Introduction
Gastric cancer has been proven to be one of the most
common and most commonly diagnosed malignant tumors worldwide
(1), with a low 5-year survival
rate (2). As is already known,
early diagnosis and effective therapeutic targets are critical
factors influencing clinical treatment and prognosis. However, the
underlying mechanisms of tumorigenesis and the progression of
gastric cancer have not been fully elucidated.
Flap endonuclease-1 (FEN1), a structure-specific 5′
nuclease (3), is a DNA
replication protein which is currently accepted as a pivotal
component of DNA molecule metabolism (4) due to its 5′-flap endonuclease
(5), 5′ exonuclease (6) and gap-endonuclease activities
(7,8). It plays a vital role in Okazaki
fragment maturation (9), the
restarting of stalled replication forks (7,10),
telomere maintenance (11,12),
long-pace base excision repair (13) and apoptosis-induced fragmentation
(7), which render it an essential
node in maintaining genomic fidelity (14) and in preventing cells from
carcinogenesis (4–14).
However, a growing body of evidence suggests that
the abnormal expression of (FEN1) is involved in the initiation and
progression of cancer and the development of disease (15–17). It has been reported that FEN1 is
overexpressed in lung cancer cell lines (18), metastatic prostate cancer cell
lines (19) and gastric cancer
cell lines (20) compared with
normal cell lines. In addition, the upregulated expression of FEN1
has been reported in tumor tissues, including prostate cancer
(21), neuroblastoma (22), pancreatic cancer (23) and breast cancer (24) at the mRNA and protein level. FEN1
has further been confirmed to be upregulated in lung cancer,
testicular cancer, glioblastoma and astrocytoma (25).
Therefore, in this study, the expression of FEN1 was
evaluated in gastric tumor tissues by RT-PCR. In addition, the
association between FEN1 expression and the clinicopathological
characteristics of gastric cancer patients was explored by
immunohistochemistry and regarded as evidence that FEN1 may be a
potential biomarker for the diagnosis of gastric cancer. The
effects of the silencing of FEN1 by siRNA on the proliferation and
apoptosis of the selected gastric cell line were also evaluated in
order to elucidate the underlying mechanisms responsible for the
proression of gastric cancer, and to determine whether FEN1 is a
potential therapeutic target in this disease.
Materials and methods
Preparation of human tissues and total
RNA
Samples of surgically resected gastric tumors and
corresponding normal tissues from 42 patients with gastric cancer
were collected from the Department of Gastrointestinal Surgery, The
First Affiliated Hospital of Chongqing Medical University,
Chongqing, China, after obtaining written informed consent from
each patient. These specimens were momentarily stored in liquid
nitrogen for further research. All the gastric cancer patients were
admitted to our hospital from February 2012 to April 2013. All 42
patients were determinately diagnosed by a pathological technique
and had not received any pre-operative chemotherapy, radiotherapy
or immunotherapy. The tumors obtained were divided into 6 groups
according to the age (>65 or ≤65 years) and gender (male or
female) of the patients, the differentiation grade (low, moderate
or high), tumor size (>3cm or ≤3cm), the lymphatic metastasis
(positive or negative) and the TNM classification (I + II or III +
IV) of tumors. The study was approved by the Ethics Committee of
The First Affiliated Hospital of Chongqing Medical University.
Total RNA was isolated from the tissues using RNAiso reagent
(Takara Bio, Inc., Shiga, Japan). The concentration of the RNA was
determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Waltham, MA, USA).
Cell culture and transfection of cells
with siRNA
Three gastric cancer cell lines, SGC-7901, MKN-28,
MGC-803, were acquired from the Molecular Medicine and Cancer
Research Center of Chongqing Medical University. All the cell lines
were maintained in RPMI-1640 medium (Invitrogen, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (Invitrogen), 1%
penicillin-streptomycin and 2.0 g/l sodium bicarbonate at 36.8°C.
The cells were used for the extraction of total RNA and protein
when they reached 80–90% confluent. The siRNA vectors targeting
FEN1 in the human gastric cells were designed and constructed by
Invitrogen. A negative control (NC) siRNA was also used. The
sequences of the siRNA were as follows: sense strand,
5′-GGACUUGUAGUCCUGCGAUTT-3′ and antisense strand,
5′-AUCGCAGGACUACAAGUCCTT-3′. The cells were plated in 6-well plates
and each well had 3×105 cells. After 24 h, the cells
were cultured in an incubator containing CO2 at 36.7°C,
the mixture of siRNA and Lipofectamine™ 2000 (Invitrogen) was added
to each well containing cells and according to the manufacturer’s
instructions. Fresh medium was used to replace the transfection
mixture after 6 h. Forty-eight hours after transfection, the cells
were used for further experiments.
Semiquantitative reverse
transcription-PCR (RT-PCR)
The mRNA levels of FEN1 in the tumor tissues and
corresponding normal tissues were examined by RT-PCR. Total mRNA
from all the tissues was reverse transcribed into complementary DNA
using a reverse transcription kit (Takara Bio, Inc.) according to
the manufacturer’s instructions. The specific oligonucleotide
primers used for FEN1 and those of the internal reference gene,
β-actin, were designed using Primer 5.0 software (Premier Biosoft,
Palo Alto, CA, USA) and synthesized by Takara Bio, Inc.. The
primers for FEN1 were as follows: forward, 5′-AGCCCGTGTATGTCTTTG-3′
and reverse, 5′-AGTCAGGTGTCGCATTAG-3′ and for β-actin forward,
5′-CCTTCTACAATGAGCTGCGT-3′ and reverse, 5′-CCTGGATAGCAACGTACATG-3′.
The PCR products were electrophoresed on 1.5% agarose gels. The
expression of FEN1 relative to β-actin was measured using a Bio-Rad
gel imaging analysis system and analyzed using Quantity One
software (Bio-Rad, Hercules, CA, USA). The RT-PCR experiments were
repeated independently 3 times.
Immunohistochemistry
Immunohistochemical staining was performed on
paraffin-embedded tissues from 42 gastric cancer patients. The
tissue sections were dewaxed with xylene and rehydrated with
alcohol after baking in the 60°C thermostat for 1 h. After rinsing
in PBS, the sections were heated with citrate buffer in a microwave
oven for antigen retrieval. Normal goat serum was then used to
block out non-specific binding after washing in PBS, the endogenous
peroxidase activity of the tissues had been already blocked by 3%
peroxide. The cells were incubated with primary anti-FEN1 antibody
(Epitomics, Burlingame, CA, USA) overnight at a dilution of 1:300
at 4°C. The sections, after a thorough cleaning in PBS, were
subjected to 1-h incubation with anti-rabbit secondary antibodies
and were then incubated with horseradish peroxidase (HRP)-labeled
streptavidin for 30 min. In addition, the tissue sections were
stained with diaminobenzidine (DAB) and then redyed by hematoxylin
counterstain. Ultimately, the treated sections were observed under
a microscope and estimated by the standard described in a previous
study (24). The staining was
scored according to two standards of FEN1 expression: the staining
intensity scored from grade 0 to 3 (0, negative; 1, pale yellow
staining; 2, moderate yellow staining; 3, brown staining) and the
percentage of positively stained cells (0, no FEN1-positive cells,
1, <10% positive cells; 2, 11–15% positive cells; 3, 51–80%
positive cells and 4, >80% positive cells). The final result of
FEN1 expression was the product of scores of the staining intensity
and the percentage of positively stained cells (negative, 0–1;
weakly positive, 1–2; moderately positive, 2–3; strongly positive,
≥3).
Western blot analysis
Western blot analysis was used to verify the
inhibitory effects of FEN1 siRNA. The 3 gastric cell lines were
lysed using cell lysis buffer for the isolation of total protein.
The bicinchoninic acid (BCA) protein assay kit was used to measured
the concentration of the extracted protein of which 30 μl was
separated on SDS-PAGE for electrophoresis and electrophoretically
transferred onto polyvinylidene fluoride (PVDF) membranes. The
membranes were blocked in 5% fat-free milk at room temperature and
incubated for 2 h with a 1:1,000 dilution of primary rabbit
monoclonal antibodies, followed by HRP-conjugated secondary
antibody. After rinsing, the blots were detected using a
chemiluminescence western blotting detection system. The western
blot analysis experiments were repeated independently 3 times.
Detection of cell proliferation by MTS
assay
The cells in the experimental group and control
group were trypsinized and placed into 96-well culture plants in
the logarithmic phase at a density of 7×103 cells/well.
The MTS kit (CellTiter 96 AQueous one solution reagent; Promega
Corp., Madison, WI, USA) was then added to each well and the cells
were incubated for a further 4 h before harvesting at 24, 48, 72
and 96 h according to the manufacture’s instructions. Optical
density was measured at 490 nm for the absorbance values. These
procedures were repeated independently 3 times.
Cell apoptosis assay
The fluorescein-isothiocyanate-labeled enhanced
AnnexinV/propidium iodide Apoptosis Detection kit (Invitrogen) was
used to determine the apoptotic rate of the cells in the
experimental group and control group according to the manufacture’s
instructions. Methotrexate (MTX; Shanghai Yuanye Bio-Technology
Co., Ltd, Shanghai, China) was used to induce cell apoptosis. The
cells were then subjected to flow cytometry within 1 h.
Statistical analysis
All the data were analyzed with a paired t-test,
Wilcoxon rank sum test and χ2 test using IBM SPSS 19.0
software (SPSS Inc., Chicago, IL, USA). A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of FEN1 gene in tumors
compared with matched normal tissues
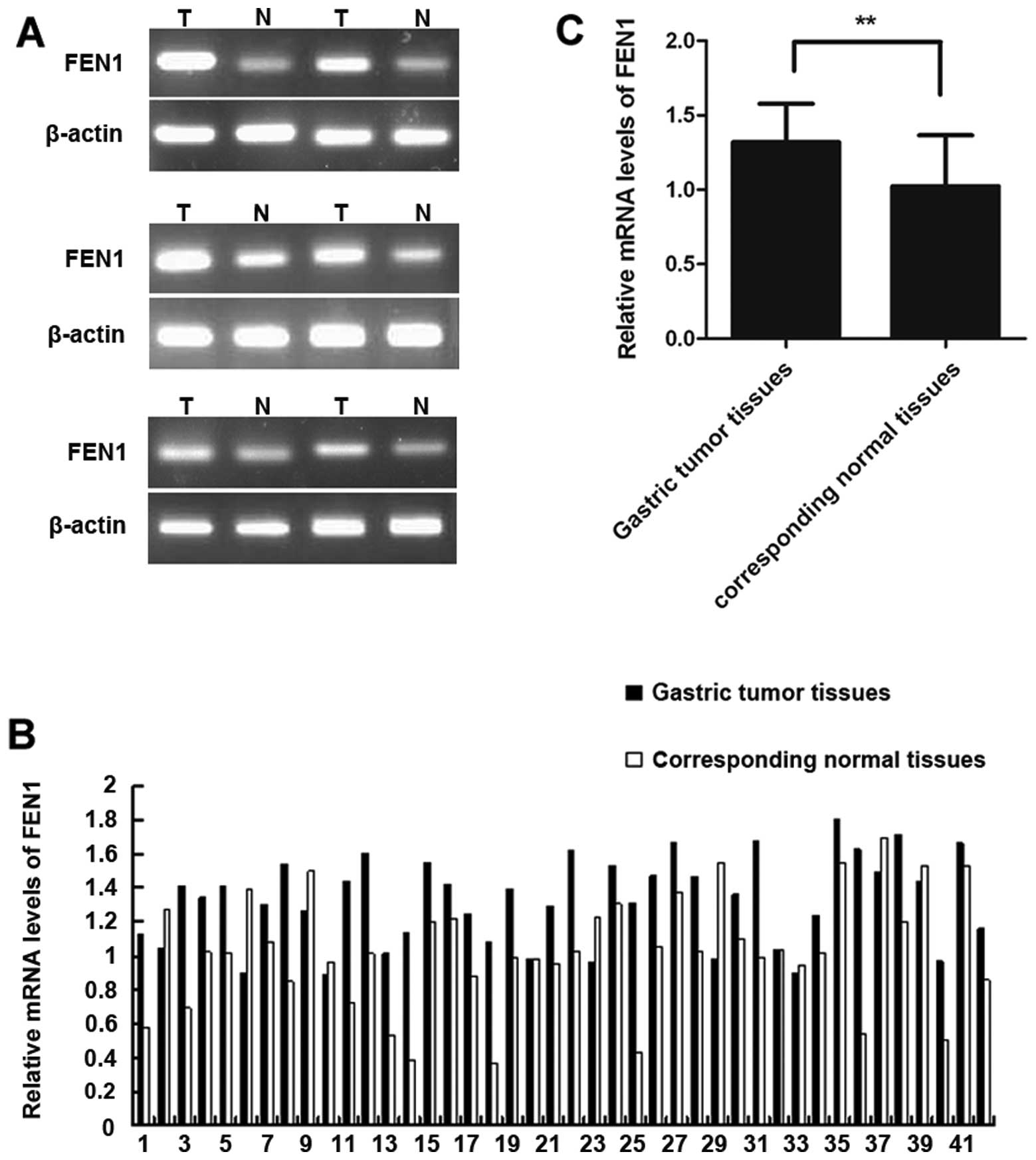
The FEN1 mRNA expression levels in the gastric tumor
and corresponding normal tissues was determined by semiquantitative
RT-PCR and the results are shown in Fig. 1. The FEN1 mRNA expression of 6 out
of 42 paired gastric tumor tissues is shown in Fig. 1A. Of the 42 samples, 32 (76%)
displayed a higher FEN1 expression in the tumor tissues compared to
the corresponding normal tissues (Fig. 1B). The relative mRNA expression of
FEN1 in the tumor tissues and matched normal tissues was 1.32±0.26
and 1.02±0.34, respectively. The difference between the 2 groups
(corresponding to normal and tumor tissues) was statistically
significant (P<0.01) as shown in Fig. 1C.
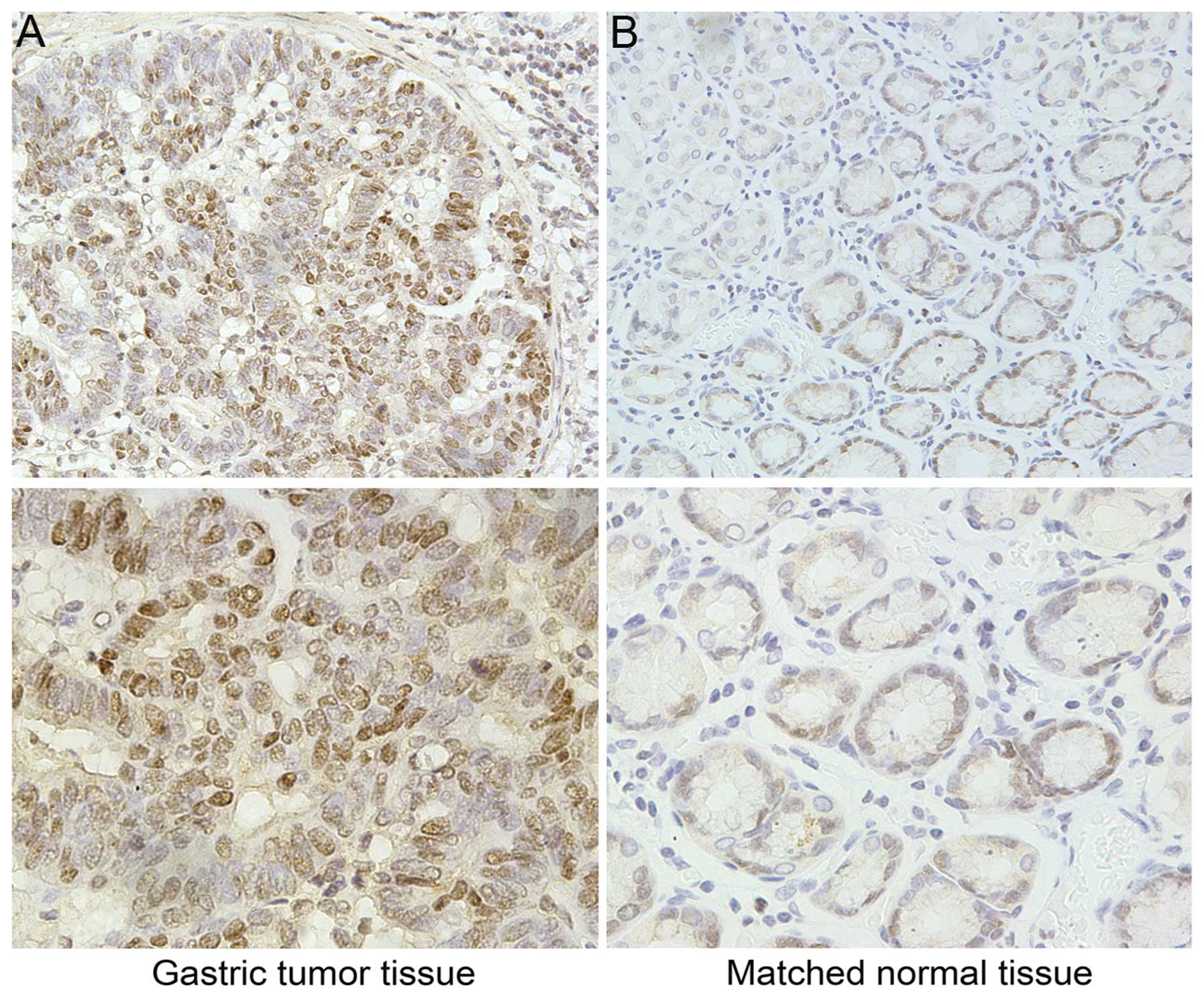
The relative protein expression of FEN1 in the 42
paired tissues was determined by immunohistochemistry. As shown in
Fig. 2, FEN1 was primarily
expressed in the nucleus and occasionally, in the cytoplasm. As
shown in Table I, 32 samples of
the 42 tumor tissues (76%) and 8 of the 42 matched normal tissues
(19%) had a positive expression of FEN1. A higher expression of
FEN1 was observed in the tumor tissues in comparison to the
corresponding normal tissues and the difference was statistically
significant as determined by the Wilcoxon rank sum test
(P<0.01).
 | Table IExpression of FEN1 in gastric cancer
and matched normal tissues. |
Table I
Expression of FEN1 in gastric cancer
and matched normal tissues.
| | FEN1 expression | |
|---|
| |
| |
|---|
| Group | Cases | Negative | Positive | P-valuea |
|---|
| Tumor tissues | 42 | 10 | 32 | P<0.01 |
| Matched normal
tissues | 42 | 34 | 8 |
Association between FEN1 expression and
clinicopathological characteristics
The results of immunohistochemical staining and the
clinicopathological characteristics of all the patients are
presented in Table II. We
observed a significant difference between the groups (positive or
negative for FEN1 expression) as regards the degree of
differentiation (P=0.027), lymphatic metastasis (P=0.001), tumor
size (P=0.026) and TNM stage (P=0.020). However, as regards age and
gender, no significant differences were observed between the groups
(P>0.05). The outcomes intimated that the expression of FEN1
positively correlates with the degree of differentiation, lymphatic
metastasis, tumor size and TNM stage in gastric cancer, but has no
significant correlation with age and gender, as shown in our 42
cases of gastric cancer.
 | Table IIAssociation between FEN1 expression
and the clinicopathological characteristics in gastric cancer
patients. |
Table II
Association between FEN1 expression
and the clinicopathological characteristics in gastric cancer
patients.
| | FEN1 | |
|---|
| |
| |
|---|
| Variable | Cases | Positive cases
(%) | Negative cases
(%) | P-valuea |
|---|
| Total cases | 42 | 32 (76.2) | 10 (23.8) | |
| Age (years) | | | | >0.05 |
| >65 | 15 | 12 (80) | 3 (20) | |
| ≤65 | 27 | 20 (74.1) | 7 (25.9) | |
| Gender | | | | >0.05 |
| Male | 26 | 20 (76.9) | 6 (23.1) | |
| Female | 16 | 12 (75) | 4 (25) | |
| Degree of
differentiation | | | | 0.027 |
| High-moderate | 16 | 9 (56.1) | 7 (43.8) | |
| Low | 26 | 23 (88.5) | 3 (11.5) | |
| Lymphatic
metastasis | | | | 0.001 |
| Negative | 18 | 9 (50) | 9 (50) | |
| Positive | 24 | 23 (95.8) | 1 (4.2) | |
| Tumor size
(cm) | | | | 0.026 |
| >3 | 23 | 21 (91.3) | 2 (8.7) | |
| ≤3 | 19 | 11 (57.9) | 8 (42.1) | |
| TNM stage | | | | 0.020 |
| I + II | 15 | 8 (53.3) | 7 (46.7) | |
| III + IV | 27 | 24 (88.9) | 3 (11.1) | |
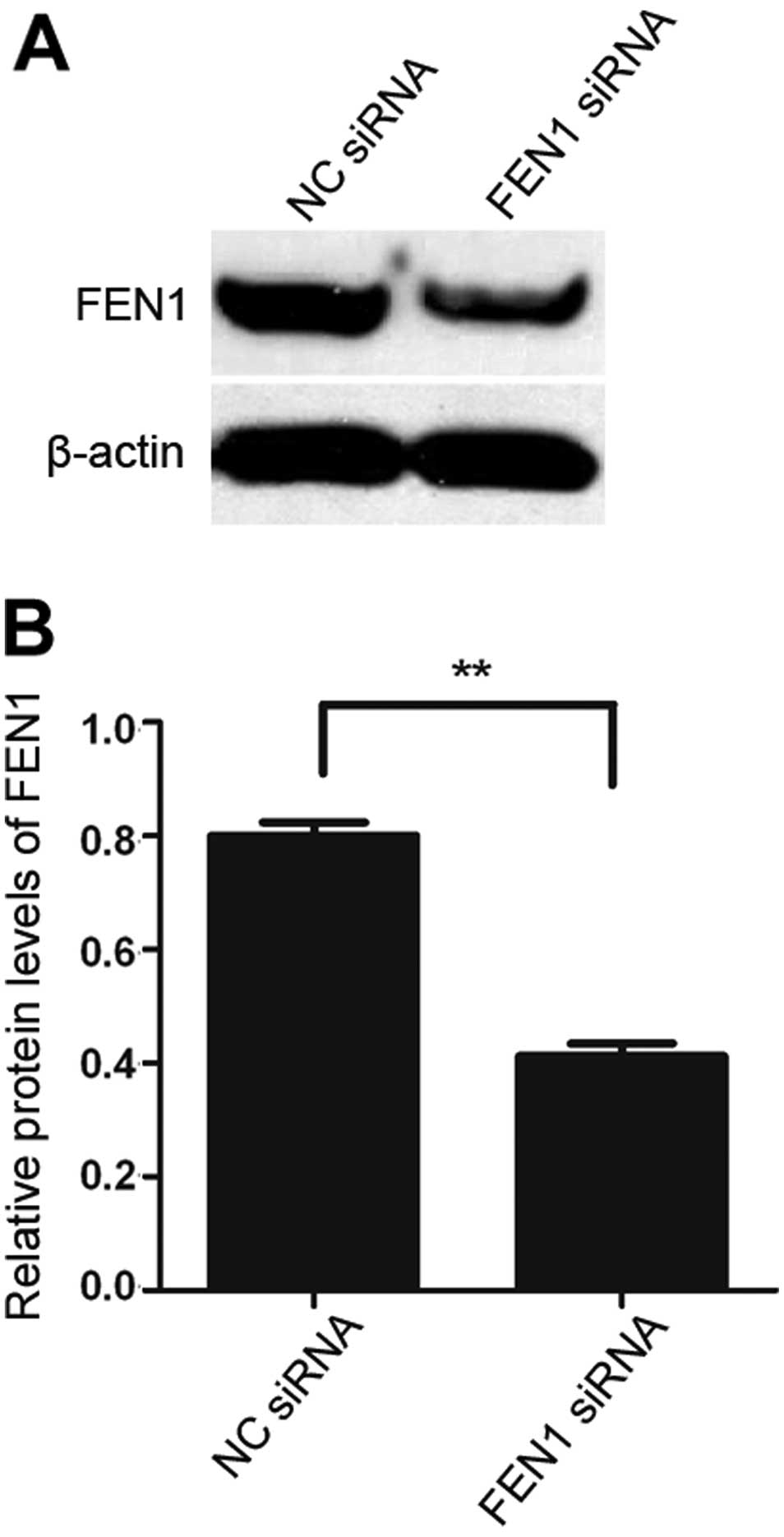
FEN1 expression is markedly inhibited by
siRNA in gastric cancer cells (SGC-7901)
In order to validate the inhibitory effects of FEN1
siRNA, the expression levels of FEN1 in the SGC-7901 cells were
determined by western blot analysis. As shown in Fig. 3, the downregulation of FEN1
expression was observed in the SGC-7901 cells transfected with FEN1
siRNA compared to the cells in the negative control group
(transfected with NC siRNA; P<0.01). These results demonstrated
that FEN1 expression in the SGC-7901 cells was markedly decreased
following transfection with FEN1 siRNA.
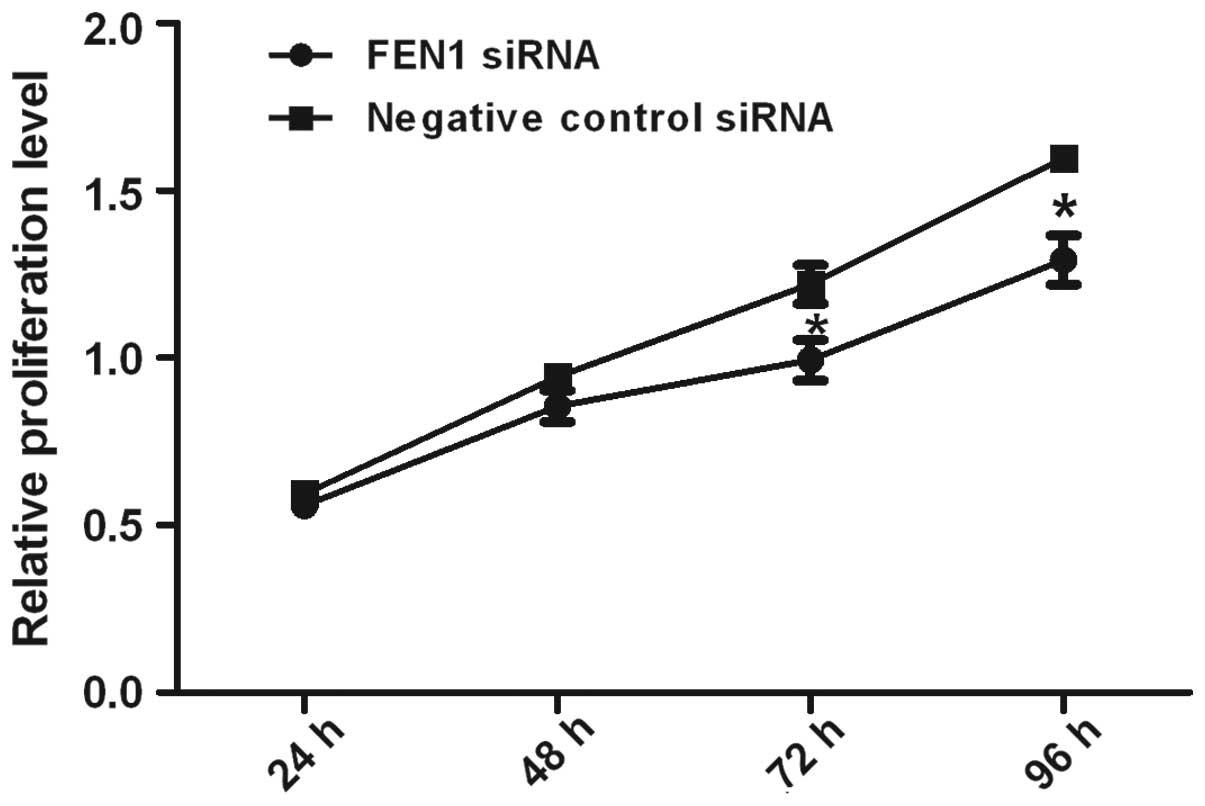
Downregulation of FEN1 expression
suppresses SGC-7901 cell proliferation
The proliferation of the SGC-7901 cells transfected
with FEN1 siRNA was determined to be markedly suppressed at 24, 48,
72 and 96 h in comparison to the cells transfected with the
negative control (NC siRNA). The data from MTS assay revealed that
the proliferation of the SGC-7901 gastric cancer cells was
suppressed following the downregulation of FEN1 gene expression by
siRNA (Fig. 4).
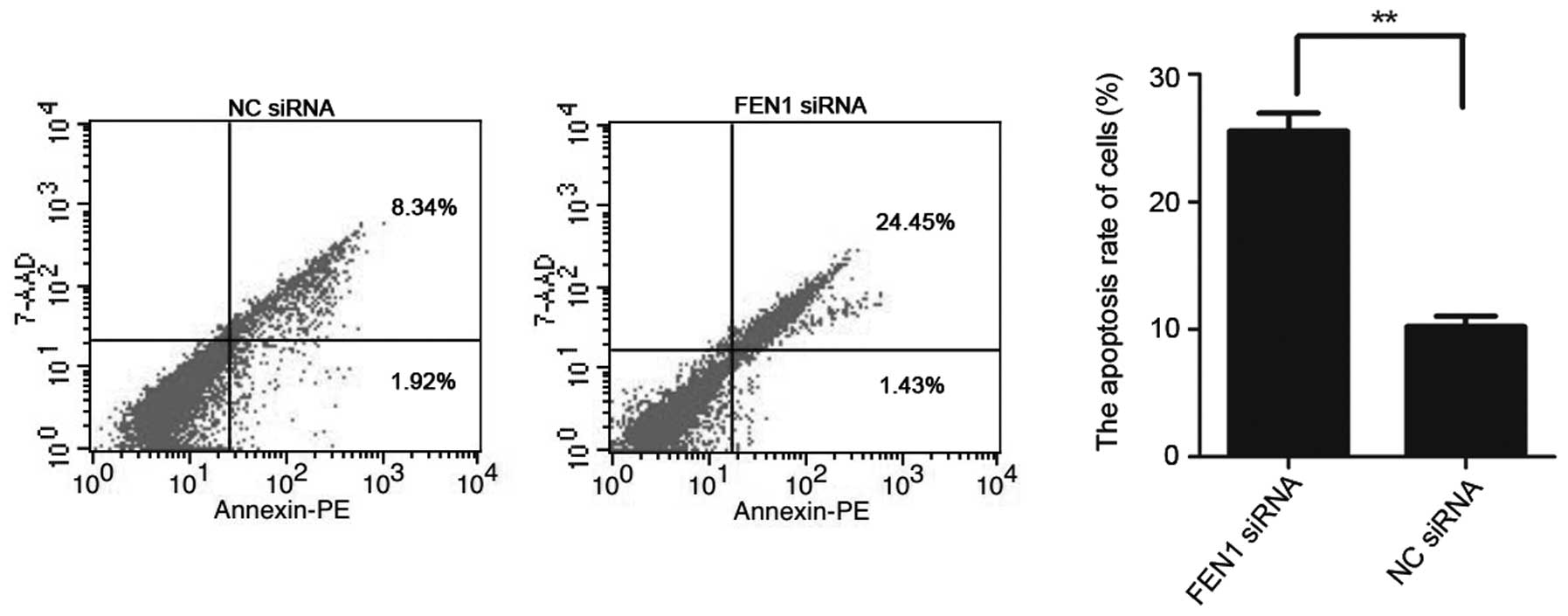
Downregulation of FEN1 expression induces
the apoptosis of SGC-7901 cells
The apoptotic rates of the 2 groups of cells were
detected by flow cytometry, and the results demonstrated that the
average apoptotis rate of the SGC-7901 cells transfected with FEN1
siRNA was markedly increased in comparison to the cells in the
negative control group (transfected with NC siRNA; P<0.01). The
outcomes intimated that the downregulation of FEN1 expression
induced the apoptosis of SGC-7901 gastric cancer cells (Fig. 5).
Discussion
As a structure-specific endonuclease (3), FEN1 has been confirmed to be a DNA
replication protein related to the coordination of various
fundamental DNA transactions (26) and the prevention of cell
carcinogenesis (4–14). Therefore, FEN1 is widely
considered as an effective tumor suppressor (9) involved in the replication and
apoptosis of DNA. However, accumulating evidence indicates the
existance of a significant association between the abnormal
expression of FEN1 and the development of cancer or the progression
of pathological transformation. Although the overexpression of FEN1
is being increasingly reported in diverse types of cancer,
including stomach tumors mentioned in a few studies (24), its correlation with the
clinicopathological characteristics of gastric cancer patienits
remains unelucidated. Thus, in this study, we first determined the
expression of FEN1 in 42 paired tumor and normal tissues at the
mRNA and protein levels to validate FEN1 expression in gastric
tumors. The results demonstrated that the expression level of FEN1
was evidently greater in gastric tumor tissues compared with the
corresponding normal tissues. This is consistent with previously
reported data from a gene expression profiling array revealing the
differences in FEN1 expression between cancer tissues and matched
normal tissues (24).
Furthermore, the data obtained in the present study demonstrate
that the overexpression of FEN1 is associated with tumor size,
lymphatic metastasis, degree of differentiation and TNM stage in
gastric cancer, whereas it is not associated with age and gender
(Table II). These data suggest
that the DNA repair gene, FEN1, may be a more precise maker for
identifying gastric cancer with clinical data and selecting
patients in different situations. Owing to its involvement in DNA
replication, restoration and degradation, the increased expression
of FEN1 is also a probable response to growing DNA damage in cancer
cells.
The increased expression of FEN1 has not only been
detected in various types of tumor tissues, but also in cancer cell
lines (18,19) including gastric cancer cells
(20). It has been reported that
FEN1 may also be conductive to telomere maintenance in human cells
(11,12,27). Thus the dysfunction of FEN1 in
telomere and genomic stability may participate in the
transformation of normal cells into cancer cells. In addition, FEN1
has been detected in all proliferative cells, but in dormant cells
it has only been detected at a very low level (28). A previous study highlighted that
FEN1 participates in the progression of mouse gastrointestinal
tract cancer in the form of haploinsufficiency (15). This suggests that FEN1 may be
necessary in the growth and progression of tumors. The expression
level of FEN1 increases with tumor grade and dedifferentiation
(21,24). In contrast to its role as an
effective tumor suppressor, it is presumed that the increased
expression of FEN1 contributes to the hyper-proliferation of tumor
cells (29).
Therefore, in this study, to further explore the
effects of FEN1 on proliferation and apoptosis in carcinoma cell
lines in vitro, we downregulated FEN1 using siRNA in the
SGC-7901 gastric cancer cell line and determined this downregution
by western blot analysis. Our results revealed that the cell growth
of FEN1-silenced cells was markedly inhibited and the apoptotic
rate of these cells was significantly greater in comparison with
the cells in the negative control group (transfected with NC
siRNA), indicating that the downregulatiion of FEN1 in SGC-7901
gastric cancer cells suppressed proliferation and induced
apoptosis. These data suggest that FEN1 may be an effective
therapeutic target in human gastric cancer. Several FEN1
inhibitors, including 2,4-diketobutyric acids and N-hydroxyurea
series, were discovered and have been developed over the years,
permitting sensitization to DNA injury agents (30,31). These may be promising treatment
methods that could enhance the traditional chemotherapeutics for
stomach and other types of cancer.
In conclusion, our results revealed that FEN1 was
overexpressed in gastric cancer in comparison with corresponding
normal tissues, and the high expression of FEN1 positively
correlated with tumor size, lymphatic metastasis, degree of
differentiation and TNM stage in gastric cancer. Moreover, the
downregulation of FEN1 suppressed the proliferation and induced the
apoptosis of SGC-7901 gastric carcinoma cells. Therefore, FEN1 may
be used as an effective biomarker for the diagnosis and treatment
of gastric cancer. To the best of our knowledge, ours is the first
study to report the association between FEN1 expression and the
clinicopathological characteristics of gastric cancer patients, as
well as the effects of silencing FEN1 on the proliferation and
apoptosis of SGC-7901 gastric cancer cells.
Acknowledgements
We appreciate the assistance provided by the
Department of Gastrointestinal Surgery, The First Affiliated
Hospital of Chongqing Medical University. This study was supported
by the Research Fund of Chongqing Municipal Health Bureau (grant
no. 2009-2-345).
References
|
1
|
Mitelman F: Catalogue of Chromosomes
Aberrations in Cancer. Cytogenet Cell Genet. 36:1–515.
1983.PubMed/NCBI
|
|
2
|
Hartgrink HH, Jansen EPM, van Grieken NCT
and van de Velde CJH: Gastric cancer-Authors’ reply. Lancet.
374:1594–1595. 2009.
|
|
3
|
Harrington JJ and Lieber MR: The
characterization of a mammalian DNA structure-specific
endonuclease. EMBO J. 13:1235–1246. 1994.PubMed/NCBI
|
|
4
|
Liu Y, Kao HI and Bambara RA: Flap
endonuclease 1: a central component of DNA metabolism. Ann Rev
Biochem. 73:589–615. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frank G, Qiu J, Somsouk M, et al: Partial
functional deficiency of E160D flap endonuclease-1 mutant in vitro
and in vivo is due to defective cleavage of DNA substrates. J Biol
Chem. 273:33064–33072. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen B, Singh P, Liu R, et al: Multiple
but dissectible functions of FEN-1 nucleases in nucleic acid
processing, genome stability and diseases. Bioessays. 27:717–729.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng L, Zhou M, Chai Q, et al: Novel
function of the flap endonuclease 1 complex in processing stalled
DNA replication forks. EMBO Rep. 6:83–89. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsutakawa SE, Classen S, Chapados BR, et
al: Human flap endonuclease structures, DNA double-base flipping,
and a unified understanding of the FEN1 superfamily. Cell.
145:198–211. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Henneke G, Friedrich-Heineken E and
Hübscher U: Flap endonuclease 1: a novel tumour suppressor protein.
Trends Biochem Sci. 28:384–390. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saharia A, Teasley DC, Duxin JP, Dao B,
Chiappinelli KB and Stewart SA: FEN1 ensures telomere stability by
facilitating replication fork re-initiation. J Biol Chem.
285:27057–27066. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saharia A, Guittat L, Crocker S, et al:
Flap endonuclease 1 contributes to telomere stability. Curr Biol.
18:496–500. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sampathi S, Bhusari A, Shen B and Chai W:
Human flap endonuclease I is in complex with telomerase and is
required for telomerase-mediated telomere maintenance. J Biol Chem.
284:3682–3690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klungland A and Lindahl T: Second pathway
for completion of human DNA base excision-repair: reconstitution
with purified proteins and requirement for DNase IV (FEN1). EMBO J.
16:3341–3348. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh P, Zheng L, Chavez V, Qiu J and Shen
B: Concerted action of exonuclease and Gap-dependent endonuclease
activities of FEN-1 contributes to the resolution of triplet repeat
sequences (CTG)n- and (GAA)n-derived secondary structures formed
during maturation of Okazaki fragments. J Biol Chem. 282:3465–3477.
2007. View Article : Google Scholar
|
|
15
|
Kucherlapati M, Yang K, Kuraguchi M, et
al: Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid
tumor progression. Proc Natl Acad Sci USA. 99:9924–9929. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng L, Dai H, Zhou M, et al: Fen1
mutations result in autoimmunity, chronic inflammation and cancers.
Nat Med. 13:812–819. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Larsen E, Kleppa L, Meza TJ, et al:
Early-onset lymphoma and extensive embryonic apoptosis in two
domain-specific Fen1 mice mutants. Cancer Res. 68:4571–4579. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sato M, Girard L, Sekine I, et al:
Increased expression and no mutation of the Flap endonuclease
(FEN1) gene in human lung cancer. Oncogene. 22:7243–7246. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
LaTulippe E, Satagopan J, Smith A, et al:
Comprehensive gene expression analysis of prostate cancer reveals
distinct transcriptional programs associated with metastatic
disease. Cancer Res. 62:4499–4506. 2002.
|
|
20
|
Kim JM, Sohn HY, Yoon SY, et al:
Identification of gastric cancer-related genes using a cDNA
microarray containing novel expressed sequence tags expressed in
gastric cancer cells. Clin Cancer Res. 11:473–482. 2005.
|
|
21
|
Lam JS, Seligson DB, Yu H, et al: Flap
endonuclease 1 is overexpressed in prostate cancer and is
associated with a high Gleason score. BJU Int. 98:445–451. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krause A, Combaret V, Iacono I, et al:
Genome-wide analysis of gene expression in neuroblastomas detected
by mass screening. Cancer Lett. 225:111–120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iacobuzio-Donahue CA, Maitra A, Olsen M,
et al: Exploration of global gene expression patterns in pancreatic
adenocarcinoma using cDNA microarrays. Am J Pathol. 162:1151–1162.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh P, Yang M, Dai H, et al:
Overexpression and hypomethylation of flap endonuclease 1 gene in
breast and other cancers. Mol Cancer Res. 6:1710–1717. 2008.
|
|
25
|
Nikolova T, Christmann M and Kaina B: FEN1
is overexpressed in testis, lung and brain tumors. Anticancer Res.
29:2453–2459. 2009.PubMed/NCBI
|
|
26
|
Balakrishnan L and Bambara RA: Flap
Endonuclease 1. Ann Rev Biochem. 82:119–138. 2013. View Article : Google Scholar
|
|
27
|
Saharia A and Stewart S: FEN1 contributes
to telomere stability in ALT-positive tumor cells. Oncogene.
28:1162–1167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim IS, Lee MY, Lee IH, Shin SL and Lee
SY: Gene expression of flap endonuclease-1 during cell
proliferation and differentiation. Biochim Biophys Acta.
1496:333–340. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng L, Jia J, Finger LD, Guo Z, Zer C
and Shen B: Functional regulation of FEN1 nuclease and its link to
cancer. Nucleic Acids Res. 39:781–794. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tumey LN, Huck B, Gleason E, et al: The
identification and optimization of 2,4-diketobutyric acids as flap
endonuclease 1 inhibitors. Bioorg Med Chem Lett. 14:4915–4918.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tumey LN, Bom D, Huck B, et al: The
identification and optimization of a N-hydroxy urea series of flap
endonuclease 1 inhibitors. Bioorg Med Chem Lett. 15:277–281. 2005.
View Article : Google Scholar : PubMed/NCBI
|