Introduction
A noteworthy property of ionizing radiation is its
ability to produce highly reactive free radicals [hydroxyl radical
(•OH)] (1). Free radicals,
whether formed by indirect or direct action, eventually damage
biologically important materials such as DNA and lead to
radiation-induced damage formation (2). This damage may be single-strand
breaks (SSBs), base damage or the most molecularly deleterious
double-strand breaks (DSBs). Effectively, the harmful damage
produced by strong oxidation can damage the body's tissues and
cells, thereby causing chronic diseases including cancer and
accelerated aging (3,4).
As with all biological systems there are methods of
protection against harmful exogenous genotoxic mutagens (5,6).
Flavonoids natural occur in plants and other leafy organisms
(7,8). Flavonoids are also commonly regarded
as antioxidant (9). New evidence
suggests that these antioxidant properties may help plants protect
themselves from harmful UV and ionizing radiation by absorbing
harmful free radicals (10,11).
Moreover, if a flavonoid is void of a glucose
entity, then they are referred to as aglycons. Naturally occurring
flavonoids hold none or few glucosyl residues coinciding with an
aglycon (7). Additionally, novel
flavonoids are synthesized by glucosylating the original glucosyl
flavonoids. Glucosylation changes many properties of a flavonoid
and specifically enhances their solubility in water (12,13). Our previous study supports that
glucosylation reduces cellular toxicity and genotoxicity in tissue
culture systems (14).
Furthermore, we suggested this may be due to a reduction of
bioavailability in cells by glucosylation or a reduction in
chemical properties, such as an inhibitory effect of PARP (15).
Previous findings suggest that a select few
flavonoids and glucosyl flavonoids possess radioprotective
properties (16). These include
glucosyl chemicals such as the glucosylation of an ascorbic acid
product, and ascorbic acid-2-glucoside, which did not alter the
radioprotective properties of ascorbic acid (17,18). However, the effect that
glucosylation has on the radioprotective properties of flavonoids
remains to be determined.
Among radiation-induced DNA damage, DNA DSBs
contribute the most towards radiation-induced cell death. DNA DSBs
can be measured by an array of assay systems such as neutral
elution (19,20), gel electrophoresis (21,22), and immunohistochemistry (22,23). For the present study 13 natural
and novel synthesized flavonoids were used. Naked Lambda DNA was
run through gel electrophoresis to visualize the suppression of DNA
DSB formation by the chemicals. In the present study, we
investigated whether glucosylation affects the antioxidant and
radical scavenging properties of flavonoids. The radioprotective
effects of novel glucosyl flavonoids were constructed visually via
a molecular combing technique (24).
Materials and methods
Chemicals
The flavonoids were obtained from the Toyo Sugar
Refining Co., Ltd. (Tokyo, Japan). Fig. 1a shows the chemical structures of
natural and novel synthetic flavonoids used in the present study.
The Quercetin group treatment consisted of Quercetin (302.23
g/mol), Isoquercetin (464.58 g/mol), Rutin (610.51 g/mol),
monoglucosyl (MG)-Rutin (772.5 g/mol), and maltooligosyl
(MO)-Isoquercetin (914.38 g/mol), MO-Rutin (1104.4 g/mol). The
Naringenin group treatment consisted of Naringenin (272.257 g/mol),
Naringin (580.54 g/mol), MG-Naringin (742.54 g/mol) and MO-Prunin
(922.54 g/mol). The Hesperetin group treatment consisted of
Hesperetin (302.28 g/mol), Hesperidin (610.57 g/mol) and
MG-Heperidin (610.57 g/mol). Flavonoids were dissolved in
double-distilled water. Their pH values at 100 μM solution
showed slightly acidic to neutral, ranging from 6.0 to 6.9.
Turbidity of solution was measured by VersaMax ELISA microplate
reader with SoftMax Pro (Molecular Devices, LLC, Sunnyvale, CA,
USA) software at 660 nm. Lambda DNA in Tris-EDTA was purchased from
Nippon Gene Co., Ltd. (Tokyo, Japan). 2,2-Diphenyl-1-picrylhydrazyl
(DPPH) was purchased from Sigma-Aldrich (St. Louis, MO, USA) and
freshly prepared for each experiment in ethanol.
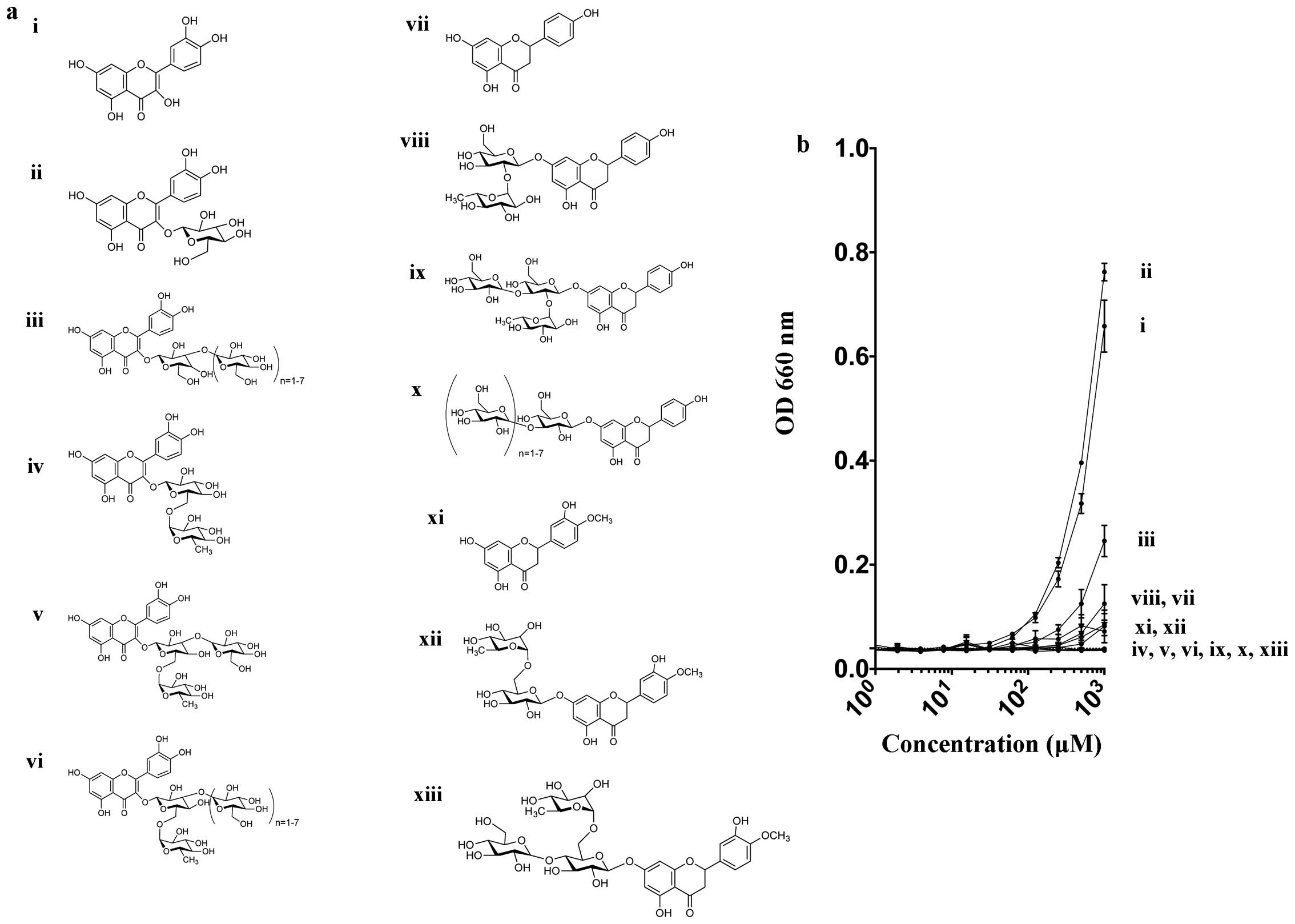 | Figure 1(a) Various chemical structures of 13
flavonoids in the present study. i) Quercetin, ii) Isoquercetin,
iii) Rutin, iv) MG-Rutin, v) MO-Isoquercetin, vi) MO-Rutin, vii)
Naringenin, viii) Naringin, ix) MG-Naringin, x) MO-Prunin, xi)
Hesperetin, xii) Hesperidin, and xiii) MG-Hesperidin. (b) Turbidity
of flavonoid solution measured at 660 nm. Dashed line indicates
background water value. Error bars indicate standard error of the
means. |
Irradiation
For gamma-ray irradiation, a J.L. Shepherd Model
Mark I-68 nominal 6000 Ci 137Cs irradiator (J.L.
Shepherd and Associates, San Fernando, CA, USA) was used at room
temperature (20°C). The dose rate was 3.9 Gy/min.
Electrophoresis and DNA DSBs
Two microliters of Lambda DNA (12
μg/μl) was mixed with 18 μl of each
concentration (0, 10, 100 μM) of flavonoids. Lambda DNA
HindIII digest (0.2 μl) was used as a marker. A total
of 20 μl of sample was exposed to gamma-rays, and run
through electrophoresis after mixing DNA with 4 μl of 6X DNA
loading dye. The Lambda DNA and the marker were immediately
immersed in a 60°C water bath for 5 min to allow for the denaturion
of DNA and the samples were placed on ice for 3 min. The gel
comprised 0.5 g agarose with 50 ml of 1X TAE buffer.
Electrophoresis was carried out at 100 V (7.63 V/cm) for 1 h in 1X
TAE buffer. After electrophoresis, the DNA was stained in ethidium
bromide solution for at least 3 h. The gel was washed with 1X TAE
buffer for 1 h. Gel images were obtained with the Molecular Imager
Gel Doc XR system with Image Lab software (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Single molecule observation
To evaluate the induction of DNA DSBs visually,
single molecule observation was conducted using a molecular combing
method (24). Before irradiation,
the 60×24 mm cover glass (no. 24601; Matsunami Glass Ind., Inc.,
Osaka, Japan) was pretreated with 30% H2O2 at
4°C for at least 4 h, and a coverslip was washed with distilled
water and 100% ethanol. After irradiation, 100 μl samples
were stained with 0.02 μl 1 mM YOYO-1 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), a fluorescent dye reagent
mixed with 0.2 μl 14.3 M 2-mercaptoethanol and 1 μl
DMSO.
The cover glass was set at 45° angles. A 50
μl Lambda DNA solution was dropped onto the top end of the
coverslip and allowed to run down to the bottom due to gravity.
Additionally, the coverslip was placed on the glass slide without a
mounting agent. Images of Lambda DNA were captured using a Zeiss
Axioplan microscope (Carl Zeiss AG, Oberkochen, Germany) with
Q-Imaging EXi Aqua CCD camera with QCapture Pro software (QImaging,
Surrey, BC, Canada). The evaluation was performed by measuring the
length of Lambda DNA using QCapture Pro software. At least 50 DNA
samples were scored for three independent experiments.
Total antioxidant capacity (TAC)
The antioxidant activity of each chemical was
measured by a Total Antioxidant Capacity kit (Sigma-MAK187)
according to the manufacturer's instructions. Cu2+
reagent was diluted 50-fold with assay diluent. Five microliters of
each concentration of flavonoid were diluted at a 1:1 ratio with
50-fold diluted Cu2+ reagent. The samples were mixed and
incubated for 90 min at room temperature. The absorbance of each
sample was measured at 570 nm with NanoDrop (Thermo Fisher
Scientific, Inc.). The values obtained with double-distilled water
were used as the control. Each data point was produced by mean of
three replicates per experiment and three independent experiments
were carried out.
DPPH antioxidant properties
DPPH analysis was performed as previously reported
(25). One hundred microliters of
100 μM DPPH, 80 μl of ethanol and 20 μl each
concentration of flavonoid were mixed. The mixtures were agitated
vigorously and allowed to stand at room temperature for 30 min.
Absorbance was measured at 517 nm using a VersaMax ELISA Microplate
Reader. DPPH scavenging activity was calculated by absorbance of
control minus absorbance of sample divided by absorbance of control
(26). Each data point was
produced by mean of triplicates per experiment and three
independent experiments were carried out.
Statistical analysis
Statistical comparison of the mean values was
performed using a t-test with GraphPad Prism 6 (GraphPad Software,
Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference. Error bars indicate the
standard error of the mean.
Results
Turbidity of each flavonoid at absorbance
at 660 nm
Our results indicated that natural flavonoids were
water insoluble and with increased glucosylation this resulted in
improved water solubility. In the Quercetin group, the natural
flavonoids, Quercetin, Isoquercetin and Rutin had poor solubility
in water, whereas monoglucosyl and maltooligosyl flavonoids had
complete solubility, even at 1 mM concentration (Fig. 1b).
Gel electrophoresis
In the gel electrophoresis assay, fragmented Lambda
DNA migrated more and was observed as a smear. Fig. 2a shows the DNA DSBs were
dose-dependent between 0 and 20 Gy for Lambda DNA in this assay.
Fig. 2b shows that flavonoids did
not produce DSBs by themselves. Fig.
2c and d shows the DSB radioprotective abilities of flavonoids
at 10 and 100 μM. For 10 μM, the Quercetin group,
particularly Isoquercetin, MG-Rutin, and MO-Isoquercetin MO-Rutin,
displayed the strongest radioprotective abilities. All the
flavonoids displayed positive radioprotective abilities at
concentrations of 100 μM, with the exception of Quercetin
and Hesperetin. Therefore, glucosylation enhanced the
radioprotective properties observed as a reduction of DSB
formation.
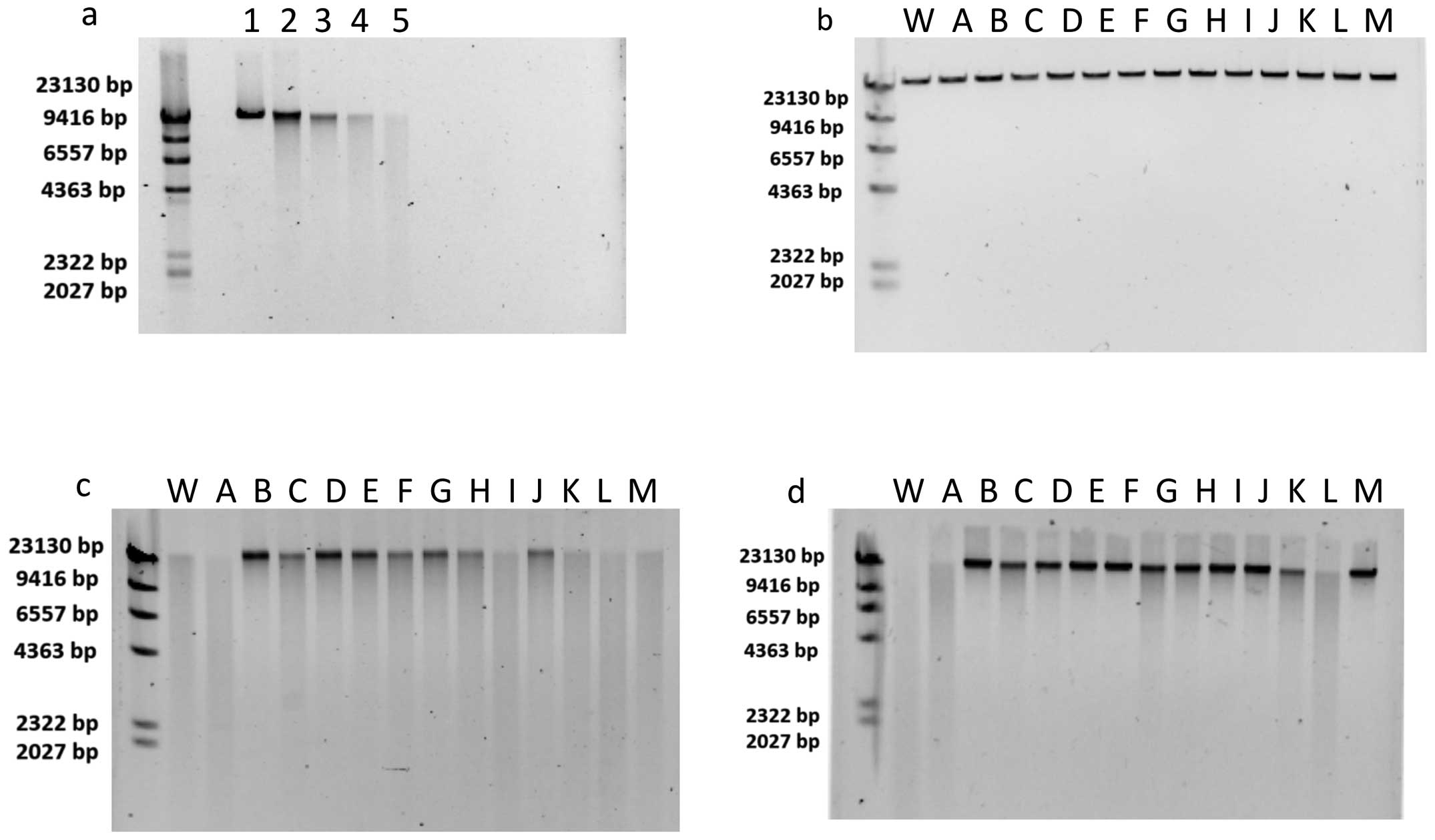 | Figure 2(a) Dose-dependent DNA double-strand
breaks (DSBs). Lane 1, 0 Gy; lane 2, 5 Gy; lane 3, 10 Gy; lane 4,
15 Gy; and lane 5, 20 Gy. (b) Effect of flavonoids without
irradiation. (c and d) Radioprotective effects of flavonoids at 10
μM with 20 Gy and radioprotective effects of flavonoids at
100 μM with 20 Gy. First left lane is Lambda
DNA-HindIII digest ladder; lane W, is negative control with
water; lane A, Quercetin; lane B, Isoquercetin; lane C, Rutin; lane
D, MG-Rutin; lane E, MO-Isoquercetin; lane F, MO-Rutin; lane G,
Naringenin; lane H, Naringin; lane I, MG-Naringin; lane J,
MO-Prunin; lane K, Hesperetin; lane L, Hesperidin; and lane M,
MG-Hesperidin. |
Molecular combing
The flavonoids Quercetin (no glucosylation, water
insoluble), MG-Rutin (glucosylated water soluble), MG-Naringin
(glucosylated water soluble) and MG-Hesperidin (glucosylated water
soluble) were tested for molecular combing. Fig. 3 shows the result of molecular
combing. Each intact Lambda DNA molecule was observed as stretched
DNA, and radiation-induced fragments of Lambda DNA were observed in
a single molecule (Fig. 3a and
b). There was a dose-dependent DSB formation of 0 to 20 Gy.
Radiation (20 Gy) produced approximately 5 DSB/Lambda DNA. After
exposure to 20 Gy, the addition of 10 μM MG-Rutin showed the
strongest reduction in the frequency of DSB among the four
flavonoids (p<0.05) (Fig. 3d),
and the addition of 100 μM of any MG-flavonoids greatly
reduced the DSB frequency (p<0.005) but not Quercetin (p=0.84).
MG-Naringin and MG-Hesperidin (10 μM) did not show
statistically significant radioprotection compared to the 20
Gy-irradiated control (p>0.1).
TAC
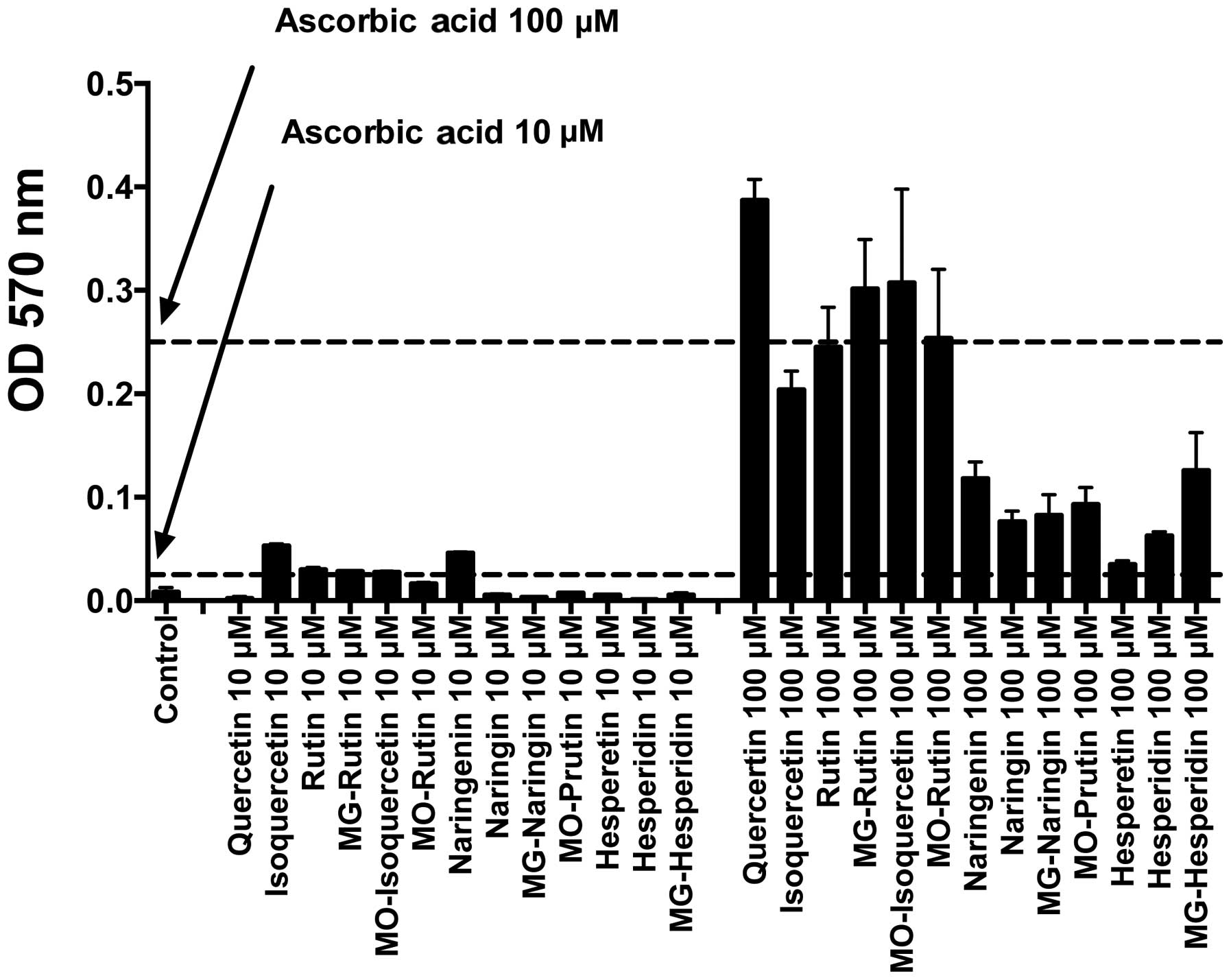
Fig. 4 shows the
total antioxidant capacity of 13 flavonoids, which was measured at
the optical density of 570 nm. Quercetin group showed good TAC at
each concentration compared with other flavonoid groups. The
Quercetin groups also exhibited antioxidant properties similar to
that of ascorbic acid at 10 and 100 μM, with the exception
of 10 μM Quercetin. The Naringenin and Hesperetin groups
were poor antioxidants compared to the ascorbic acid at the tested
concentrations. No clear correlation between glucosylation and
total TAC was observed.
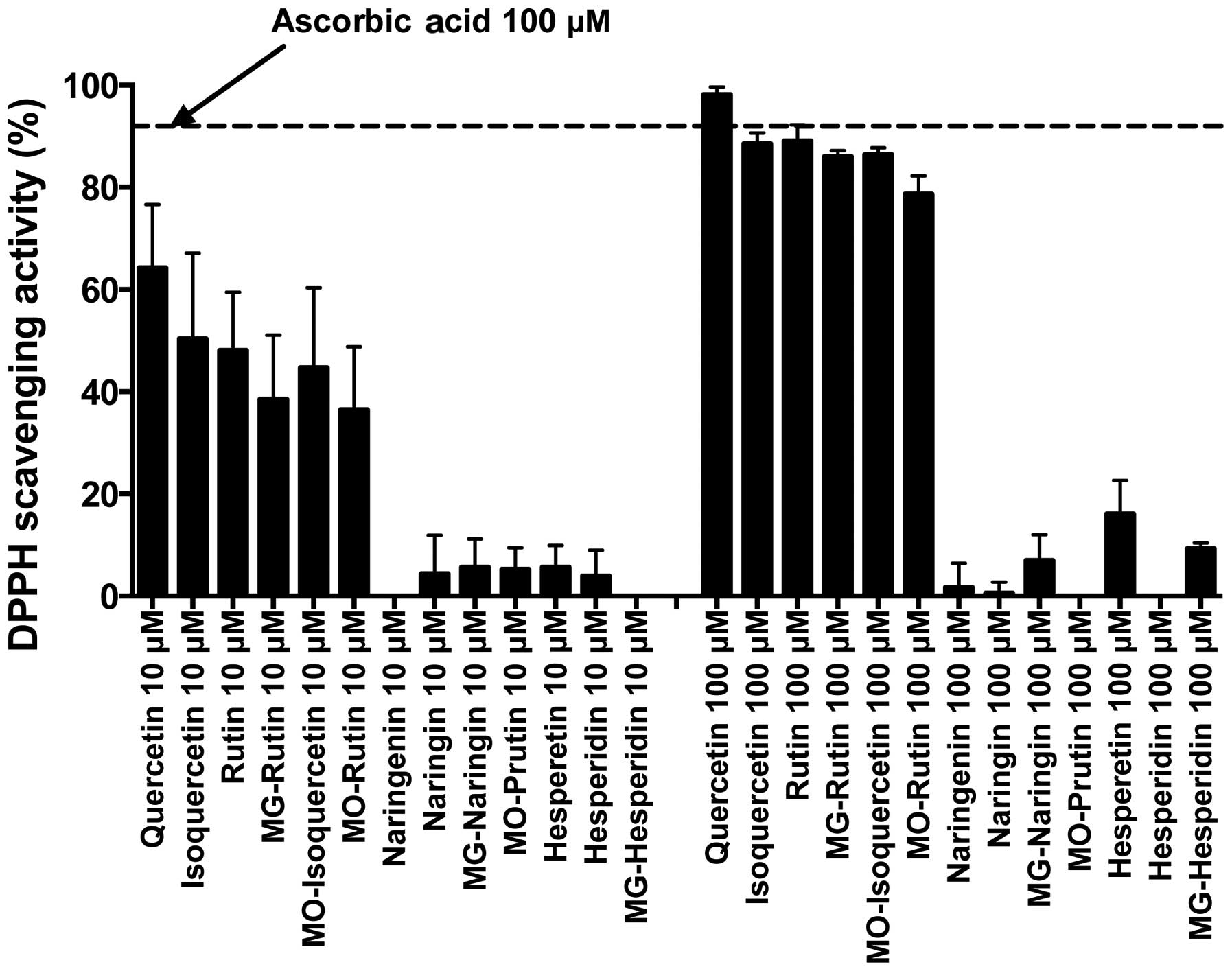
DPPH radical scavenging properties
DPPH is a stable free radical. Fig. 5 shows the result of DPPH radical
scavenging properties. We used ascorbic acid as a control to
compare the radical scavenging ability of 13 flavonoids. The
Quercetin group showed much better radical scavenging ability than
the other two groups. The result showed glucosylation slightly
reduced free radical scavenging ability in the Quercetin group at
10 and 100 μM. The Naringenin and Hesperetin groups were
poor radical scavengers at 10 and 100 μM.
Discussion
The results of the present study displayed that
radioprotective properties for DSBs were dependent on flavonoid
groups at a low concentration. All the Quercetin derivatives showed
radioprotection after 20 Gy, except natural Quercetin. On the other
hand, the majority of flavonoids showed radioprotection at 100
μM. Therefore, an increased uptake of flavonoids may protect
cells from DSB formation induced by radiation exposure. Better
radioprotective properties in DSB reduction for the glucosylated
Quercetin group were matched with total antioxidant capacity and
radical scavenging ability. Although Quercetin showed strong
radical scavenging and antioxidant properties, Quercetin did not
show DSB reduction even at 100 μM. This may be associated
with their poor water solubility or poor interaction with DNA
molecules (27). Since Quercetin,
Isoquercetin, and Rutin displayed poor water solubility, we
suggested that MG-Rutin, MO-Isoquercetin, and MO-Rutin are better
radioprotective agents and easily bioavailable with increased water
solubility. Recently, enhanced solubility of Quercetin by forming
composite particles with glucosyl flavonoids was reported (28). Enhanced solubility can be produced
by the glucosylation of flavonoids. The formation of composite
particles may be an alternative strategy to enhanced solubility and
may lead to radioprotection.
Furthermore, lambda DNA is a double-stranded DNA
with 48,502 base pairs. This size is separable with simple gel
electrophoresis and is readily obsevable miscroscopically when
using the molecular combing method. Therefore, Lambda DNA is a good
test subject for screening purposes such as that identified in the
present study. Moreover, DNA DSBs lead to serious biological
consequences, including mutations, carcinogenesis, or apoptosis
(29). We used two assays to
measure DSBs. The gel electrophoresis assay is convenient and can
be applied with multiple samples. On the other hand, the molecular
combing method is time-consuming and labor intense. However, in the
molecular combing method one can visualize individual DNA DSB in a
single molecule. DNA fragment size can be directly measured in this
method. Therefore, gel electrophoresis is suitable for screening
purposes and detailed analysis and confirmation can be achieved
using the molecular combing method.
However, there were some potential pitfalls in our
procedures. The molecular combing method may have some difficulties
for reproducibility without careful sample handling. It is possible
that different sizes of Lambda DNA do not attach onto cover glass
equally. We found that the length of the Lambda DNA decreased
sharply with vigorous pipetting, as previously reported (30). We used a 60×24 mm coverslip as
previously reported (24) after
we tested several different sizes of coverslips. Interestingly,
55×24 and 45×24 mm coverslips from the same company did not produce
a better quality of Lambda DNA stretch. It is not known why Lambda
DNA stretched well at 60×24 mm, while other size slides did not,
but it may be associated with the different surface tension.
In conclusion, antioxidant and scavenging capacity
are well related to DNA DSB formation reduction. Glucosylation
affected antioxidant and free radical scavenging abilities to some
degree. Although some flavonoids, especially glucosylated ones such
as MG-Rutin and MO-Rutin, have good protection for in vitro
double-stranded DNA, it may be limited to in vitro
radioprotective effects. Further in vitro cell culture
studies and most importantly in vivo studies are required to
prove glucosyl flavonoids are good radioprotectors.
Acknowledgments
The present study was partially supported by the
Colorado State University Start Up Fund (TAK), the Collaborative
Research Fund from Toyo Sugar Refining Co., Ltd. (MU), and Contract
Research Fund from Toyo Sugar Refining Co., Ltd. (TAK).
References
|
1
|
Harman D: Aging: a theory based on free
radical and radiation chemistry. J Gerontol. 11:298–300. 1956.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ward JF: DNA damage produced by ionizing
radiation in mammalian cells: identities, mechanisms of formation,
and reparability. Prog Nucleic Acid Res Mol Biol. 35:95–125. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lobo V, Patil A, Phatak A and Chandra N:
Free radicals, antioxidants and functional foods: impact on human
health. Pharmacogn Rev. 4:118–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Best BP: Nuclear DNA damage as a direct
cause of aging. Rejuvenation Res. 12:199–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andersson DI, Slechta ES and Roth JR:
Evidence that gene amplification underlies adaptive mutability of
the bacterial lac operon. Science. 282:1133–1135. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baumann P and West SC: Role of the human
RAD51 protein in homologous recombination and double-stranded-break
repair. Trends Biochem Sci. 23:247–251. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar S and Pandey AK: Chemistry and
biological activities of flavonoids: an overview. Scientific World
Journal. 2013:1627502013. View Article : Google Scholar
|
|
8
|
Harborne JB and Paxman GJ: Genetics of
anthocyanin product in the radish. Hered Edinb. 19:505–506. 1964.
View Article : Google Scholar
|
|
9
|
Thompson M, Williams CR and Elliot GE:
Stability of flavonoid complexes of copper(II) and flavonoid
antioxidant activity. Anal Chim Acta. 85:375–381. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Svobodová A, Psotová J and Walterová D:
Natural phenolics in the prevention of UV-induced skin damage. A
review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub.
147:137–145. 2003. View Article : Google Scholar
|
|
11
|
Rahman K: Studies on free radicals,
antioxidants, and co-factors. Clin Interv Aging. 2:219–236.
2007.PubMed/NCBI
|
|
12
|
Plumb GW, De Pascual-Teresa S,
Santos-Buelga C, Cheynier V and Williamson G: Antioxidant
properties of catechins and proanthocyanidins: effect of
polymerisation, galloylation and glycosylation. Free Radic Res.
29:351–358. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimoi K, Yoshizumi K, Kido T, Usui Y and
Yumoto T: Absorption and urinary excretion of Quercetin, Rutin, and
alphaG-Rutin, a water soluble flavonoid, in rats. J Agric Food
Chem. 51:2785–2789. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Engen A, Maeda J, Wozniak DE, Brents CA,
Bell JJ, Uesaka M, Aizawa Y and Kato TA: Induction of cytotoxic and
genotoxic responses by natural and novel Quercetin glycosides.
Mutat Res Genet Toxicol Environ Mutagen. 784–785:15–22. 2015.
View Article : Google Scholar
|
|
15
|
Maeda J, Roybal EJ, Brents CA, Uesaka M,
Aizawa Y and Kato TA: Natural and glucosyl flavonoids inhibit
poly(ADP-ribose) polymerase activity and induce synthetic lethality
in BRCA mutant cells. Oncol Rep. 31:551–556. 2014.
|
|
16
|
Sunada S, Fujisawa H, Cartwright IM, Maeda
J, Brents CA, Mizuno K, Aizawa Y, Kato TA and Uesaka M:
Monoglucosyl-Rutin as a potential radioprotector in mammalian
cells. Mol Med Rep. 10:10–14. 2014.PubMed/NCBI
|
|
17
|
Chandrasekharan DK, Kagiya TV and Nair
CKK: Radiation protection by 6-palmitoyl ascorbic acid-2-glucoside:
studies on DNA damage in vitro, ex vivo, in vivo and oxidative
stress in vivo. J Radiat Res (Tokyo). 50:203–212. 2009. View Article : Google Scholar
|
|
18
|
Kobashigawa S, Kashino G, Suzuki K,
Yamashita S and Mori H: Ionizing radiation-induced cell death is
partly caused by increase of mitochondrial reactive oxygen species
in normal human fibroblast cells. Radiat Res. 183:455–464. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kemp LM, Sedgwick SG and Jeggo PA: X-ray
sensitive mutants of Chinese hamster ovary cells defective in
double-strand break rejoining. Mutat Res. 132:189–196.
1984.PubMed/NCBI
|
|
20
|
Wlodek D and Olive PL: Physical basis for
detection of DNA double-strand breaks using neutral filter elution.
Radiat Res. 124:326–333. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Olive PL, Wlodek D and Banáth JP: DNA
double-strand breaks measured in individual cells subjected to gel
electrophoresis. Cancer Res. 51:4671–4676. 1991.PubMed/NCBI
|
|
22
|
Kato TA, Okayasu R, Jeggo PA and Fujimori
A: ASPM influences DNA double-strand break repair and represents a
potential target for radiotherapy. Int J Radiat Biol. 87:1189–1195.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fujii Y, Genet MD, Roybal EJ, Kubota N,
Okayasu R, Miyagawa K, Fujimori A and Kato TA: Comparison of the
bromodeoxyuridine-mediated sensitization effects between low-LET
and high-LET ionizing radiation on DNA double-strand breaks. Oncol
Rep. 29:2133–2139. 2013.PubMed/NCBI
|
|
24
|
Oshige M, Yamaguchi K, Matsuura S, Kurita
H, Mizuno A and Katsura S: A new DNA combing method for biochemical
analysis. Anal Biochem. 400:145–147. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karioti A, Hadjipavlou-Litina D, Mensah
ML, Fleischer TC and Skaltsa H: Composition and antioxidant
activity of the essential oils of Xylopia aethiopica (Dun) A. Rich.
(Annonaceae) leaves, stem bark, root bark, and fresh and dried
fruits, growing in Ghana. J Agric Food Chem. 52:8094–8098. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bansal V, Sharma A, Ghanshyam C and Singla
ML: Coupling of chromatographic analyses with pretreatment for the
determination of bioactive compounds in Emblica officinalis juice.
Anal Methods. 6:410–418. 2014. View Article : Google Scholar
|
|
27
|
Solimani R: The flavonols Quercetin, Rutin
and morin in DNA solution: UV-vis dichroic (and mid-infrared)
analysis explain the possible association between the biopolymer
and a nucleophilic vegetable-dye. Biochim Biophys Acta.
1336:281–294. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fujimori M, Kadota K, Shimono K, Shirakawa
Y, Sato H and Tozuka Y: Enhanced solubility of Quercetin by forming
composite particles with transglycosylated materials. J Food Eng.
149:248–254. 2015. View Article : Google Scholar
|
|
29
|
Khanna KK and Jackson SP: DNA
double-strand breaks: signaling, repair and the cancer connection.
Nat Genet. 27:247–254. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoo HB, Lim HM, Yang I, Kim SK and Park
SR: Flow cytometric investigation on degradation of macro-DNA by
common laboratory manipulations. J Biophys Chem. 2:102–111. 2011.
View Article : Google Scholar
|