. |
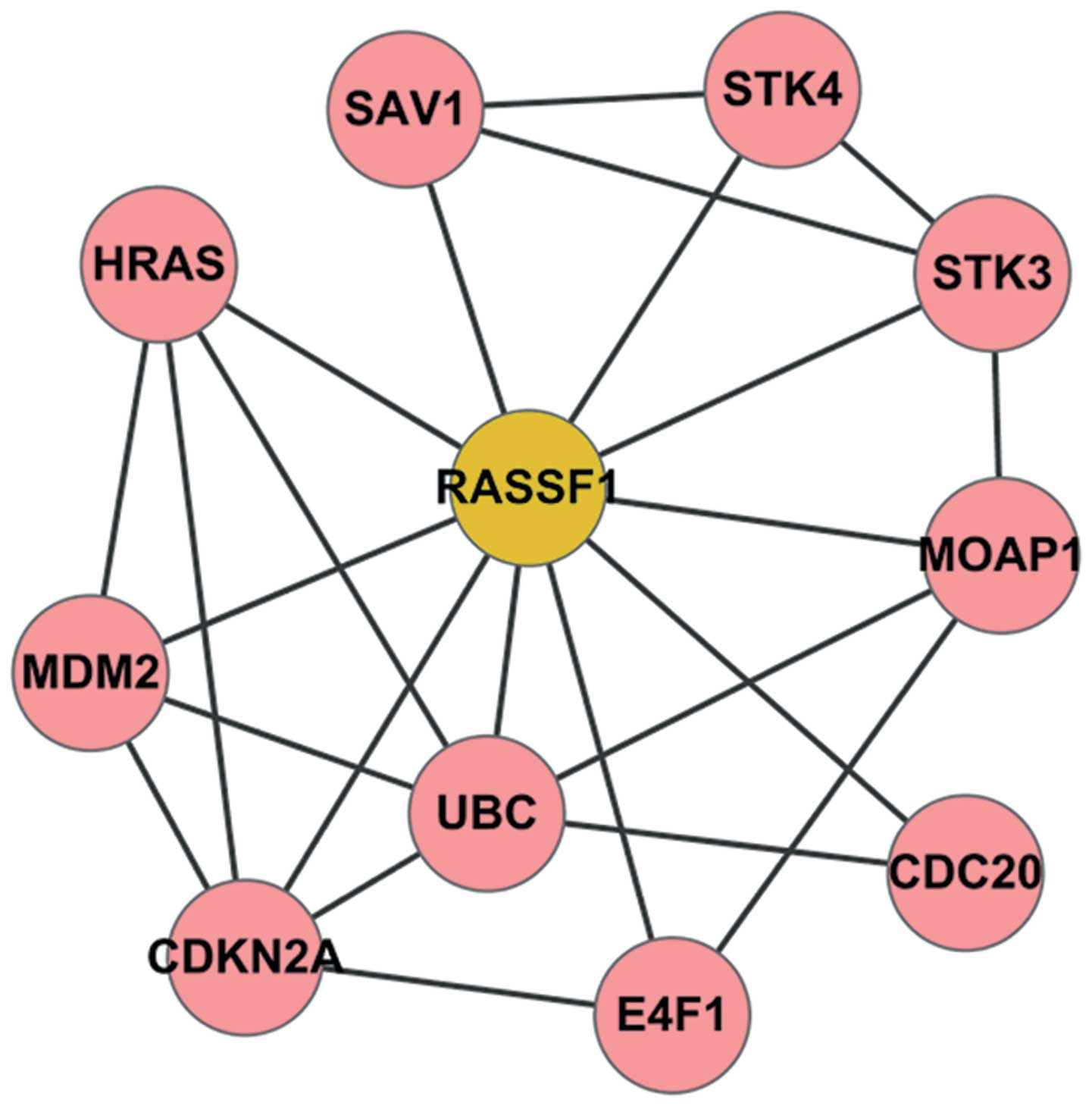
PPI network
The GSE14407 gene expression data were analyzed and
the DEGs between the OSE and CEPI samples were identified. However,
the RASSF1A gene was discovered not to be in the list of DEGs in
our study. Based on STRING and Cytoscape software, a
RASSF1A-related PPI network was constructed (Fig. 6). In this network, a total of 10
genes, including serine/threonine kinase (STK)4, Harvey rat sarcoma
viral oncogene homolog (HRAS), cell division cycle 20 (CDC20),
STK3, modulator of apoptosis 1 (MOAP1), salvador homolog 1
(Drosophila) (SAV1), E4F1, ubiquitin C (UBC), murine double
minute 2 (MDM2) and DKN2A was found to be closely associated with
RASSF1A (Table I).
 | Table IProtein-protein interactions and their
combined scores. |
Table I
Protein-protein interactions and their
combined scores.
| Node 1 | Node 2 | Combined score | Node 1 | Node 2 | Combined score |
|---|
| MDM2 | UBC | 0.999 | RASSF1 | SAV1 | 0.956 |
| MDM2 | CDKN2A | 0.999 | RASSF1 | E4F1 | 0.953 |
| STK3 | SAV1 | 0.997 | RASSF1 | UBC | 0.95 |
| STK4 | SAV1 | 0.996 | CDKN2A | RASSF1 | 0.947 |
| STK4 | RASSF1 | 0.996 | CDKN2A | E4F1 | 0.94 |
| RASSF1 | HRAS | 0.993 | MDM2 | HRAS | 0.923 |
| RASSF1 | CDC20 | 0.991 | STK3 | STK4 | 0.9 |
| STK3 | RASSF1 | 0.985 | UBC | HRAS | 0.858 |
| RASSF1 | MOAP1 | 0.976 | CDKN2A | HRAS | 0.752 |
| UBC | CDC20 | 0.975 | UBC | MOAP1 | 0.507 |
| MDM2 | RASSF1 | 0.974 | STK3 | MOAP1 | 0.502 |
| CDKN2A | UBC | 0.961 | E4F1 | MOAP1 | 0.401 |
Pathway enrichment analysis
KEGG pathway analysis was conducted using DAVID and
a total of 8 pathways were identified with values of P<0.05
(Table II). The RASSF1A gene was
shown to participate in 3 significant pathways, including bladder
cancer (P=1.79E-05), non-small cell lung cancer (P=3.85E-05) and
pathways in cancer (P=5.09E-04). In addition, all the enriched
pathways were associated with cancer.
 | Table IISignificant pathways in the
RASSF1A-related protein-protein interaction network. |
Table II
Significant pathways in the
RASSF1A-related protein-protein interaction network.
| Term | Description | Count | P-value | Genes |
|---|
| hsa05219 | Bladder cancer | 4 | 1.79E-05 | HRAS, CDKN2A,
RASSF1, MDM2 |
| hsa05223 | Non-small cell lung
cancer | 4 | 3.85E-05 | HRAS, CDKN2A,
RASSF1, STK4 |
| hsa05200 | Pathways in
cancer | 5 | 5.09E-04 | HRAS, CDKN2A,
RASSF1, MDM2, STK4 |
| hsa05214 | Glioma | 3 | 3.05E-03 | HRAS, CDKN2A,
MDM2 |
| hsa05218 | Melanoma | 3 | 3.86E-03 | HRAS, CDKN2A,
MDM2 |
| hsa05220 | Chronic myeloid
leukemia | 3 | 4.30E-03 | HRAS, CDKN2A,
MDM2 |
| hsa04110 | Cell cycle | 3 | 1.16E-02 | CDKN2A, MDM2,
CDC20 |
| hsa04010 | MAPK signaling
pathway | 3 | 4.84E-02 | HRAS, STK4,
STK3 |
Discussion
In spite of great efforts that have been made by
researchers to enable the early diagnosis of ovarian cancer, the
results have not been satisfactory and the pathogenesis of the
disease is not yet fully understood (27). In this study, the RASSF1A gene was
found to be present in HO8910 and HO8910PM cells, and absent in
SKOV-3 and OVCAR-3 cells. The SKOV-3 cells were then transfected by
an adenovirus with or without RASSF1A, and RASSF1A expression in
the differently treated SKOV-3 cells was analyzed by qRT-PCR.
Moreover, the morphology and structure, as well as the apoptotic
and proliferative ability of the SKOV-3 cells transfected with
adenovirus carrying RASSF1A were altered significantly compared
with the untreated SKOV-3 cells. Furthermore, the RASSF1A gene was
shown to induce apoptosis and suppress cell proliferation through
several cancer-related pathways by interacting with other
genes.
Firstly, the expression of RASSF1A in 4 types of
human ovarian cancer cell lines, including HO8910, HO8910PM, SKOV-3
and OVCAR-3, was detected by qRT-PCR. The result revealed that
RASSF1A is absent in SKOV-3 and OVCAR-3 cells, while it is present
in the HO8910 and HO8910PM cells. Our findings are consistent with
those of a previous study showing that SKOV-3 and OVCAR-3 cells
represent an ovarian cancer, referred to as serous adenocarcinoma
(28). We subsequently
transfected SKOV-3 cells with an adenovirus with or without RASSF1A
and the expression of RASSF1A in the differently treated SKOV-3
cells was analyzed. The expression of RASSF1A was observed in the
SKOV-3 cells transfected with RASSF1A and no expression was
observed in the untreated SKOV-3 cells. This finding verifies that
SKOV-3 cells do not express RASSF1A and suggests that the
transfection efficiency was very high.
Secondly, the morphology, structure, apoptosis and
cell cycle progression of SKOV-3 cells before and after
transfection with RASSF1A were examined under a fluorescence
microscope, TEM and flow cytometer, respectively. Compared with the
untreated cells, the transfected SKOV-3 cells were round in shape,
their volumes were increased, and the nuclei were vacuolated. In
addition, fluorescence was observed in the cytoplasm and was mainly
distributed in the cytoskeleton. Moreover, the nuclear chromatin of
the transfected cells displayed showed marked shrinkage,
condensation and formed apoptotic bodies. All these changes in
morphology and structure are in accordance with those observed in a
previous study (29). In
addition, the apoptosis of the SKOV-3 cells transfected with
RASSF1A was significantly higher compared with the other
(untreated) cells. Our result indicated that the SKOV-3 cells
transfected with adenovirus carrying RASSF1A were mainly located at
the S phase of the cell cycle; these results are consistent with
those of a previous study reporting that RASSF1A overexpression
induces G2/M cell cycle arrest (30). These data indicate that RASSF1A
modulates the cell cycle, as was also shown in the study by
Matallanas et al (31),
who reported that RASSF1A promotes cell cycle arrest and
apoptosis.
In this study, the expression of
proliferation-related proteins, including cyclin D1, survivin, p27
and caspase-3 was also examined in the SKOV-3 cells before and
after transfection with adenorius with or without RASSF1A by
western blot analysis. Cyclin D1 is a component of the core cell
cycle machinery and is usually overexpressed in human cancers
(32). As a member of the
inhibitor of apoptosis protein (IAP) family, survivin is
upregulated in almost all human tumors (33). Our results demonstrated that the
expression of cyclin D1 and survivin was decreased in the SKOV-3
cells transfected with RASSF1A, which indicates that RASSF1A may
promote cancer cell apoptosis. Additionally, the cyclin-dependent
kinase (Cdk) inhibitor, p27, regulates cell proliferation, cell
motility and apoptosis (34).
Caspase-3 is a frequently activated death protease by catalyzing
the specific cleavage of several key cellular proteins (35). In this study, the expression of
p27 and caspase-3 was increased in the cells transfected with
adenovirus carrying RASSF1A, which suggests that RASSF1A suppresses
cell proliferation.
Finally, a bioinformatics approach was applied to
explore the mechanisms responsible for the effects of RASSF1A on
ovarian cancer cells. Between the ovarian cancer and normal
samples, the expression of RASSF1A changed insignificantly. The
RASSF1A-related PPI network was constructed containing 10
associated genes, including STK4, STK3, HRAS and CDC20. Moreover,
these genes were shown to be enriched in 8 cancer-associated
pathways, such as bladder cancer, non-small cell lung cancer and
pathways in cancer. The members of STKV, STK3 and STK4 localize to
the microtubules and interact with RASSF1A to regulate apoptosis
(36). The HRAS gene encodes a
protein which is involved primarily in the regulation of cell
growth, division and apoptosis (37). It has been previously reported
that RASSF1A controls mitotic progression by binding to and
inhibiting CDC20, which is an activator of the anaphase-promoting
complex (38). Of note, our
findings are consistent with those of previous studies which have
been mentioned above.
Furthermore, these identified genes were involved in
several cancer-related pathways, such as bladder cancer, non-small
cell lung cancer and pathways in cancer. These results indicate
that RASSF1A and its associated genes may suppress the
proliferation and promote the apoptosis of cancer cells by
affecting these pathways in the development of bladder cancer,
non-small cell lung cancer and ovarian cancer. Kim et al
(39) identified RASSF1A as a
promising prognostic marker in recurrent non-muscle invasive
bladder cancer. Senchenko et al (13) reported that 3 tumor suppressor
genes, RASSF1A, RBSP3 and NPRL2, which were identified in the
3p21.3 region, are involved in the development of several types of
cancer, including non-small cell lung cancer. Moreover, RASSF1A has
been shown to prevent hypertrophy by disrupting the MAPK signaling
pathway in cardiac myocytes (40). Therefore, our findings are
consistent with those of previous studies and provide some
knowledge of the association between RASSF1A and ovarian
cancer.
In conclusion, in this study, the expression of
RASSF1A was detected in different ovarian cancer cell lines, and
its effects on apoptosis and proliferation were examined.
Furthermore, the underlying mechanisms were investigated using a
bioinformatics approach. Our data demonstrate that RASSF1A promotes
apoptosis and suppresses the proliferation of ovarian cancer cells.
Our findings provide a biomarker for the early diagnosis and
treatment of ovarian cancer. However, further research is required
to verify our findings.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (30100104).
References
|
1
|
Yap TA, Carden CP and Kaye SB: Beyond
chemotherapy: targeted therapies in ovarian cancer. Nat Rev Cancer.
9:167–181. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buys SS, Partridge E, Black A, et al:
Effect of screening on ovarian cancer mortality: the prostate,
lung, colorectal and ovarian (PLCO) cancer screening randomized
controlled trial. JAMA. 305:2295–2303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vaughan S, Coward JI, Bast RCJC, et al:
Rethinking ovarian cancer: recommendations for improving outcomes.
Nat Rev Cancer. 11:719–725. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moore RG, Miller MC, Eklund EE, Lu KH,
Bast RC Jr and Lambert-Messerlian G: Serum levels of the ovarian
cancer biomarker HE4 are decreased in pregnancy and increase with
age. Am J Obstet Gynecol. 206:349.e341–e347. 2012.PubMed/NCBI
|
|
5
|
Nosov V, Su F, Amneus M, et al: Validation
of serum biomarkers for detection of early-stage ovarian cancer. Am
J Obstet Gynecol. 200:639.e631–e635. 2009.PubMed/NCBI
|
|
6
|
Weiland F, Martin K, Oehler MK and
Hoffmann P: Deciphering the molecular nature of ovarian cancer
biomarker CA125. Int J Mol Sci. 13:10568–10582. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yee KS, Grochola L, Hamilton G, et al: A
RASSF1A polymorphism restricts p53/p73 activation and associates
with poor survival and accelerated age of onset of soft tissue
sarcoma. Cancer Res. 72:2206–2217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Donninger H, Barnoud T, Nelson N, et al:
RASSF1A and the rs2073498 cancer associated SNP. Front oncol.
1:542011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kassler S, Donninger H, Birrer MJ and
Clark GJ: RASSF1A and the taxol response in ovarian cancer. Mol
Biol Int. 2012:2632672012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu L, Kron KJ, Pethe VV, et al:
Association of tissue promoter methylation levels of APC, TGFβ2,
HOXD3 and RASSF1A with prostate cancer progression. Int J Cancer.
129:2454–2462. 2011.PubMed/NCBI
|
|
11
|
Brait M, Loyo M, Rosenbaum E, et al:
Correlation between BRAF mutation and promoter methylation of
TIMP3, RARβ2 and RASSF1A in thyroid cancer. Epigenetics. 7:710–719.
2012.PubMed/NCBI
|
|
12
|
Kloten V, Becker B, Winner K, et al:
Promoter hypermethylation of the tumor-suppressor genes ITIH5,
DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer
screening. Breast Cancer Res. 15:R42013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Senchenko VN, Anedchenko EA, Kondratieva
TT, et al: Simultaneous down-regulation of tumor suppressor genes
RBSP3/CTDSPL, NPRL2/G21 and RASSF1A in primary non-small cell lung
cancer. BMC cancer. 10:752010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yi M, Yang J, Chen X, et al: RASSF1A
suppresses melanoma development by modulating apoptosis and
cell-cycle progression. J Cell Physiol. 226:2360–2369. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Amin KS and Banerjee PP: The cellular
functions of RASSF1A and its inactivation in prostate cancer. J
Carcinog. 11:32012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Laird PW: Principles and challenges of
genomewide DNA methylation analysis. Nat Rev Genet. 11:191–203.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oh JH, Craft JM, Townsend R, Deasy JO,
Bradley JD and El Naqa I: A bioinformatics approach for biomarker
identification in radiation-induced lung inflammation from limited
proteomics data. J Proteome Res. 10:1406–1415. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goertsches RH, Zettl UK and Hecker M:
Sieving treatment biomarkers from blood gene-expression profiles: a
pharmacogenomic update on two types of multiple sclerosis therapy.
Pharmacogenomics. 12:423–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kruger NJ: The Bradford method for protein
quantitation. The Protein Protocols Handbook. Springer; New York,
NY: pp. 17–24. 2009, View Article : Google Scholar
|
|
20
|
Chang C, Zhang C, Zhao X, Kuang X, Tang H
and Xiao X: Differential regulation of mitogen-activated protein
kinase signaling pathways in human with different types of mitral
valvular disease. J Surg Res. 181:49–59. 2013. View Article : Google Scholar
|
|
21
|
Bowen NJ, Walker LD, Matyunina LV, et al:
Gene expression profiling supports the hypothesis that human
ovarian surface epithelia are multipotent and capable of serving as
ovarian cancer initiating cells. BMC Med Genomics. 2:712009.
View Article : Google Scholar
|
|
22
|
Russell LJ, Capasso M, Vater I, et al:
Deregulated expression of cytokine receptor gene, CRLF2, is
involved in lymphoid transformation in B-cell precursor acute
lymphoblastic leukemia. Blood. 114:2688–2698. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Franceschini A, Kuhn M, et
al: The STRING database in 2011: functional interaction networks of
proteins, globally integrated and scored. Nucleic Acids Res.
39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shannon P, Markiel A, Ozier O, et al:
Cytoscape: a software environment for integrated models of
biomolecular interaction networks. Genome Res. 13:2498–2504. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
26
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kipps E, Tan DS and Kaye SB: Meeting the
challenge of ascites in ovarian cancer: new avenues for therapy and
research. Nat Rev Cancer. 13:273–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patankar NA, Pritchard J, Van Grinsven M,
Osooly M and Bally MB: Topotecan and doxorubicin combination to
treat recurrent ovarian cancer: the influence of drug exposure time
and delivery systems to achieve optimum therapeutic activity. Clin
Cancer Res. 19:865–877. 2013. View Article : Google Scholar
|
|
29
|
Pan Y, Du Zhen-Wu D, Zhou JW, et al:
MicroRNA-mediated silencing of RhoC inhibits tumor invasion and
increases chemosensitivity to paclitaxel in SKOV3 cells in vitro.
Chem Res Chin Univ. 27:70–74. 2011.
|
|
30
|
Hergovich A and Hemmings BA: Hippo
signalling in the G2/M cell cycle phase: lessons learned from the
yeast MEN and SIN pathways. Semin Cell Dev Biol. 23:794–802. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matallanas D, Romano D, Yee K, et al:
RASSF1A elicits apoptosis through an MST2 pathway directing
proapoptotic transcription by the p73 tumor suppressor protein. Mol
Cell. 27:962–975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jirawatnotai S, Hu Y, Michowski W, et al:
A function for cyclin D1 in DNA repair uncovered by protein
interactome analyses in human cancers. Nature. 474:230–234. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cheung CH, Cheng L, Chang KY, Chen HH and
Chang JY: Investigations of survivin: the past, present and future.
Front Biosci (Landmark Ed). 16:952–961. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chu IM, Hengst L and Slingerland JM: The
Cdk inhibitor p27 in human cancer: prognostic potential and
relevance to anticancer therapy. Nat Rev Cancer. 8:253–267. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Larsen BD, Rampalli S, Burns LE, Brunette
S, Dilworth FJ and Megeney LA: Caspase 3/caspase-activated DNase
promote cell differentiation by inducing DNA strand breaks. Proc
Natl Acad Sci USA. 107:4230–4235. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bennani-Baiti IM: Epigenetic and
epigenomic mechanisms shape sarcoma and other mesenchymal tumor
pathogenesis. Epigenomics. 3:715–732. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun XF, Li L, Li XJ and Shen W:
Methylation pattern of oncogene HRAS gene promoter region and its
clinical relevance to urocystic tumorigenesis. Mol Biol Rep.
39:8431–8437. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu L, Baier K, Dammann RH and Pfeifer GP:
The tumor suppressor RASSF1A does not interact with Cdc20, an
activator of the anaphase-promoting complex. Cell Cycle.
6:1663–1665. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim JS, Chae Y, Ha YS, et al: Ras
association domain family 1A: a promising prognostic marker in
recurrent nonmuscle invasive bladder cancer. Clin Genitourin
Cancer. 10:114–120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Del Re DP and Sadoshima J: RASSF1A
Signaling in the heart: novel functions beyond tumor suppression.
Mol Biol Int. 2012:1542832012.PubMed/NCBI
|