Introduction
The nuclear factor erythroid 2-related factor 2
(Nrf2)-mediated signaling pathway is a major cellular defense
pathway against oxidative or electrophilic stress (1). Nrf2, a cap ‘n’ collar (CNC) basic
leucine zipper transcription factor, can bind to antioxidant
response element (ARE) in the nucleus, subsequently initiating the
expression of genes encoding antioxidant and phase II
drug-metabolizing enzymes (2–4).
These genes include glutathione S-transferase (GST) (5,6),
NAD(P)H: quinine reductases (NQO1) (7,8)
and heme oxygenase (HO-1) (9,10),
which contribute to detoxification and elimination of reactive
oxidants and electrophilic agents. It has been documented that
certain synthetic or natural compounds can activate the
Nrf2-mediated signaling pathway and provide protection against
environmental or chemical insults (11–14). The knockout of Nrf2 in mice has
been shown to reduce not only basal, but also the inducible
expression of antioxidant and phase II drug-metabolizing enzymes
(15,16). Silencing of the Nrf2 gene
increases susceptibility to various oxidative stress-related
pathologies, including chemical carcinogenesis, acetaminophen
toxicity and hyperoxia (17–19). Therefore, Nrf2 has also been
considered as a potential target for preventing
inflammation-associated and chemical-induced carcinogenesis
(20,21).
However, the regulatory mechanisms involved in Nrf2
activation are not yet fully understood. Although it is well
established that Nrf2 activity is controlled, at least in part, by
the cytosolic protein, Kelch-like ECH-associated protein 1 (Keap1),
the mechanisms by which Keap1 acts to repress Nrf2 activity remain
to be fully characterized. Previously, it was considered that most
of the Nrf2 proteins were bound to Keap1 and sequestered in
cytoplasm under homeostatic conditions (22,23). In response to oxidative stress or
Nrf2 inducers/activators, the Nrf2 protein can dissociate from
Keap1, transfer from the cytoplasm to the nucleus, bind to ARE and
lead to the subsequent transcription of ARE-regulated genes
(24,25). Thus, Nrf2 activation seems to be
the result of its nuclear translocation. Nevertheless, a number of
studies have suggested a different possible mechanism. Nrf2 was
found to be primarily a nuclear protein. Under homeostatic
conditions, Keap1 transiently shuttles the Nrf2 protein from the
nucleus to the cytoplasm, prompts Nrf2 protein ubiquitination and
degradation in the cytoplasm, maintains the intracellular Nrf2
protein at a basal level, thus repressing its activity (26). Oxidative stress or Nrf2
inducers/activators regulate the interaction between Nrf2 and
Keap1, inhibiting Nrf2 protein degradation and increasing its
stability, which leads to Nrf2 protein accumulation in stressed
cells. Therefore, Nrf2 activation has been suggested to be
dependent on increasing Nrf2 protein stability (1,14,27).
Mangiferin (MA),
2-C-β-D-glucopyranosyl-1,3,6,7-tetrahydroxyxanthone, is a compound
monomer extracted from certain plants of the Anacardiaceae and
Gentianaceae families, including Mangifera indica L.
(mango), particularly in their leaves and bark (28). MA is widely used as a nutritional
supplement, and as a cosmetic and phytomedicine in Southeast Asia
and South America (29). This
natural xanthone derivative has been reported to have various
bioactivities, such as antioxidant, anti-tumor, anti-viral,
anti-diabetic, anti-inflammatory, anti-allergic and
immunomodulatory activities (28). Among these pharmacological
activities, the antioxidant and cytoprotective properties of MA
have been well elucidated. MA presents cardio-, hepato- and
neuroprotective activities, as well as radioprotective activities
against radiation (14,28,29).
Previous stuides have revealed that MA induces
Nrf2-mediated antioxidant response, which provides an explanation
for its antioxidant activity (29,30). It has been reported that MA
enhances the expression of several detoxification and antioxidant
enzymes, including NQO1, GST, HO-1, superoxide dismutase (SOD) and
uridin 5′-diphosphate-glucuronosyl transferase (UDP-GT) (33–36). Furthermore, MA has been shown to
increase Nrf2 expression in D-galactosamine intoxicated rat liver
(29). Moreover, in our previous
study, we found that MA activated Nrf2-ARE signaling in human HL60
myeloid leukemia cells (30).
However, to the best of our knowledge, the mechanisms by which MA
increases Nrf2 expression have not been documented to date.
In this study, we demonstrate that MA increases Nrf2
expression, but not transcription in human HL60 myeloid leukemia
cells. We provide evidence that MA prolongs the half-life of the
Nrf2 protein by inhibiting its ubiquitination and degradation.
Materials and methods
Reagents
MA (C19H18O11;
molecular weight, 422.34) and cycloheximide (CHX, C15H23NO4;
molecular weight, 281.4) were purchased from Sigma-Aldrich (St.
Louis, MO, USA). MG132
(C26H42N3O5; molecular
weight, 475.6) was obtained from Enzo Life Sciences (Farmingdale,
NY, USA). MA and MG132 were initially dissolved in dimethyl
sulfoxide (DMSO), stored at −20°C and thawed on ice prior to use.
CHX was dissolved in ultrapure water, stored at −80°C and thawed on
ice prior to use. RNAiso plus, PrimeScript RT Master Mix and SYBR
Premix Ex Taq were purchased from Takara Bio, Inc. (Otsu, Japan).
The primers used for real-time PCR were designed and synthesized by
Takara Bio, Inc.. Rabbit polyclonal antibody against human Nrf2
(C-20) was obtained from Santa Cruz Biotechnology (Dallas, TX,
USA). Mouse monoclonal antibody against human ubiquitin was
obtained from Merck Millipore (Billerica, MA, USA). The BCA protein
assay kit was from Pierce Biotechnology, Inc. (Rockford, IL, USA).
The Image Lab enhanced chemiluminescence (ECL) detection system was
from Bio-Rad (Hercules, CA, USA). Cell culture medium, RPMI-1640
and fetal abovine serum (FBS) were from HyClone (Logan, UT,
USA).
Cell culture
The human HL60 myeloid leukemia cell line was kindly
provided by Professor Jianfeng Zhou (Cancer Biology Research
Center, Tongji Hospital, Wuhan, China), and then maintained in
RPMI-1640 medium supplemented with 10% fetal bovine serum at 37°C
in a humidified incubator containing 5% CO2 in air.
Real-time PCR
Total RNA was isolated using RNAiso Plus according
to the manufacturer’s instructions. Total RNA was isolated using
RNAiso Plus, and then 400 ng total RNA were reverse transcripted
into cDNA. Subsequently, 2 μl cDNA were amplified with SYBR-Green
Universal PCR Master mix in triplicate on a real-time PCR system
(CFX96, Bio-Rad). Nrf2 mRNA levels related to β-actin levels were
calculated using the ΔCt (cycle threshold) method. The primer
sequences for human Nrf2 were: forward,
5′-ACTCCGGCATTTCACTAAACACAAG-3′ and reverse,
5′-CTGAGGCCAAGTAGTGTGTCTCCA-3′. The primer sequences for human
β-actin were: forward, 5′-GCCCAGTCCTCTCCCAAGTC-3′ and reverse,
5′-GGCACGAAGGTCATCATTC-3′. Control cells were processed in an
identical manner apart from MA treatment.
Western blot analysis
For immunoblotting, whole-cell lysates were prepared
using lysis buffer for 30 min on ice. Supernatants were collected
as samples following centrifuged at 15,000 rpm for 10 min at 4°C.
Protein concentrations were determined using a BCA protein assay
kit (Pierce Biotechnology, Inc.). Following denaturation, equal
amounts of the protein extracts were resolved by 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a polyvinylidene difluoride (PVDF) membrane. The
membrane was blocked in phosphate-buffered saline (PBS) containing
5% non-fat milk for 1 h at room temperature. It was subsequently
incubated with anti-Nrf2 antibody (dilution 1:1,000) at 4°C
overnight, followed by washing and treatment with HRP-labeled
secondary antibody (Pierce Biotechnology, Inc.) for 2 h at room
temperature. The blots were incubated with ECL reagent for 5 min,
and the signals were then detected with a chemiluminescence
detection system (Bio-Rad). After stripping, the membrane was
re-probed with human anti-β-actin antibody as a control for equal
protein loading and protein integrity.
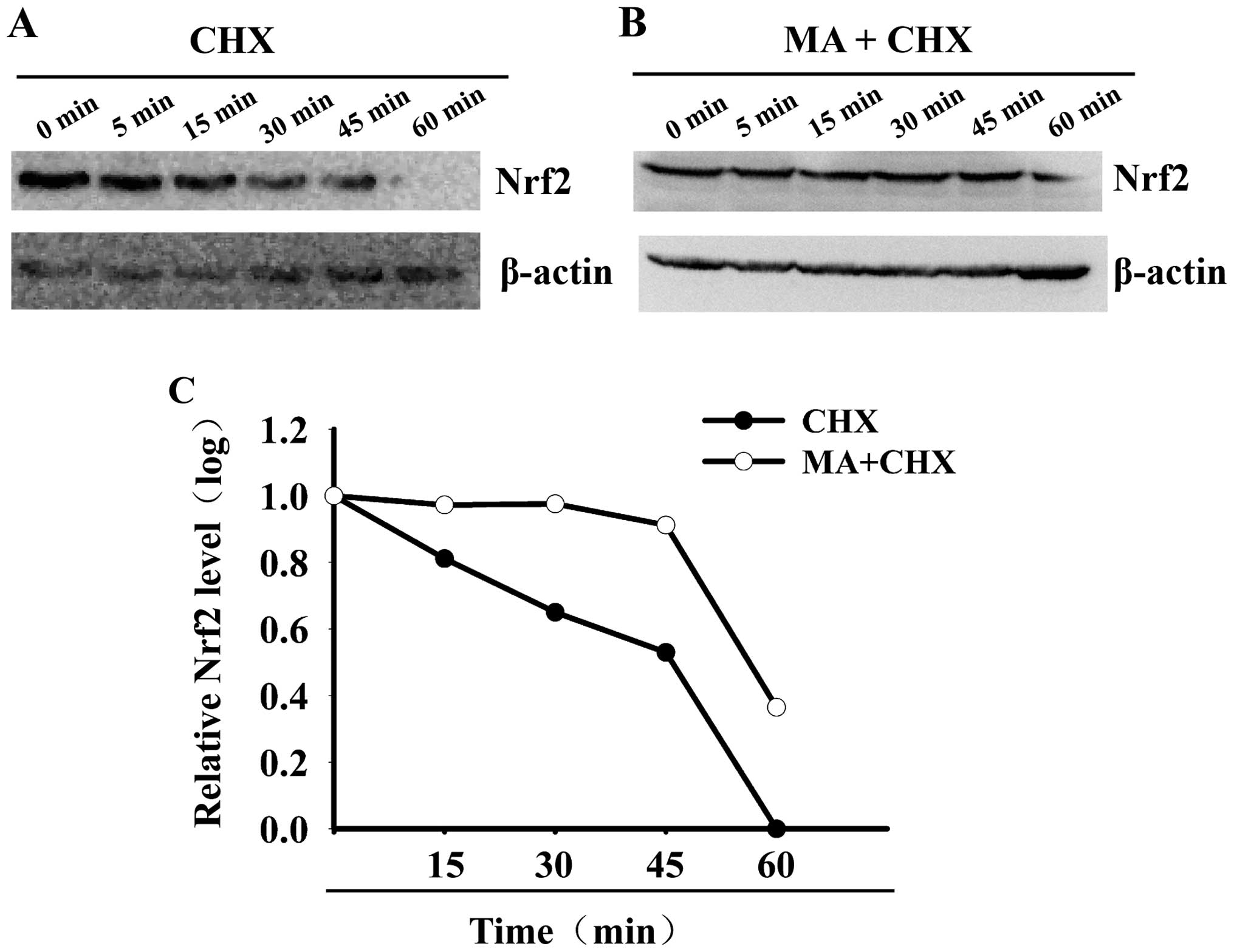
CHX-chase analysis
Nrf2 protein degradation was analyzed by CHX-chase
analysis. The HL60 cells were pre-incubated with or without 50 μM
MA for 4 h. Subsequently, 100 μg/ml CHX were added to inhibit
protein synthesis. The cells were collected at 0, 5, 15, 30, 45 and
60 min after following treatment with CHX, as previously described
(31). Total protein (60 μg) was
fractionated on SDS-PAGE and blotted with anti-Nrf2 and
anti-β-actin antibodies. The results from western blot analysis
were quantified by densitometry. Nrf2 protein half-lives were
calculated from the slope of the semi-logarithmically transformed
best fit line. The decay curves were analyzed individually using
linear regression of protein amount, and expressed as a percentage
of protein remaining vs. time, as previously described (39).
Immunoprecipitation (IP)
The HL60 cells were treated with 50 μM MA, 10 μM
MG132, or a combination of 50 μM MA and 10 μM MG132 for 4 h. The
cells were washed twice with ice-cold PBS. The cells were then
prepared in lysis/IP buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 1%
Nonidet P-40, 0.5% DOC, 0.1% SDS, 50 mM NaF, 1 mM
Na3VO4, 20 mM β-glycerophosphate and 1 mM
okadaic acid] on ice for 30 min, then centrifuged at 14,000 rpm for
15 min. Cell lysates were incubated with anti-Nrf2 antibody at 4°C
for 16 h. The immune complexes were then precipitated with protein
A-Sepharose beads at 4°C for an additional 2 h. Subsequently, the
precipitates were washed extensively with IP buffer, fractionated
by SDS-PAGE and immunobloted with anti-ubiquitin antibody, as
previously described (27).
Control cells were processed in an identical manner apart from
treatment with MA or/and MG132.
Statistical analysis
Data are expressed as the mean ± SD of at least 3
independent experiments and processed by SPSS 17.0 statistical
software for Windows. One-way ANOVA and Student-Newman-Keuls tests
were applied for comparisons between each group. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
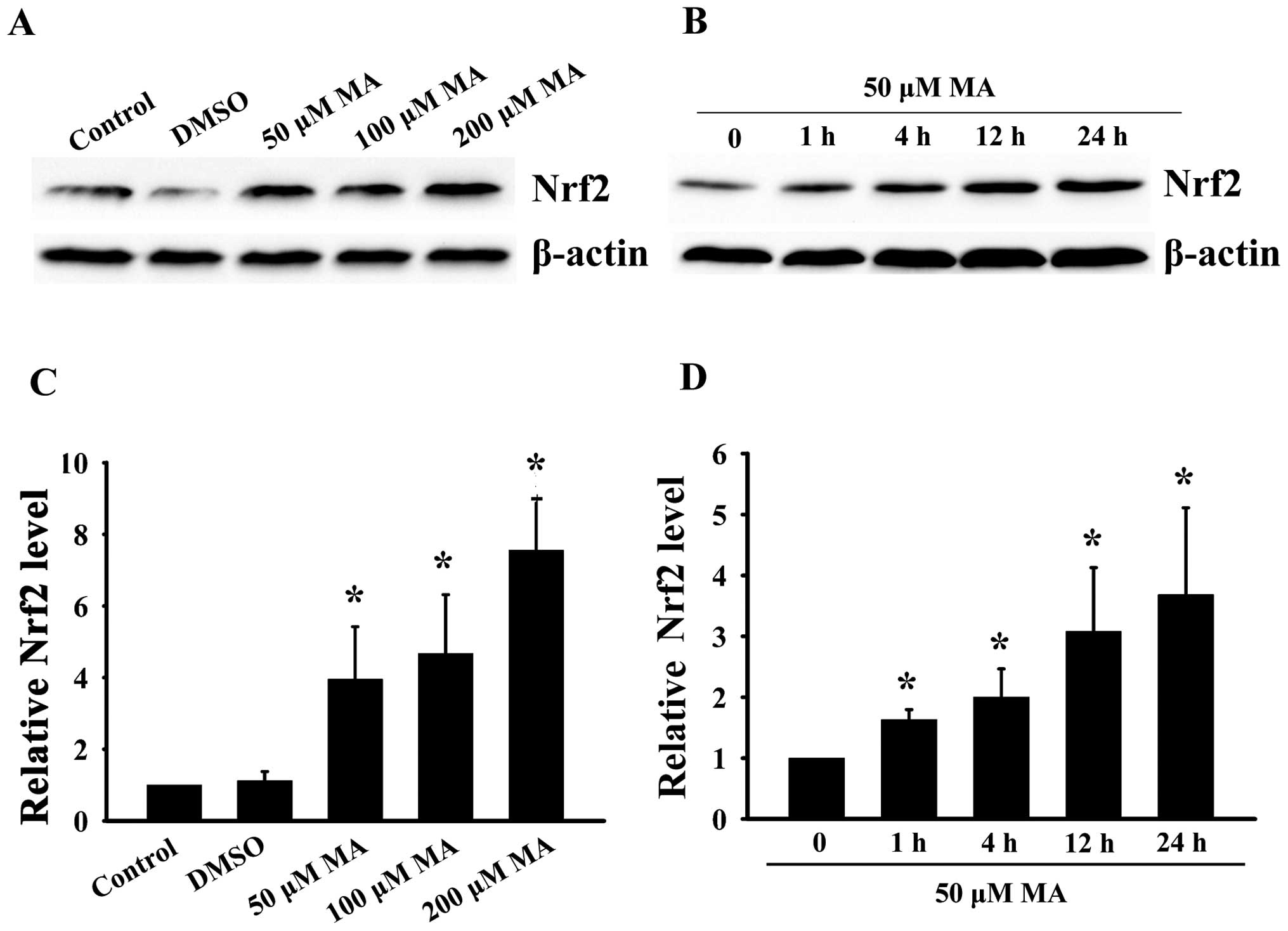
MA markedly increases Nrf2 expression in
HL60 cells
To determine the effects of MA on Nrf2 expression,
the HL60 cells were treated with MA in dose-and time-course
experiments. The Nrf2 protein levels in whole cell lysate were then
detected by western blot analysis. As shown in Fig. 1, the HL60 cells were treated with
50, 100 or 200 μM MA for 24 h. These treatments resulted in a
dose-dependent increase in the Nrf2 protein level. The Nrf2 protein
level increased to 3.95-fold of the basal value following treatment
with 50 μM MA, and increased to nearly 7.56-fold of the basal level
following treating with 200 μM MA for 24 h. In the time-course
experiments, Nrf2 expression also markedly increased in a
time-dependent manner when the HL60 cells were treated with 50 μM
MA for 1, 4, 12 or 24 h.
MA does not affect Nrf2 transcription in
HL60 cells
To determine the effects of MA on Nrf2
transcription, the HL60 cells were treated with MA in dose-and
time-course experiments as described above, and the Nrf2 mRNA
levels were then detected by real-time PCR. As shown in Fig. 2, the Nrf2 mRNA levels did not
differ significantly following treatment with MA, neither in a
dose-dependent nor in a time-dependent manner.
MA prolongs the half-life of Nrf2 protein
in HL60 cells
To determine the effects of MA on Nrf2 protein
stability, the half-life of Nrf2 protein was calculated in the
MA-treated and non-MA-treated HL60 cells. As shown in Fig. 3, the Nrf2 protein level decreased
by ~50% within 20 min in the non-MA-treated HL60 cells. Its
half-life was only 20 min. However, the Nrf2 protein level
decreased by ~50% after 50 min in the MA-treated cells. Its
half-life was significantly prolonged to 58 min. These data
indicate that MA increases Nrf2 protein stability and prolongs its
half-life.
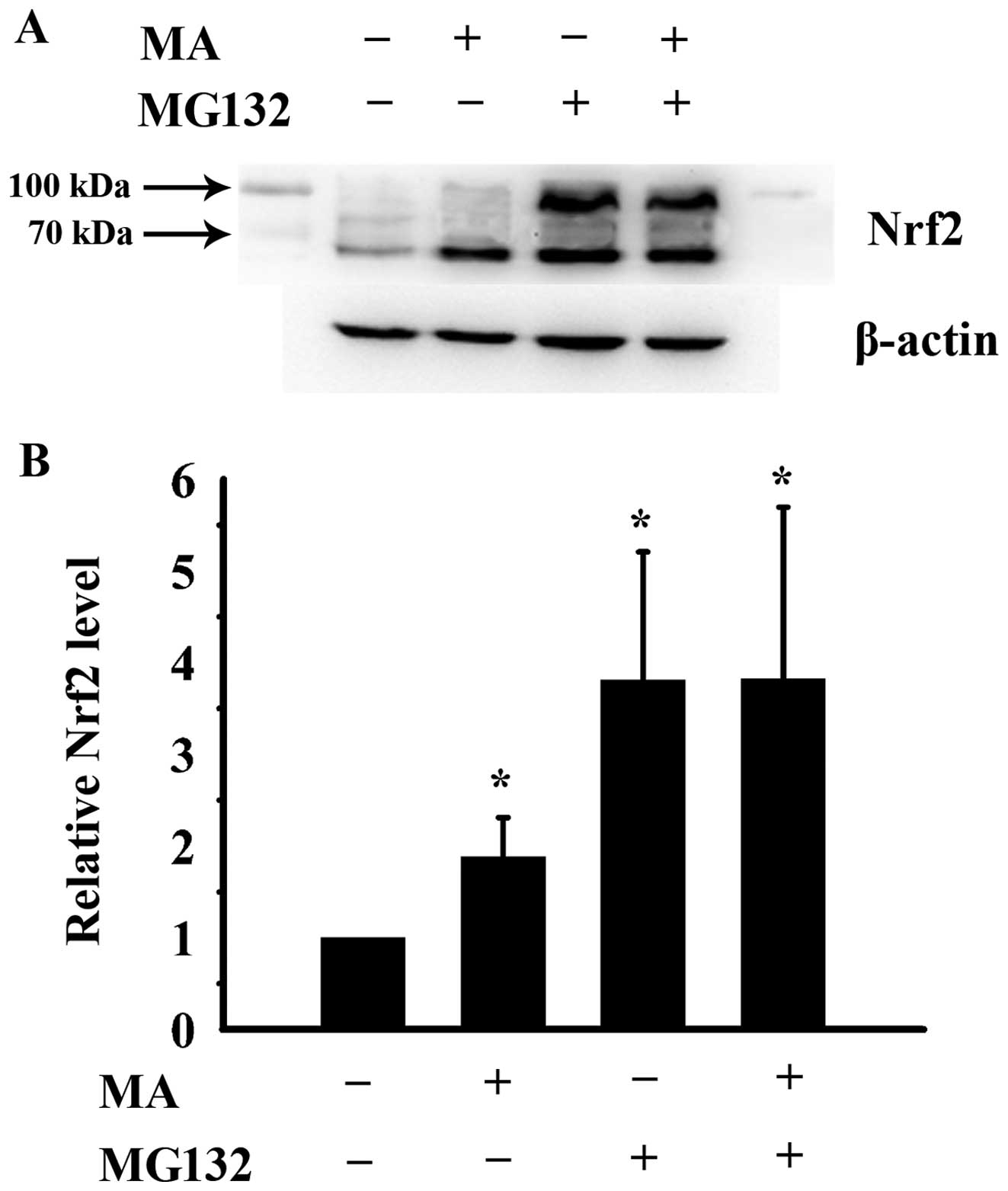
MA increases Nrf2 stability by
interfering with the ubiquitin-proteasome protein degradation
pathway
To investigate the mechanisms by which MA increases
Nrf2 protein stability, the HL60 cells were treated with 50 μM MA,
10 μM of the proteasome inhibitor, MG132, or a combination of 50 μM
MA and 10 μM MG132 for 4 h. The cells were then prepared for the
analysis of the Nrf2 level by western blot analysis. The molecular
weights of Nrf2 and poly-ubiquitinated Nrf2 are 57 and 100 kDa,
respectively, according to the Nrf2 antibody data sheet (Santa Cruz
Biotechnology). As shown in Fig.
4, the Nrf2/poly-ubiquitinated Nrf2 protein level significantly
increased to 1.88-, 3.81- and 3.82-fold of that of the control
cells following treatment with MA, MG132 or the combination
treatment, respectively. The total Nrf2 level following treatment
with MG132 alone did not differ significantly from that observed
following combined treatment with MA and MG132. Of note, MA mainly
increased the non-ubiquitinated Nrf2 protein level. However, MG132
enhanced both the non-ubiquitinated and poly-ubiquitinated Nrf2
protein levels. These results indicate that MA increases Nrf2
protein stability by interfering with the ubiquitin-proteasome
protein degradation pathway. However, the mechanisms involved
differ from those of the proteasome inhibitor, MG132. MA inhibits
ubiquitination, while MG132 suppresses proteasome activity.
MA inhibits Nrf2 ubiquitination in HL60
cells
To investigate whether MA suppresses Nrf2
ubiquitination, we performed IP experiments to pull down Nrf2 from
the cell lysis of HL60 cells, and then identified ubiquitinated
Nrf2 by immunoblot analysis with anti-ubiquitin monoclonal
antibody. As shown in Fig. 5, the
10 kDa band represents ubiquitinated Nrf2; the levels of
ubiquitinated Nrf2 decreased when the HL60 cells were treated with
MA. On the contrary, these conjugated proteins significantly
accumulated when the cells were incubated with the proteasome
inhibitor, MG132. These results suggest that MA decreases Nrf2
degradation by inhibiting its ubiquitination.
Discussion
The present study demonstrates that the natural
antioxidant, mangiferin (MA), increases Nrf2 expression through
post-translational mechanisms in hematopoietic cells. MA enhances
the Nrf2 protein level, but does not affect its transcription. MA
reduces protein degradation and prolongs the half-life of Nrf2 by
inhibiting its ubiquitination, which leads to its intracellular
accumulation. These results provide evidence that MA enhances Nrf2
expression by interfering with the ubiquitin-proteasome protein
degradation pathway and increasing its protein stability. As
mentioned above, MA activates Nrf2 and this activation may result
from the increased protein stability and subsequent intracellular
accumulation (1,27). Therefore, our data suggest that MA
activates the Nrf2-mediated signaling pathway by increasing Nrf2
stability.
Previous studies have confirmed that Nrf2 is
targeted for rapid degradation by the ubiquitin-proteasome pathway.
Nrf2 is a highly unstable protein and its half-life is only 15–30
min in unstressed cells (32–36). In our study, the Nrf2 half-life
was ~20 min in the HL60 cells and MA increased its half-life to 58
min, which suggests that MA increases Nrf2 protein stability. Under
homeostatic conditions, Nrf2 binds to its repressor, Keap1. Keap1
brings Nrf2 to the Cul3-E3 ubiquitin ligase and targets the 26S
proteasome for protein degradation (26). However, oxidative stress can
antagonize the Keap1-Nrf2 interaction, increasing Nrf2 stability,
leading to Nrf2 accumulation within cells (32,33,35). Therefore, an increase in Nrf2
stability can be a result of the interference with the Keap1-Nrf2
interaction, inhibiting Nrf2 ubiquitination or reducing 26S
proteasome activity.
In the present study, MA markedly inhibited Nrf2
ubquitination, which may explain the increase in Nrf2 stability.
Certain natural or synthetic Nrf2 inducers/activators can also
increase Nrf2 protein stability; however, the mechanisms involved
are not completely similar to those of MA (11–14,43). Treatment with
tert-butylhydroquinone (tBHQ) has been shown to prolong the
half-life of Nrf2 protein in human neural stem cells. However, tBHQ
increases ubiquitinated Nrf2, bu MA decreases ubiquitinated Nrf2
(14). This Nrf2 activator seems
to increase Nrf2 protein stability by stabilizing ubiquitinated
Nrf2, which differs from MA. Oridonin, a diterpenoid purified from
the Chinese medicinal herb, Rabdosia rubescens, was found to
suppress Nrf2 ubiquitination and enhance Keap1 ubiquitination in
human MDAMB-231 breast carcinoma cells (12). Thus, oridonin may induce a shift
in ubiquitination from the substrate, Nrf2, to the substrate
adaptor, Keap1. Ajoene, a stable garlic by-product, has been shown
to inhibit the Nrf2-Keap1 interaction and decrease Nrf2
ubiquitination in HepG2 cells (13). The latter two Nrf2 activators
increase Nrf2 stability through at least partly similar mechanisms
to those of MA. Another Nrf2 activator 1, 2-dithiole-3-thione (D3T)
has also been shown to significantly reduce the degradation of Nrf2
protein in PC12 cells. Nevertheless, it was unexplored as to which
step of the ubiquitin-proteasome degradation of Nrf2 protein was
affected by D3T (11). Therefore,
Nrf2 activators can increase Nrf2 protein stability by interfering
with the ubiquitin-proteasome pathway; however, the specific
mechanisms involved may differ. The mechanisms through which MA
inhibits Nrf2 ubiquitinationt require further investigation.
As a newly identified Nrf2 activator, MA may be a
potential cytoprotective agent for hematopoietic cells, as well as
a chemopreventive agent against leukemia. It has been well
documented that Nrf2 activators/inducers exert cytoprotective
effects through antioxidant mechanisms (37–39). Our previous study also
demonstrated that MA relieved etoposide-induced DNA damage by
activating Nrf2-mediated signaling and increasing NQO1 expression
in mononuclear human umbilical cord blood (MNC hUCB) cells (Li S,
et al, ASH Annual Meeting Abstracts 118: abs. 4626, 2011).
This suggests that MA protects hematopoietic cells against injury
induced by chemotherapy. Moreover, Nrf2 is a key target for
chemoprevention against carcinogenesis (40–42). One major molecular mechanism is
the induction of detoxification cytoprotective enzymes by Nrf2
activation (2). NQO1, a
well-known detoxification and cytoprotective enzyme, is subject to
a genetic polymorphism (C609T) leading to the impairment of
intermediate NQO1 activity. It has been reported that the NQO1
C609T polymorphism significantly increases the risk of
treatment-related leukemia and myelodysplastic leukemia (43,44). It has also been noted that
oxidative stress is strongly associated with the relapse of acute
myeloid leukemia and a poor prognosis (45,46). Therefore, there is a possibility
that MA may protect hematopoietic cells from leukemia genesis and
relapse by activating the Nrf2-mediated antioxidant response; this,
however, requires further investigation.
In conclusion, our study confirms that MA inhibits
Nrf2 ubiquitination and increases its stability. This may be one of
the mechanims through which it induces Nrf2 cellular accumulation
and activates Nrf2-mediated signaling. MA may be a potential
cytoprotective agent for hematopoietic cells and a chemopreventive
agent against leukemia, which warrants further study.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 30900632 and no.
81372541). The authors would like to thank the Department of
Central Laboratory, Union Hospital, Tongji Medical College,
Huazhong University of Science and Technology, Wuhan, China, for
providing relevant experimental facilities and technical
support.
References
|
1
|
Nguyen T, Nioi P and Pickett CB: The
Nrf2-antioxidant response element signaling pathway and its
activation by oxidative stress. J Biol Chem. 284:13291–13295. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eggler AL, Gay KA and Mesecar AD:
Molecular mechanisms of natural products in chemoprevention:
induction of cytoprotective enzymes by Nrf2. Mol Nutr Food Res.
52(Suppl 1): S84–S94. 2008.PubMed/NCBI
|
|
3
|
Huang HC, Nguyen T and Pickett CB:
Regulation of the antioxidant response element by protein kinase
C-mediated phosphorylation of NF-E2-related factor 2. Proc Natl
Acad Sci USA. 97:12475–12480. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nguyen T, Huang HC and Pickett CB:
Transcriptional regulation of the antioxidant response element.
Activation by Nrf2 and repression by MafK. J Biol Chem.
275:15466–15473. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friling RS, Bensimon A, Tichauer Y, et al:
Xenobiotic-inducible expression of murine glutathione S-transferase
Ya subunit gene is controlled by an electrophile-responsive
element. Proc Natl Acad Sci USA. 87:6258–6262. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Casalino E, Calzaretti G, Landriscina M,
et al: The Nrf2 transcription factor contributes to the induction
of alpha-class GST isoenzymes in liver of acute cadmium or
manganese intoxicated rats: comparison with the toxic effect on
NAD(P)H:quinone reductase. Toxicology. 237:24–34. 2007. View Article : Google Scholar
|
|
7
|
Liang L, Gao C, Luo M, et al:
Dihydroquercetin (DHQ) induced HO-1 and NQO1 expression against
oxidative stress through the Nrf2-dependent antioxidant pathway. J
Agric Food Chem. 61:2755–2761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim HJ, Zheng M, Kim SK, et al: CO/HO-1
induces NQO-1 expression via Nrf2 activation. Immune Netw.
11:376–382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee YM, Auh QS, Lee DW, et al: Involvement
of nrf2-mediated upregulation of heme oxygenase-1 in
mollugin-induced growth inhibition and apoptosis in human oral
cancer cells. Biomed Res Int. 2013:2106042013.PubMed/NCBI
|
|
10
|
Maruyama A, Mimura J, Harada N, et al:
Nrf2 activation is associated with Z-DNA formation in the human
HO-1 promoter. Nucleic Acids Res. 41:5223–5234. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong J, Yan D and Chen SY: Stabilization
of Nrf2 protein by D3T provides protection against ethanol-induced
apoptosis in PC12 cells. PLoS One. 6:e168452011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du Y, Villeneuve NF, Wang XJ, et al:
Oridonin confers protection against arsenic-induced toxicity
through activation of the Nrf2-mediated defensive response. Environ
Health Perspect. 116:1154–1161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kay HY, Won Yang J, Kim TH, et al: Ajoene,
a stable garlic by-product, has an antioxidant effect through
Nrf2-mediated glutamate-cysteine ligase induction in HepG2 cells
and primary hepatocytes. J Nutr. 140:1211–1219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J: Stabilization of Nrf2 by tBHQ
confers protection against oxidative stress-induced cell death in
human neural stem cells. Toxicol Sci. 83:313–328. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kurzatkowski DM and Trombetta LD: Maneb
causes pro-oxidant effects in the hippocampus of Nrf2 knockout
mice. Environ Toxicol Pharmacol. 36:427–436. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mukhopadhyay S, Sekhar KR, Hale AB, et al:
Loss of NRF2 impairs gastric nitrergic stimulation and function.
Free Radic Biol Med. 51:619–625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yates MS, Tran QT, Dolan PM, et al:
Genetic versus chemoprotective activation of Nrf2 signaling:
overlapping yet distinct gene expression profiles between Keap1
knockout and triterpenoid-treated mice. Carcinogenesis.
30:1024–1031. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ni HM, Boggess N, McGill MR, et al:
Liver-specific loss of Atg5 causes persistent activation of Nrf2
and protects against acetaminophen-induced liver injury. Toxicol
Sci. 127:438–450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reddy NM, Kleeberger SR, Kensler TW, et
al: Disruption of Nrf2 impairs the resolution of hyperoxia-induced
acute lung injury and inflammation in mice. J Immunol.
182:7264–7271. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kundu JK and Surh YJ: Nrf2-Keap1 signaling
as a potential target for chemoprevention of
inflammation-associated carcinogenesis. Pharm Res. 27:999–1013.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao CR, Gao ZH and Qu XJ: Nrf2-ARE
signaling pathway and natural products for cancer chemoprevention.
Cancer Epidemiol. 34:523–533. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tong KI, Kobayashi A, Katsuoka F, et al:
Two-site substrate recognition model for the Keap1-Nrf2 system: a
hinge and latch mechanism. Biol Chem. 387:1311–1320.
2006.PubMed/NCBI
|
|
23
|
McMahon M, Thomas N, Itoh K, et al:
Dimerization of substrate adaptors can facilitate cullin-mediated
ubiquitylation of proteins by a ‘tethering’ mechanism: a two-site
interaction model for the Nrf2-Keap1 complex. J Biol Chem.
281:24756–24768. 2006.PubMed/NCBI
|
|
24
|
Dhakshinamoorthy S and Jaiswal AK:
Functional characterization and role of INrf2 in antioxidant
response element-mediated expression and antioxidant induction of
NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 20:3906–3917. 2001.
View Article : Google Scholar
|
|
25
|
Malhotra D, Portales-Casamar E, Singh A,
et al: Global mapping of binding sites for Nrf2 identifies novel
targets in cell survival response through ChIP-Seq profiling and
network analysis. Nucleic Acids Res. 38:5718–5734. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nguyen T: Nrf2 controls constitutive and
inducible expression of ARE-driven genes through a dynamic pathway
involving nucleocytoplasmic shuttling by Keap1. J Biol Chem.
280:32485–32492. 2005. View Article : Google Scholar
|
|
27
|
Nguyen T: Increased protein stability as a
mechanism that enhances Nrf2-mediated transcriptional activation of
the antioxidant response element. Degradation of Nrf2 by the 26 S
proteasome. J Biol Chem. 278:4536–4541. 2002. View Article : Google Scholar
|
|
28
|
Luo F, Lv Q, Zhao Y, et al: Quantification
and purification of mangiferin from Chinese mango (Mangifera
indica L.) cultivars and its protective effect on human
umbilical vein endothelial cells under
H2O2-induced stress. Int J Mol Sci.
13:11260–11274. 2012.PubMed/NCBI
|
|
29
|
Das J, Ghosh J, Roy A and Sil PC:
Mangiferin exerts hepatoprotective activity against D-galactosamine
induced acute toxicity and oxidative/nitrosative stress via
Nrf2-NFκB pathways. Toxicol Appl Pharmacol. 260:35–47.
2012.PubMed/NCBI
|
|
30
|
Zhang BP, Zhao J, Li SS, et al: Mangiferin
activates Nrf2-antioxidant response element signaling without
reducing the sensitivity to etoposide of human myeloid leukemia
cells in vitro. Acta Pharmacol Sin. 35:257–266. 2014. View Article : Google Scholar
|
|
31
|
He X and Ma Q: NRF2 cysteine residues are
critical for oxidant/electrophile-sensing, Kelch-like
ECH-associated protein-1-dependent ubiquitination-proteasomal
degradation, and transcription activation. Mol Pharmacol.
76:1265–1278. 2009. View Article : Google Scholar
|
|
32
|
Stewart D, Killeen E, Naquin R, et al:
Degradation of transcription factor Nrf2 via the
ubiquitin-proteasome pathway and stabilization by cadmium. J Biol
Chem. 278:2396–2402. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gu B, Johnston VK, Gutshall LL, et al:
Arresting initiation of hepatitis C virus RNA synthesis using
heterocyclic derivatives. J Biol Chem. 278:16602–16607. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McMahon M, Itoh K, Yamamoto M, et al:
Keap1-dependent proteasomal degradation of transcription factor
Nrf2 contributes to the negative regulation of antioxidant response
element-driven gene expression. J Biol Chem. 278:21592–21600. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Itoh K, Wakabayashi N, Katoh Y, et al:
Keap1 regulates both cytoplasmic-nuclear shuttling and degradation
of Nrf2 in response to electrophiles. Genes Cells. 8:379–391. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang DD and Hannink M: Distinct cysteine
residues in Keap1 are required for Keap1-dependent ubiquitination
of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and
oxidative stress. Mol Cell Biol. 23:8137–8151. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shehzad A and Lee YS: Molecular mechanisms
of curcumin action: signal transduction. Biofactors. 39:27–36.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dinkova-Kostova AT: Chemoprotection
against cancer by isothiocyanates: a focus on the animal models and
the protective mechanisms. Top Curr Chem. 329:179–201. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang S, Penchala S, Prabhu S, et al:
Molecular basis of traditional Chinese medicine in cancer
chemoprevention. Curr Drug Discov Technol. 7:67–75. 2010.
View Article : Google Scholar
|
|
40
|
Hayes JD, McMahon M, Chowdhry S, et al:
Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2
pathway. Antioxid Redox Signal. 13:1713–1748. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hybertson BM, Gao B, Bose SK, et al:
Oxidative stress in health and disease: the therapeutic potential
of Nrf2 activation. Mol Aspects Med. 32:234–246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Slocum SL and Kensler TW: Nrf2: control of
sensitivity to carcinogens. Arch Toxicol. 85:273–284. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Naoe T, Takeyama K, Yokozawa T, et al:
Analysis of genetic polymorphism in NQO1, GST-M1, GST-T1, and
CYP3A4 in 469 Japanese patients with therapy-related
leukemia/myelodysplastic syndrome and de novo acute myeloid
leukemia. Clin Cancer Res. 6:4091–4095. 2000.
|
|
44
|
Larson RA, Wang Y, Banerjee M, et al:
Prevalence of the inactivating 609C-->T polymorphism in the
NAD(P)H:quinone oxidoreductase (NQO1) gene in patients with primary
and therapy-related myeloid leukemia. Blood. 94:803–807.
1999.PubMed/NCBI
|
|
45
|
Li W and Kong AN: Molecular mechanisms of
Nrf2-mediated antioxidant response. Mol Carcinog. 48:91–104. 2009.
View Article : Google Scholar
|
|
46
|
Zhou FL, Zhang WG, Wei YC, et al:
Involvement of oxidative stress in the relapse of acute myeloid
leukemia. J Biol Chem. 285:15010–15015. 2010. View Article : Google Scholar : PubMed/NCBI
|