Introduction
Deep vein thrombosis (DVT) refers to the formation
of a blood clot within a deep vein, predominantly in the legs. DVT
and pulmonary embolism (PE) constitute a single disease process,
termed venous thromboembolism (VTE), which is the third most common
vascular disease in the United States (1). As the disease has an insidious onset
and there is no obvious clinical symptom or sign in the early
stages, the rate of misdiagnosis is high. When timely diagnosis and
effective therapy are missed, DVT may lead to the abnormal swelling
and ulceration of lower limbs, post-thrombotic syndrome and PE
(1,2). The incidence of VTE is >1,000 per
year in the United States (3).
Thus, DVT is a major cause of mortality and leads to significant
morbidity. In forensic practice, PE is a major cause of sudden
death and is attributable primarily to DVT (4). Studies of DVT improve the
understanding of the disease, as well as increase the successful
rescue rate and detection rate, which is significant in clinical
diagnosis treatment and forensic identification.
In recent years, with the deepening of theoretical
research into thrombosis, DVT has been recognized as a disease that
involves multiple factors and systems (5). Due to the complexity of the disease,
traditional Northern blotting and quantitative polymerase chain
reaction methods have been unable to fully elucidate its
mechanisms. To develop a more comprehensive understanding of DVT,
high-throughput microRNA (miRNA) and messenger RNA (mRNA)
microarray technology were used in the present study.
miRNAs represent a class of non-coding small RNAs
that post-transcriptionally suppress their target genes (6). There is increasing evidence that
miRNA expression patterns change in many vascular diseases
(7–10). Although DVT has been investigated
extensively, the molecular mechanisms underlying the
pathophysiological changes remain to be defined. Furthermore,
information on changes in miRNA expression within the vessel
tissues is limited and, to date, to the best of our knowledge,
there are no studies describing miRNA-mRNA interactions in DVT.
In the present study, miRNA and mRNA expression in
vessel tissue samples from rat DVT models were assessed by
microarray. Furthermore, bioinformatics analyses were used to build
and analyze the miRNA-mRNA network. The present findings provide
systematic and comprehensive insights into the molecular mechanisms
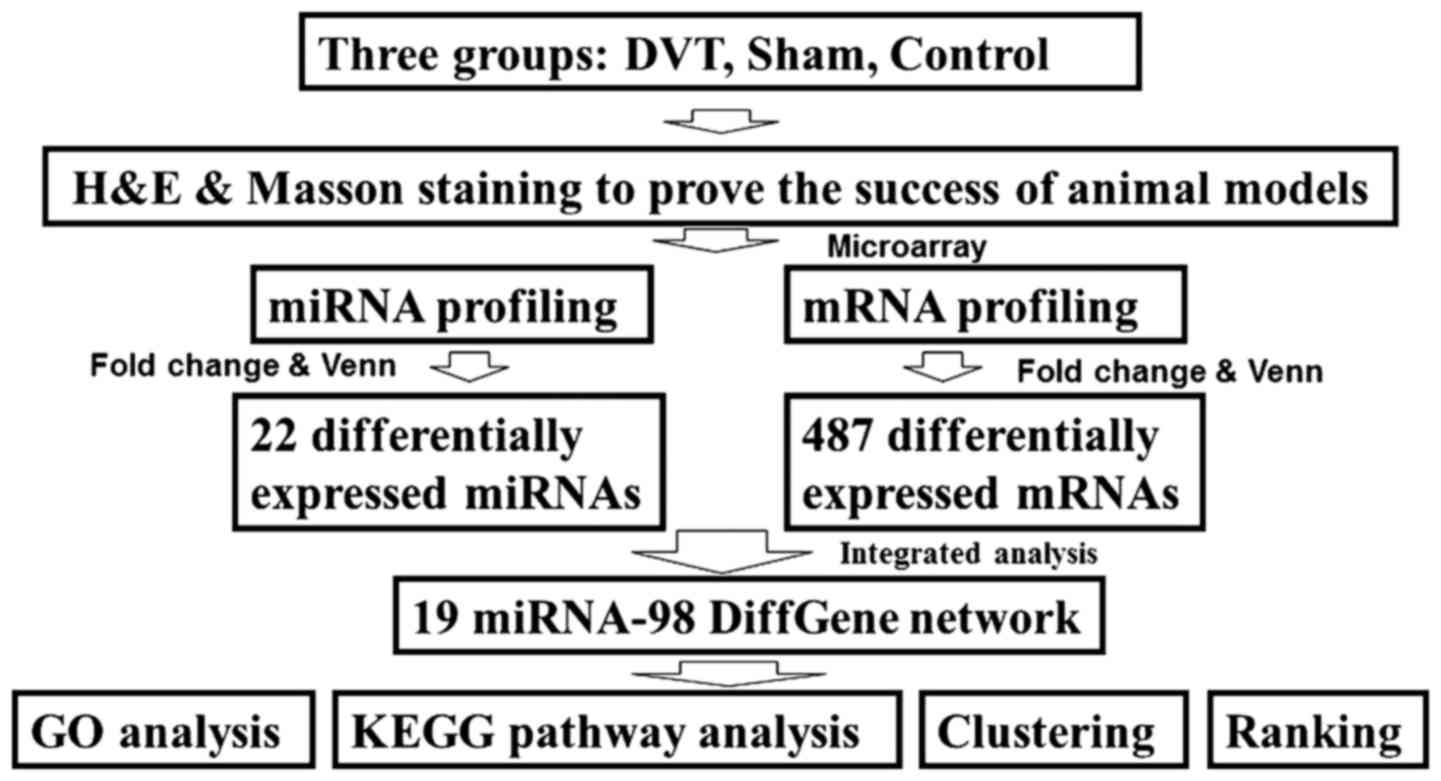
of DVT. The study design is presented in Fig. 1.
Materials and methods
Animal model of venous thrombosis
All experiments were reviewed and approved by the
ethics committee of the Institute of Laboratory Animal Science of
Shanxi Medical University (Taiyuan, China).
Adult male Sprague Dawley (SD) rats (n=36; Shanxi
Laboratory Animal Center, Taiyuan, China) 8–10 weeks of age and
weighing 280–300 g were used in the present study. The rats were
divided into three groups as follows: DVT, sham and control (n=12).
The rats were anesthetized by 10% chloral hydrate. A midline
laparotomy was performed. The inferior vena was explored by moving
the small intestine out of the way, and all side branches were
ligated. IVC was ligated just below the left renal vein. A
microvascular clamp was attached to the confluence of iliac veins
for 15 min. The skin was sutured by 3–0 Prolene suture and
penicillin powder (Sigma, Washington, DC, USA) covering the
incision evenly was used. The sham-surgery rats received anesthesia
and all surgical procedures, but without IVC ligation or clamping.
The control group received no treatment.
Tissue harvesting
The rats were sacrificed at day 3 after ligation.
The IVC with thrombus was carefully harvested. One part of the
tissue was fixed in 10% formalin solution for histological analysis
and the rest was stored in RNAsafety (Shanghai Biotechnology Corp.,
Shanghai, China) for microarray analyses.
Histological analysis
For histological examinations, the IVC tissue was
fixed with 4% paraformaldehyde for 48 h and embedded in paraffin
wax. The tissues were then cut into 4-μm-thick sections and
the sections were dewaxed in xylene twice at 37°C for 15 min each
time, rehydrated through decreasing concentrations of ethanol, and
washed in distilled water at room temperature for 5 min each time.
They were finally stained with hematoxylin and eosin (H&E) or
Masson's trichrome, both following the company's instructions
(Nanjing Jiancheng Bioengineering Institute, Nanjing, China). For
H&E staining, the slides were dipped into the jar containing
hematoxylin for 5 min and with eosin for 30 sec with agitation at
room temperature. For Masson staining, slides were stained at room
temperature with nuclear staining solution for 1 min, with
cytoplasmic stain solution for 45 sec, then washed with
phosphomolybdic acid for 6 min and last counterstained for 5 min.
After staining, specimens were observed under a light microscope
(Panoramic SCAN II; 3DHISTECH Kft., Budapest, Hungary) to evaluate
the histomorphology of the venous walls.
RNA isolation and quantification
Total RNA was extracted and purified using an Ambion
mirVana miRNA isolation kit (cat. no. AM1561; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and RNA integrity was assessed
using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.,
Santa Clara, CA, USA). The RNA samples were used for microarrays.
To avoid differences between individuals, 200 ng total RNA isolated
from three rats per group was pooled into a single sample.
Microarray experiments were repeated to produce three independent
biological replicates.
miRNA microarray experimental set-up and
initial data analysis
miRNA expression analysis was performed using nine
Agilent rat miRNA (8×60K) V21.0 microarrays (design ID: 70154;
Agilent Technologies, Inc.) at the National Engineering Center for
Biochip, Shanghai Biotechnology, Corp. (Shanghai, China) miRNA was
labeled using the miRNA complete labeling and hybridization kit
(cat. no. p/n 5190-0456; Agilent Technologies, Inc.). The kit was
used to hybridize each slide with Cy3-labeled RNA in a
hybridization oven. Subsequent to hybridization, slides were washed
and scanned using a Microarray Scanner (cat. no. G2565CA; Agilent
Technologies, Inc.) using Feature Extraction software (version
10.7; Agilent Technologies, Inc.).
Raw data were normalized using the quantile
algorithm in the GeneSpring software (version 12.6; Agilent
Technologies, Inc.). The results of signal values are presented as
means ± standard deviation. Student's t-test was used to identify
differences between groups using the R package. Furthermore, the
fold change was the ratio of the mean values of two comparative
groups. miRNAs with fold differences ≥2.0 and P≤0.05 were
considered to indicate a statistically significant difference.
mRNA microarray experimental set-up and
initial data analysis
mRNA expression analysis was performed using nine
Agilent Whole Rat Genome Microarrays 4×44K (design ID:014879;
Agilent Technologies, Inc.) at the National Engineering Center for
Biochip, Shanghai Biotechnology Corp. Total RNA was amplified and
labeled using the Low Input Quick Amp Labeling kit, One-Color (cat.
no. 5190-2305; Agilent Technologies, Inc.). Labeled cRNA was
purified using an RNeasy mini kit (cat. no. 74106; Qiagen GmbH,
Hilden, Germany). The left process was similar to miRNA microarray.
mRNAs with fold differences ≥2.0 and P≤0.05 were considered to
indicate a statistically significant difference.
Integrated analysis of miRNA and mRNA
expression profiles
Five prediction tools: TargetMiner (http://www.isical.ac.in/~bioinfo_miu/targetminer20.htm),
miRDB (http://mirdb.org/miRDB/index.html), microRNA
(http://www.microrna.org/microrna/home.do), TarBase
(http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index),
and RNA22 (https://cm.jefferson.edu/rna22/) quoted by miRBase
database, were used to obtain the target genes of differentially
expressed miRNAs. All of the tools were assembled into an online
tool (http://www.shbio.com/analysis.html).
The predicted mRNA targets were then compared with
experimentally determined mRNAs by microarray. Subsequently,
according to Pearson's correlation coefficients and the
associations of mRNAs in STRING (http://string-db.org/), a regulatory network was
determined, which was comprised of 19 miRNAs and 98 mRNAs.
These 98 mRNAs were uploaded to Database for
Annotation, Visualization and Integrated Discovery (version 6.7;
https://david.ncifcrf.gov/) for Gene
Ontology (GO) functional annotation. The Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway database (http://www.genome.jp/kegg/kegg1.html) was used to
identify the enriched pathways of targets. Additionally, 19 miRNAs
were subjected to a two-way unsupervised hierarchical clustering
analysis using MultiExperiment Viewer software 4.7.4, a part of the
TM4 Microarray software suite (Dana-Farber Cancer Institute,
Boston, MA, USA) and ranked according to fold change and degrees in
the network.
Statistical analysis
Data are presented as the means ± SD. Statistical
difference (P≤0.05) was calculated by Student's t-test and
correlations were evaluated by Pearson's correlation coefficients
using the R package. The P-value <0.05 was considered
statistically significant.
Results
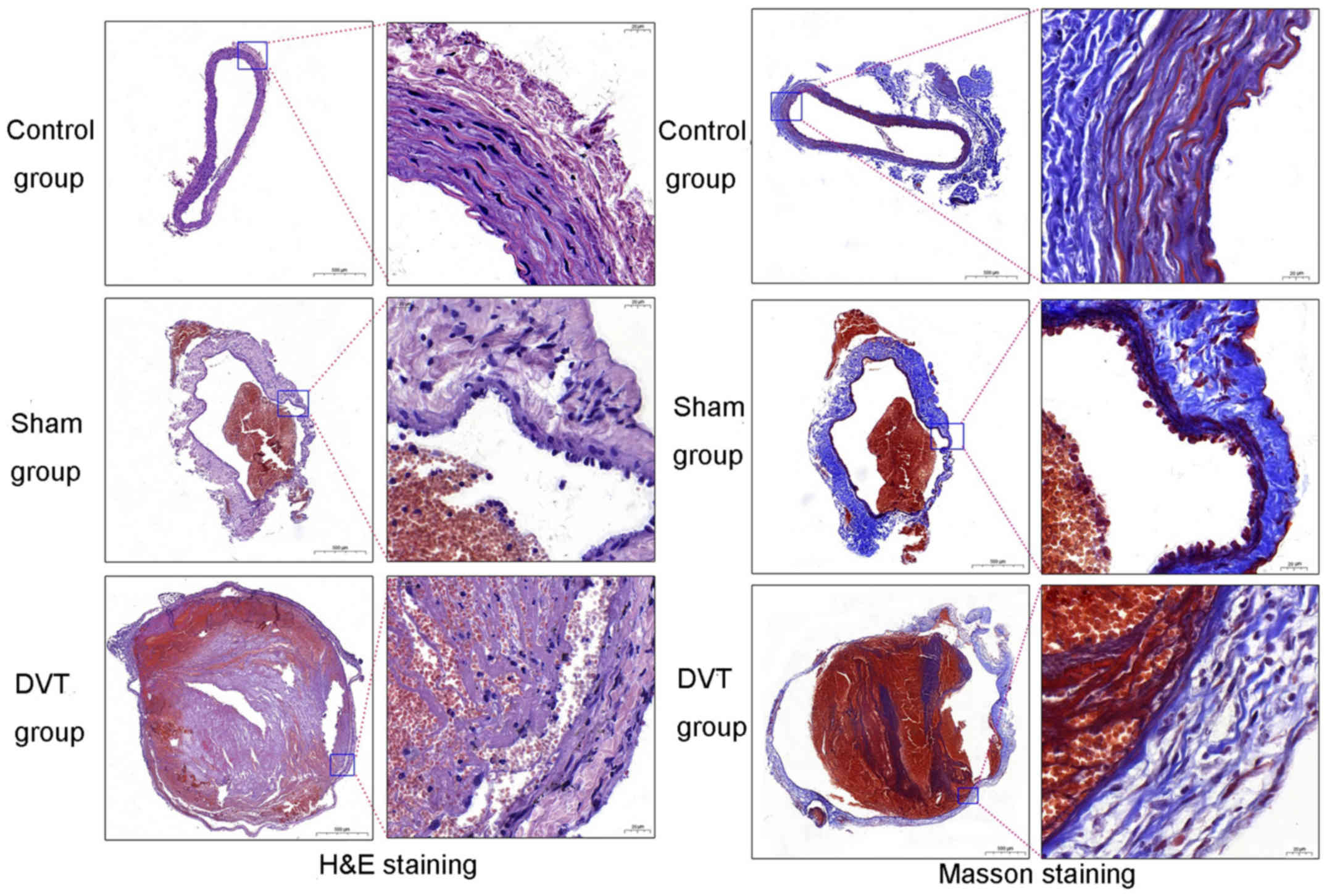
Histological changes in veins following
ligation
Sections of the IVC were stained with H&E and
Masson's trichrome and observed under a light microscope. Clear
differences were observed among the three groups (Fig. 2). With discontinuous vascular
endothelial cells (ECs), numerous inflammatory cells infiltrated
and entered the blood vessels, demonstrating a stable mixed
thrombus following treatment in the DVT group. The staining results
indicated the success of the SD DVT models.
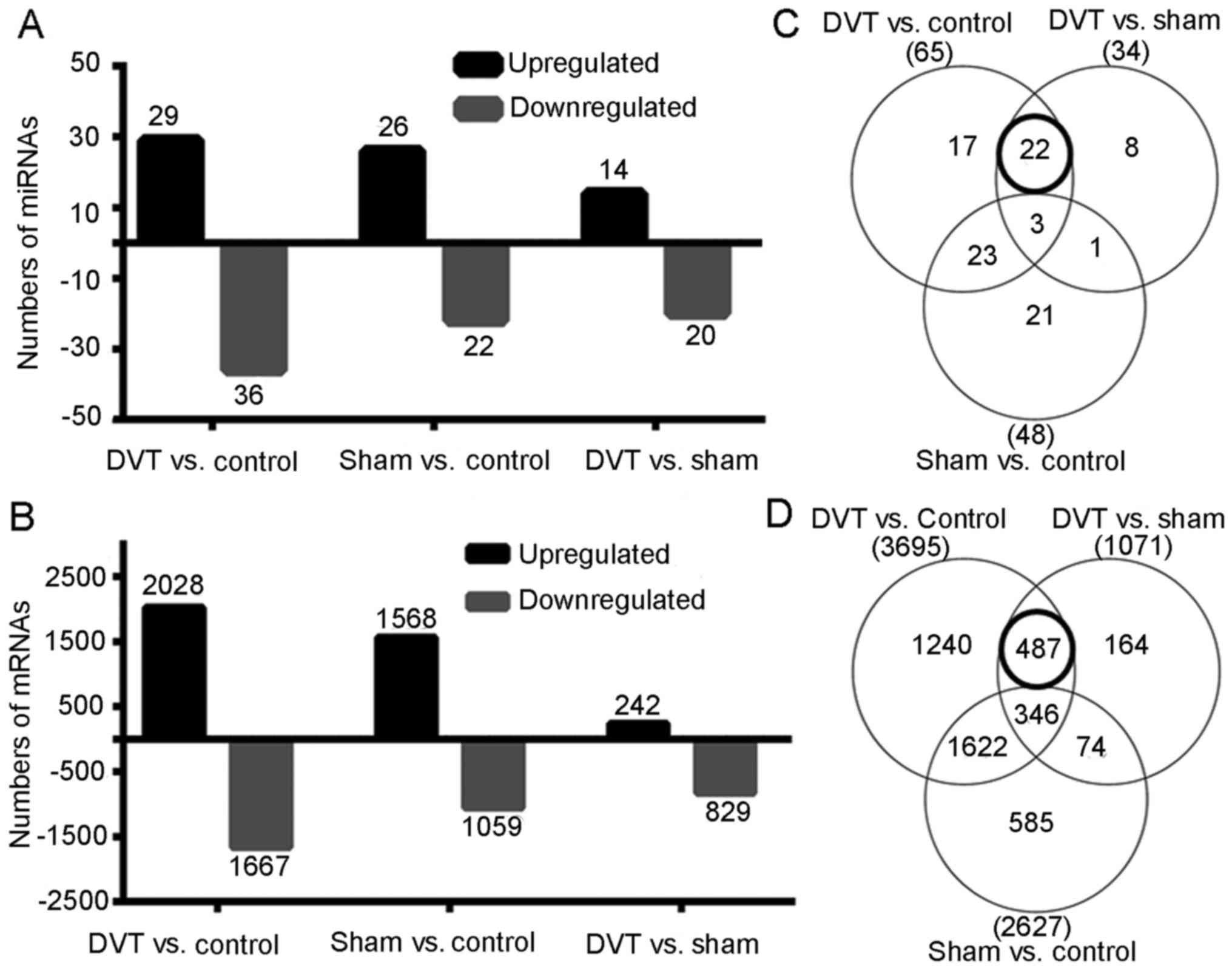
miRNA expression profiling
To evaluate the role of miRNAs in thrombosis, the
miRNA profiling in the DVT animal models were investigated, along
with the sham-surgery and control rats (n=9) by microarray. Among
758 detectable miRNAs, 65 miRNAs (29 upregulated and 36
downregulated) significantly difference between the DVT and control
groups, 34 miRNAs (14 upregulated and 20 downregulated) between the
DVT and sham groups, and 48 miRNAs (26 upregulated and 22
downregulated) between the sham and control groups using a cut-off
with an adjusted P≤0.05 and a fold-change of ≥2.0 (Fig. 3A). To exclude any effects of the
surgical procedure, only 22 differentially expressed miRNAs in the
overlapping areas (Fig. 3C) were
focused upon, indicating differences between the DVT and the two
other groups, but no difference between the control and sham
groups. These 22 miRNAs were subjected to further analyses.
mRNA expression profiling
More than 41,000 rat genes and transcripts were
investigated in the present study. Of them, 3,695 genes (2,028
upregulated and 1,667 downregulated) differed significantly between
the DVT and control groups, 2,627 genes (1,568 upregulated and
1,059 downregulated) between the sham and control groups, and 1,071
genes (242 upregulated and 829 downregulated) between the DVT and
sham groups using a cut off with an adjusted P≤0.05 and a
fold-change of ≥2.0 (Fig. 3B).
Similarly, 487 differentially expressed genes (DEGs) in overlapping
areas (Fig. 3D) were selected for
further analyses.
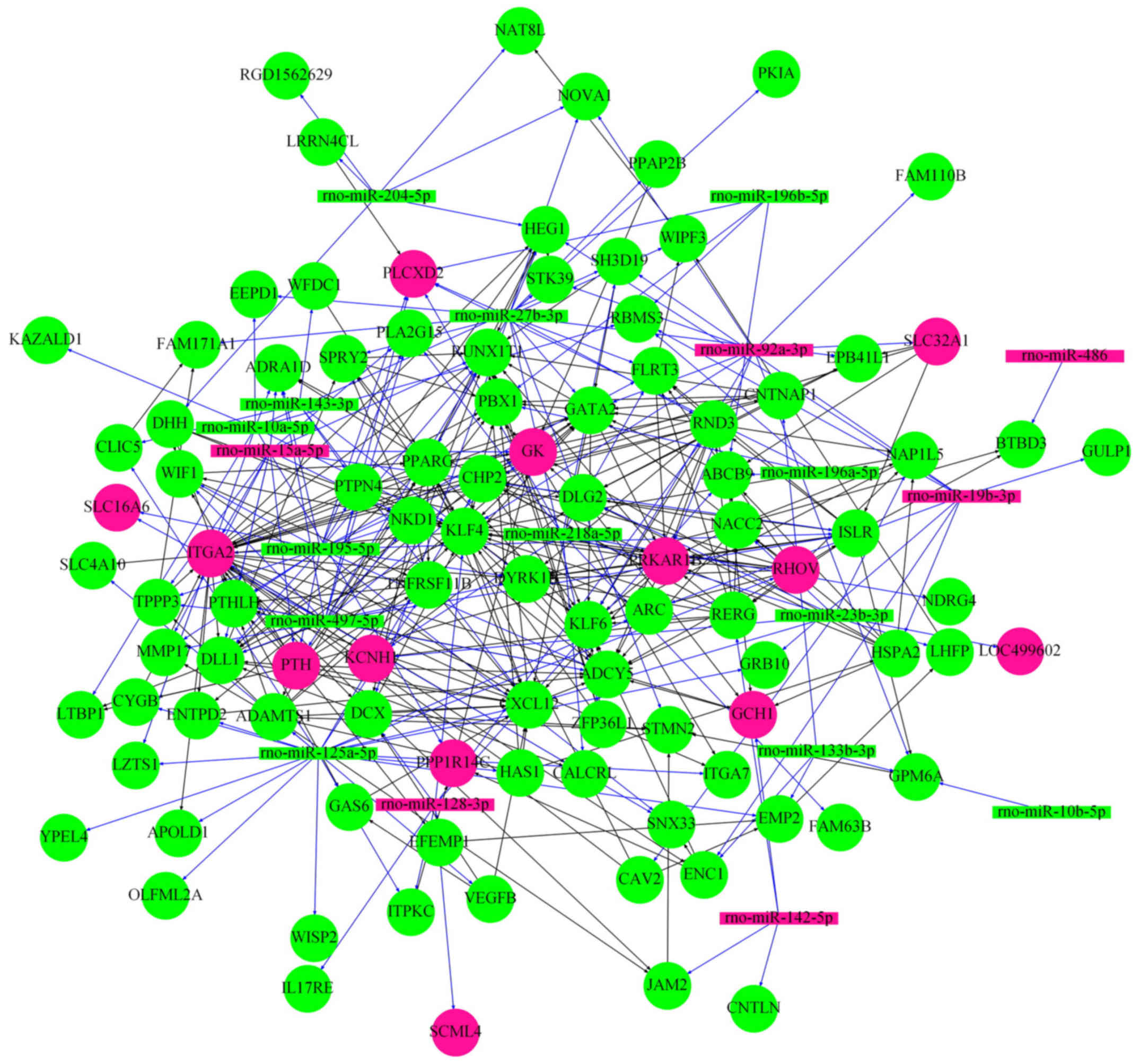
miRNA-mRNA network
The 22 miRNAs were integrated to the experimentally
determined 487 DEGs to obtain the miRNA-mRNA network (Fig. 4), which was composed of 19 miRNAs
(6 upregulated and 13 downregulated) and 98 differentially
expressed mRNAs (13 upregulated and 85 downregulated).
Functional classification of 98
genes
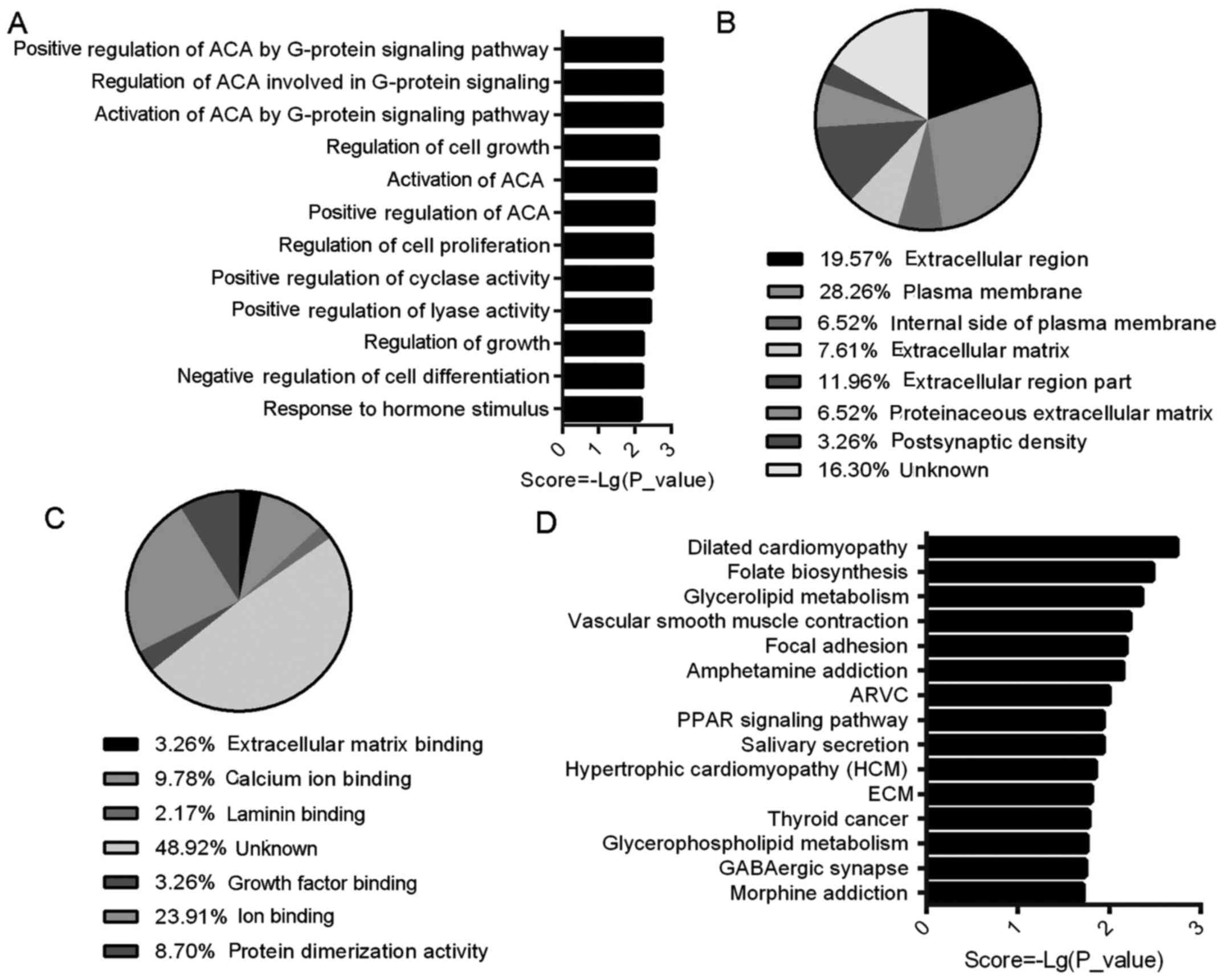
GO assigns biological processes (BP), cellular
components (CC) and molecular function (MF). The BP analysis
involved 83 GO terms; five of the top 10 were associated with
adenylate cyclase activity, and two were associated with cell
growth (Fig. 5A). Seven were
found in CC, indicating that the DEGs were predominantly
extracellular genes (45.66%) and plasma membrane genes (34.78%)
(Fig. 5B). In seven MF
annotations, the majority of genes belonged to binding activity
genes (18.4%), while active protein genes represented 8.7%
(Fig. 5C).
To evaluate the roles of the target genes in DVT,
six specific terms (angiogenesis, cell proliferation, adhesion,
inflammatory response, apoptosis and hypoxia) mediating vascular
function were extracted from multiple levels within the GO
hierarchy (Table I). Notably, the
majority of these genes were closely associated with endothelial
function (marked in bold in Table
I) and the majority demonstrated a decreased expression level
in DVT. For example, the expression levels of EC proliferative and
anti-apoptosis genes, growth arrest specific 6 (GAS6), vascular
endothelial growth factor B (VEGFB), the most well-known inducer of
angiogenesis, and endothelial inflammation factor, Kruppel like
factor 4 (KLF4) were reported to be reduced in atherosclerosis
(11). In addition,
adhesion-associated genes, including junctional adhesion molecule 2
(JAM2), which affects EC junctions (12), C-X-C motif chemokine ligand 12
(CXCL12), which mediates recruitment of inflammatory and thrombotic
cells (13), and apolipoprotein L
domain containing 1 (APOLD1) (14), a novel EC early response protein
associated with hypoxia, were equally reduced in DVT.
 | Table IGenes associated with specific GO
terms (angiogenesis, cell proliferation, adhesion, inflammatory
response, apoptosis and hypoxia) in DVT. Genes related to
endothelial cells are marked in bold. |
Table I
Genes associated with specific GO
terms (angiogenesis, cell proliferation, adhesion, inflammatory
response, apoptosis and hypoxia) in DVT. Genes related to
endothelial cells are marked in bold.
| Specific Go
terms | Upregulated
genes | Downregulated
genes |
|---|
| Angiogenesis | - | APOLD1,
CALCRL, KLF4, DLL1, CXCL12,
EMP2, GATA2, VEGFB |
| Cell
proliferation | PTH,
KCNH1, ITGA2 | ARC,
CALCRL, CAV2, CHP2, CXCL12, DLL1,
EFEMP1, EMP2, GAS6, GATA2, KLF4,
NACC2, NDRG4, PBX1, PPARG, PTHLH, RERG,
SPRY2, TNFRSF11B, VEGFB, WFDC1,
WISP2, ZFP36L1 |
| Adhesion | ITGA2 | PPAP2B,
HSPA2, GAS6, ZFP36L1, CXCL12,
JAM2, KLF4, EMP2, FLRT3, WISP2,
DLL1, CAV2, HAS1, ITGA, RND3,
CNTNAP1 |
| Inflammatory
response | ITGA2 | PPARG,
CALCRL, WFDC1, STK39, TNFRSF11B,
IL17RE |
| Apoptosis | PTH | ARC,
HSPA2, GAS6, TNFRSF11B, NACC2, PPARG,
SPRY2, CXCL12, VEGFB, GULP1,
KLF4, ZFP36L1, DLL1, STK39 |
| Hypoxia | ITGA2 | APOLD1,
CYGB, ARC, CXCL12 |
KEGG pathway analysis demonstrated that four of the
top 15 signal pathways were associated with cardiovascular disease,
and three to metabolism. Among them, the following three had
enriched gene numbers >3: dilated cardiomyopathy, vascular
smooth muscle contraction and focal adhesion pathways (Fig. 5D).
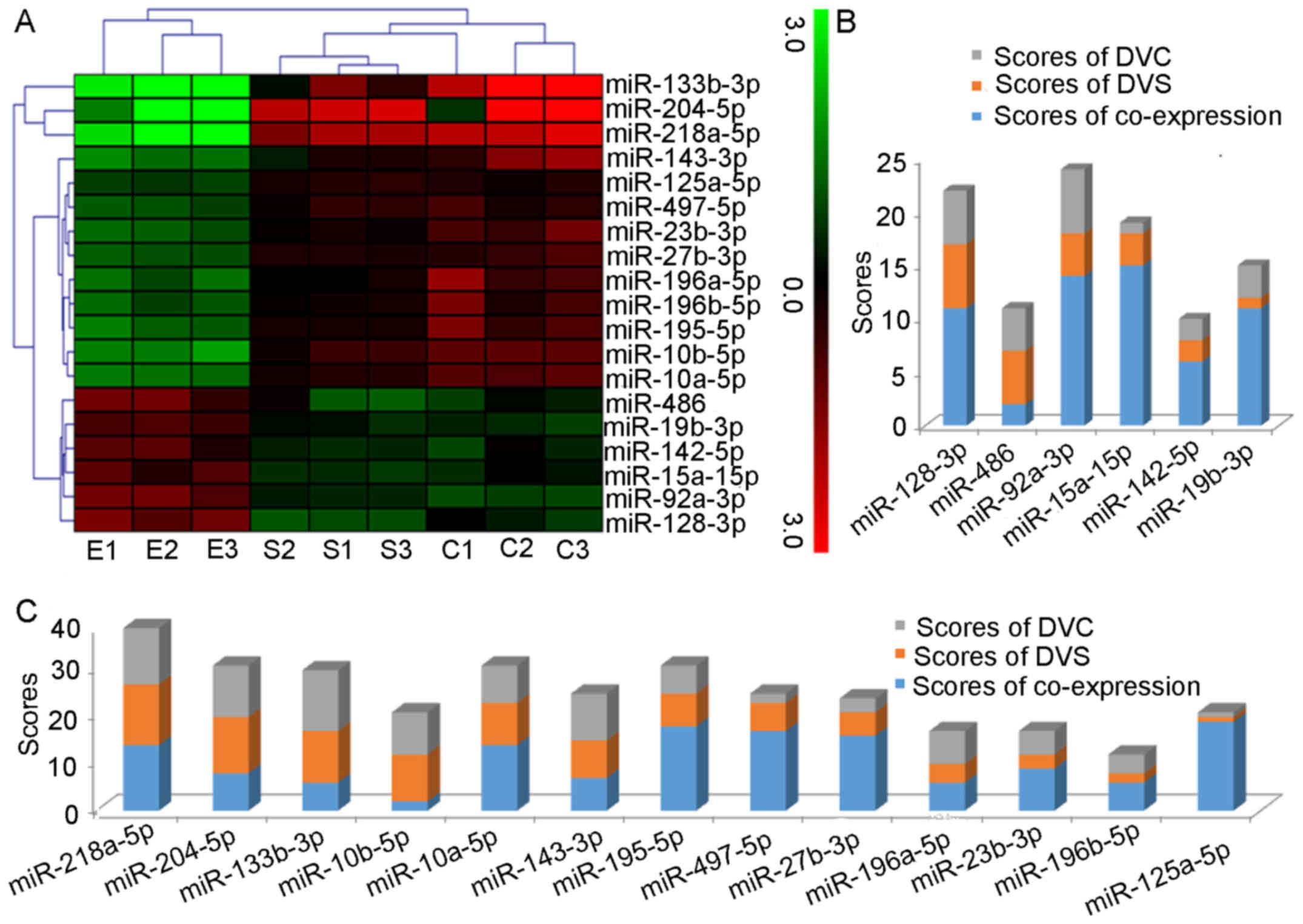
Clustering of 19 miRNA expression
The clustering analysis of the nine samples and 19
differentially expressed miRNAs revealed a distinct miRNA signature
during DVT (Fig. 6A). The six
upregulated miRNAs grouped together and the 13 downregulated miRNAs
were divided into two categories, in which miR-133b-3p, miR-218a-5p
and miR-204-5p were grouped together, while the remaining 10 miRNAs
all gather in a second cluster.
Ranking
To demonstrate the value of the 19 miRNAs in DVT,
they were positioned according to the degree of co-expression from
network (Table II) and fold
change form profiling (Tables
III and IV). The
co-expression was ranked according to the degree of co-expression,
the highest degree scored 19 points, and the lowest 1 point. The
fold change score was ranked in the same way. The most
differentially expressed upregulated miRNA received 6 points (for a
total of 6 upregulated miRNAs) and downregulated miRNAs received 13
points (for a total of 13 downregulated miRNAs), and the lowest 1
point (data not shown). Bar charts of the rankings are presented in
Fig. 6B and C. The top
upregulated miRNA was miR-92a-3p, while miR-218a-5p was ranked
first among the downregulated miRNAs.
 | Table IIDegrees of differential expression
and targets of the 19 differentially expressed miRNAs. |
Table II
Degrees of differential expression
and targets of the 19 differentially expressed miRNAs.
| miRNA | Degree | Targets |
|---|
| miR-125a-5p | 22 | ENTPD2, CXCL12,
GAS6, YPEL4, ARC, CHP2, GRB10, ITGA7, CYGB, SLC4A10, HAS1, WISP2,
OLFML2A, LZTS1, TNFRSF11B, ADAMTS1, EMP2, APOLD1, RHOV, VEGFB,
CLIC5, ITPKC |
| miR-195-5p | 20 | NKD1, DLL1, PLCXD2,
ADRA1D, GK, SLC16A6, WIF1, ITGA2, SNX33, PLA2G15, FLRT3, TPPP3,
MMP17, ADCY5, RUNX1T1, ISLR, DYRK1B, PTHLH, PTPN4, PTH |
| miR-497-5p | 19 | DLL1, ITGA2, NKD1,
GK, PTH, ADRA1D, ISLR, PLA2G15, PTHLH, RUNX1T1, SNX33, TPPP3,
MMP17, SLC16A6, PLCXD2, ADCY5, DYRK1B, WIF1, PTPN4 |
| miR-27b-3p | 18 | PPARG, RND3,
ZFP36L1,PPAP2B, FAM171A1, EPB41L1, HEG1, FLRT3, GATA2, NOVA1,
STK39, EEPD1, WIPF3, SPRY2, PLCXD2, STMN2, DCX, PKIA |
| miR-15a-5p | 12 | ISLR, RUNX1T1,
DLL1, PTH, PTHLH, TPPP3, GK, NKD1, ITGA2, ADRA1D, PLA2G15,
MMP17 |
| miR-10a-5p | 9 | ADRA1D, RBMS3,
LZTS1, KAZALD1, SH3D19, KLF4, EEPD1, LTBP1, PPP1R14C |
| miR-218a-5p | 9 | FLRT3, SH3D19,
PLA2G15, CALCRL, KCNH1, PLCXD2, NDRG4, FAM63B, LHFP |
| miR-92a-3p | 9 | FAM110B, GATA2,
NOVA1, SLC32A1, STK39, KLF4, PRKAR1B, KLF6, GPM6A |
| miR-128-3p | 8 | EFEMP1, DCX, SCML4,
RND3, IL17RE, ITPKC, KLF4, ABCB9 |
| miR-19b-3p | 8 | GULP1, SH3D19,
HSPA2, RBMS3, HEG1, GRB10, ENC1, ARC |
| miR-23b-3p | 7 | KCNH1, DLG2,
CXCL12, LOC499602, CAV2, NACC2, NAP1L5 |
| miR-204-5p | 6 | NAT8L, LRRN4CL,
DHH, NOVA1, HEG1, RGD1562629 |
| miR-143-3p | 5 | DYRK1B, CXCL12,
CLIC5, WFDC1, PTPN4 |
| miR-133b-3p | 4 | EMP2, ENC1,
CNTNAP1, GPM6A |
| miR-142-5p | 4 | RERG, JAM2, CNTLN,
GCH1 |
| miR-196a-5p | 4 | PLCXD2, RBMS3,
PBX1, ABCB9 |
| miR-196b-5p | 4 | RBMS3, PLCXD2,
PBX1, ABCB9 |
| miR-10b-5p | 1 | GPM6A |
| miR-486 | 1 | BTBD3 |
 | Table IIIDifferential expression of 6
upregulated miRNAs in the DVT, the control and the sham group. |
Table III
Differential expression of 6
upregulated miRNAs in the DVT, the control and the sham group.
| miRNA | Signal values
(means ± SD)
| Fold change
(P-value)
|
|---|
| DVT group | Control group | sham group | DVT/control | DVT/sham |
|---|
| miR-19b-3p | 573.02±42.12 | 229.30±24.47 | 273.82±34.41 | 2.50
(1.14E-03) | 2.09
(5.09E-03) |
| miR-142-5p | 104.09±22.38 | 45.03±11.02 | 42.85±2.04 | 2.31
(2.52E-02) | 2.43
(3.16E-02) |
| miR-15a-5p | 185.14±32.59 | 89.34±11.84 | 72.66±56.24 | 2.07
(1.40E-02) | 2.55
(1.44E-02) |
| miR-92a-3p | 769.03±111.47 | 190.20±9.65 | 258.05±14.72 | 4.04
(3.17E-03) | 2.98
(5.12E-03) |
| miR-486 | 826.37±182.19 | 303.27±55.64 | 263.43±112.57 | 2.72
(1.21E-02) | 3.14
(2.93E-02) |
| miR-128-3p | 396.74±52.95 | 52.95±25.23 | 88.78±4.53 | 2.91
(3.89E-03) | 4.47
(1.61E-03) |
 | Table IVDifferential expression of 13
downregulated miRNAs in the DVT, the vontrol and the sham
group. |
Table IV
Differential expression of 13
downregulated miRNAs in the DVT, the vontrol and the sham
group.
| miRNA | Signal values
(means ± SD)
| Fold change
(P-value)
|
|---|
| DVT group | Control group | Sham group | DVT/Control | DVT/sham |
|---|
| miR-218a-5p | 1.03±0.73 | 58.88±9.69 | 40.44±6.74 | 56.94
(2.92E-02) | 39.11
(3.41E-02) |
| miR-204-5p | 0.73±0.87 | 29.89±18.58 | 28.45±2.91 | 40.75
(3.01E-02) | 38.79
(3.93E-02) |
| miR-133b-3p | 1.79±1.71 |
1631.01±1898.14 | 47.74±22.25 | 912.16
(1.00E-02) | 26.70
(4.85E-02) |
| miR-10b-5p | 75.88±9.55 | 488.90±0.00 | 333.18±53.56 | 6.44
(2.48E-03) | 4.39
(9.00E-04) |
| miR-10a-5p | 113.96±6.70 | 569.35±34.06 | 365.225±26.42 | 5.00
(1.11E-05) | 3.20
(9.26E-05) |
| miR-143-3p | 807.53±91.71 |
5490.83±1852.49 | 2341.94±438.29 | 6.80
(1.54E-02) | 2.90
(6.51E-03) |
| miR-195-5p | 169.80±23.48 | 793.40±215.98 | 456.61±0.00 | 4.67
(5.20E-03) | 2.69
(1.06E-02) |
| miR-497-5p | 121.73±10.93 | 331.66±56.00 | 308.19±39.63 | 2.72
(4.81E-03) | 2.53
(2.33E-03) |
| miR-27b-3p | 245.33±14.03 | 768.64±108.02 | 605.37±8.52 | 3.13
(2.67E-03) | 2.47
(1.26E-03) |
| miR-196a-5p | 32.02±5.81 | 157.60±58.56 | 74.12±5.96 | 4.92
(1.21E-02) | 2.31
(9.65E-03) |
| miR-23b-3p | 421.90±49.45 |
1712.503±358.92 | 972.26±53.57 | 4.06
(2.97E-03) | 2.30
(3.16E-03) |
| miR-196b-5p | 31.71±4.44 | 120.13±38.66 | 70.53±3.39 | 3.79
(1.75E-02) | 2.22
(8.43E-03) |
| miR-125a-5p | 70.06±2.92 | 142.10±10.40 | 150.30±9.76 | 2.03
(1.14E-03) | 2.15
(4.62E-04) |
Discussion
DVT and PE are significant public health concerns,
representing major sources of mortality and morbidity. Animal
models are important for understanding the pathophysiology of these
thrombogenesis-associated diseases (15). To minimize the error caused by
surgery, a rat IVC ligation model was used in the present study,
which provides a total stasis environment and a consistent thrombus
size after 3 days of ligation. This model has been widely used in
previous studies (1,2,16–19).
Although miRNA screening methodologies have become
widely available, and large studies of the role of miRNAs were
performed in the pathogenesis of various cardiovascular diseases
(20–24), to the best of our knowledge, only
three studies reported miRNA profiling in vein thrombosis. Xiao
et al (25) identified
markedly higher plasma levels of various miRNAs, (including
miR-134, miR-410 and miR-520 amongst others) in patients suffering
from acute PE (25). The miR-320a
and miR-320b were upregulated in plasma samples from VTE patients
compared with healthy controls (26). Qin et al (27) demonstrated an increased serum
level of three miRNAs (miR-582, miR-195 and miR-532) in patients
with postoperative DVT versus control subjects (27).
All previous studies identified differentially
expressed miRNAs in the thrombosis group, consistent with the
present study, but the specific miRNA profiles differed, which may
be associated with differences in the subsets of selected diseases,
species and types of samples that were evaluated. Beyond this, all
of the known miRNA profiles associated with thrombosis were
detected in plasma or serum as potential markers, while, to the
best of our knowledge, there are no studies evaluating the miRNA
profiles in venous tissue. The miRNA profiles from biological fluid
are used primarily for diagnoses, while miRNA profiles in venous
tissues may be more conducive to investigating the underlying
mechanism of DVT, which has rarely been the focus of study.
Therefore, the present study was designed to investigate this
further.
By integrative methodology, miRNA/mRNA pairs in DVT
were used to construct the regulatory network, indicating that
these miRNAs may be important in DVT by regulating their target
genes. Further GO and KEGG analyses of 98 genes indicated that DVT
was associated with specific biological processes, such as
angiogenesis or inflammation.
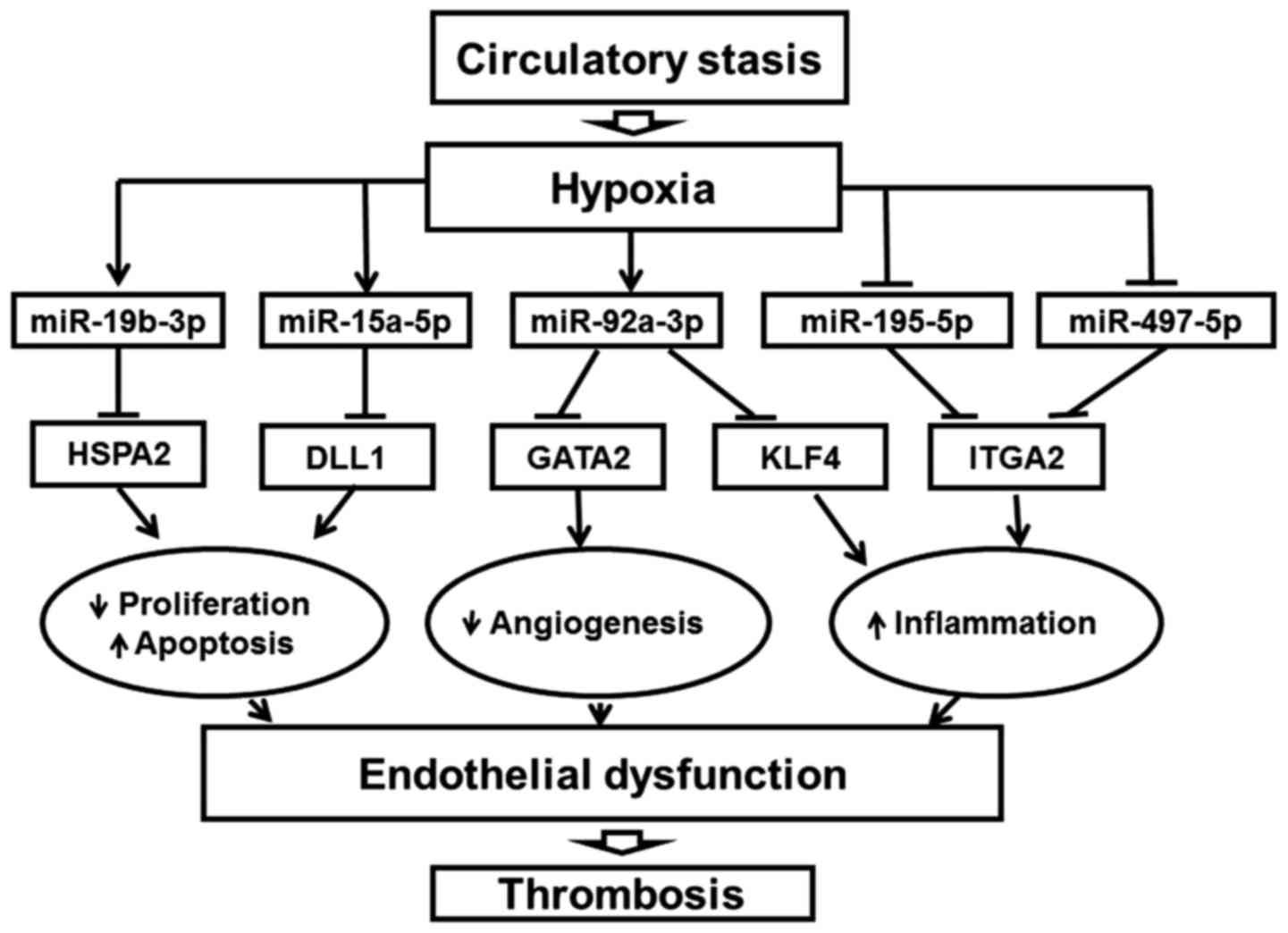
It is worth noting that the changes in these genes
were reported to affect endothelial function (28,29). For example, heat shock protein
family A (Hsp70) member 2 (HSPA2) in the adult corneal endothelium
were demonstrated to be sensitized to mediators of cell death
(30). In addition, delta like
canonical Notch ligand 1 (DLL1) is an essential Notch ligand in the
vascular endothelium, and activates Notch1 to maintain arterial
integrity (31). Another example
is the angiogenic factor, GATA binding protein 2 (GATA2). Previous
studies (32,33) have indicated that GATA2 is
important for vascular integrity. In addition, KLF4 functions as an
important regulator of EC inflammation (34). Integrin subunit α2 (ITGA2), the
major collagen-binding α-integrin subunit in ECs, is involved in
inflammatory processes and cell invasion (35,36). Increased expression levels of
ITGA2 increases joint inflammation (36).
Clustering analysis of 19 miRNAs revealed a distinct
miRNA signature during DVT. Notably, the majority of the 19 miRNAs
have also been reported in the endothelium. For example, miR-10b-5p
and miR-195 have been identified as diagnostic markers of VTE
(26,27). In addition, miR-92a, miR-15a,
miR-196a/196b and miR-19b have been reported to be involved in the
processes of EC dysfunction, proliferation, apoptosis, migration
and angiogenesis, which ultimately influence diseases, such as
thrombosis and atherosclerosis, and tumors (37–41).
Maintenance of the functional integrity of the
endothelium is important to preserve blood flow and prevent
thrombosis (42). The endothelium
secretes factors that control vascular relaxation and contraction,
thermogenesis, fibrinolysis, and platelet activation and inhibition
(43). While EC injury and
dysfunction are considered to be the initial events in the
development of thromboembolism, atherosclerosis, postangioplasty
restenosis and plaque erosion contribute to macrovascular
complications (44). The present
findings indicate that endothelial dysfunction is particularly
significant in ligation-induced thrombosis. A flowchart of the role
of endothelial miRNAs in DVT was generated by integrating the
expression and regulation of endothelial-associated miRNAs and
target genes (Fig. 7).
In conclusion, profiling of miRNA and mRNA
expression was performed in parallel to integrate the two
biological levels to better understand DVT. The use of this
integrated approached elucidates the associations between the
expression levels of miRNAs and mRNAs. This novel systematic study
provides information on miRNA target regulation and provides a
foundation for future studies.
Acknowledgments
The authors would like to thank Binbin Zou for
assistance with bioinformatics analysis. The present study was
supported by grants from the National Natural Science Foundation of
China (grant no. 81470088).
References
|
1
|
Tapson VF and Humbert M: Incidence and
prevalence of chronic thromboembolic pulmonary hypertension: From
acute to chronic pulmonary embolism. Proc Am Thorac Soc. 3:564–567.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang W, Li C, Li W, Kong L, Qian A, Hu N,
Meng Q and Li X: MiR-150 enhances the motility of EPCs in vitro and
promotes EPCs homing and thrombus resolving in vivo. Thromb Res.
133:590–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heit JA: The epidemiology of venous
thromboembolism in the community. Arterioscler Thromb Vasc Biol.
28:370–372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nosaka M, Ishida Y, Kimura A, Hama M,
Kawaguchi T, Yamamoto H, Kuninaka Y, Shimada E and Kondo T:
Immunohistochemical detection of intrathrombotic IL-6 and its
application to thrombus age estimation. Int J Legal Med.
129:1021–1025. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tamaki H and Khasnis A: Venous
thromboembolism in systemic autoimmune diseases: A narrative review
with emphasis on primary systemic vasculitides. Vasc Med.
20:369–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Sundquist K, Elf JL, Strandberg K,
Svensson PJ, Hedelius A, Palmer K, Memon AA, Sundquist J and Zöller
B: Diagnostic potential of plasma microRNA signatures in patients
with deep-vein thrombosis. Thromb Haemost. 116:328–336. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tan M, Yan HB, Li JN, Li WK, Fu YY, Chen W
and Zhou Z: Thrombin stimulated platelet-derived exosomes inhibit
platelet-derived growth factor receptor-beta expression in vascular
smooth muscle cells. Cell Physiol Biochem. 38:2348–2365. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feinberg MW and Moore KJ: MicroRNA
regulation of atherosclerosis. Circ Res. 118:703–720. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chistiakov DA, Orekhov AN and Bobryshev
YV: The role of miR-126 in embryonic angiogenesis, adult vascular
homeostasis, and vascular repair and its alterations in
atherosclerotic disease. J Mol Cell Cardiol. 97:47–55. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang Y and Davies PF: Site-specific
microRNA-92a regulation of Kruppel-like factors 4 and 2 in
atherosusceptible endothelium. Arterioscler Thromb Vasc Biol.
32:979–987. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ebnet K, Aurrand-Lions M, Kuhn A, Kiefer
F, Butz S, Zander K, Meyer zu Brickwedde MK, Suzuki A, Imhof BA and
Vestweber D: The junctional adhesion molecule (JAM) family members
JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: A
possible role for JAMs in endothelial cell polarity. J Cell Sci.
116:3879–3891. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rafii S and Nolan D: Cholesterol activates
vascular niche and hematopoiesis. Blood. 115:3857–3858. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Regard JB, Scheek S, Borbiev T, Lanahan
AA, Schneider A, Demetriades AM, Hiemisch H, Barnes CA, Verin AD
and Worley PF: Verge: A novel vascular early response gene. J
Neurosci. 24:4092–4103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Diaz JA, Obi AT, Myers DD Jr, Wrobleski
SK, Henke PK, Mackman N and Wakefield TW: Critical review of mouse
models of venous thrombosis. Arterioscler Thromb Vasc Biol.
32:556–562. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Diaz JA, Ballard-Lipka NE, Farris DM,
Hawley AE, Wrobleski SK, Myers DD, Henke PK, Lawrence DA and
Wakefield TW: Impaired fibrinolytic system in ApoE gene-deleted
mice with hyperlipidemia augments deep vein thrombosis. J Vasc
Surg. 55:815–822. 2012. View Article : Google Scholar
|
|
17
|
Nakata N and Kira Y: Effects of
preoperative glycyrrhizin infusion for the prevention of venous
thrombosis on the tissue expression of antithrombin in a rat model.
Ann Vasc Dis. 9:95–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Oboshi M, Naito Y, Sawada H, Hirotani S,
Iwasaku T, Okuhara Y, Morisawa D, Eguchi A, Nishimura K, Fujii K,
et al: Temporary dietary iron restriction affects the process of
thrombus resolution in a rat model of deep vein thrombosis. PLoS
One. 10:e01266112015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yin M, Tian S, Huang X, Huang Y and Jiang
M: Role and mechanism of tissue plasminogen activator in venous
wall fibrosis remodeling after deep venous thrombosis via the
glycogen synthase kinase-3 beta signaling pathway. J Surg Res.
184:1182–1195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yepes S, López R, Andrade RE,
Rodriguez-Urrego PA, López-Kleine L and Torres MM: Co-expressed
miRNAs in gastric adenocarcinoma. Genomics. 108:93–101. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Setién-Olarra A, Bediaga NG, Acha-Sagredo
A, Marichalar-Mendia X, de Pancorbo MM and Aguirre-Urizar JM:
Genomewide miRNA profiling of oral lichenoid disorders and oral
squamous cell carcinoma. Oral Dis. 22:754–760. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Louwies T, Vuegen C, Panis LI, Cox B,
Vrijens K, Nawrot TS and De Boever P: miRNA expression profiles and
retinal blood vessel calibers are associated with short-term
particulate matter air pollution exposure. Environ Res. 147:24–31.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yentrapalli R, Azimzadeh O, Kraemer A,
Malinowsky K, Sarioglu H, Becker KF, Atkinson MJ, Moertl S and
Tapio S: Quantitative and integrated proteome and microRNA analysis
of endothelial replicative senescence. J Proteomics. 126:12–23.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Murphy MS, Casselman RC, Tayade C and
Smith GN: Differential expression of plasma microRNA in
preeclamptic patients at delivery and 1 year postpartum. Am J
Obstet Gynecol. 213:367.e361–369. 2015. View Article : Google Scholar
|
|
25
|
Xiao J, Jing ZC, Ellinor PT, Liang D,
Zhang H, Liu Y, Chen X, Pan L, Lyon R, Liu Y, et al: MicroRNA-134
as a potential plasma biomarker for the diagnosis of acute
pulmonary embolism. J Transl Med. 9:1592011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Starikova I, Jamaly S, Sorrentino A,
Blondal T, Latysheva N, Sovershaev M and Hansen JB: Differential
expression of plasma miRNAs in patients with unprovoked venous
thromboembolism and healthy control individuals. Thromb Res.
136:566–572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qin J, Liang H, Shi D, Dai J, Xu Z, Chen
D, Chen X and Jiang Q: A panel of microRNAs as a new biomarkers for
the detection of deep vein thrombosis. J Thromb Thrombolysis.
39:215–221. 2015. View Article : Google Scholar
|
|
28
|
Johnson KE and Wilgus TA: Vascular
endothelial growth factor and angiogenesis in the regulation of
cutaneous wound repair. Adv Wound Care (New Rochelle). 3:647–661.
2014. View Article : Google Scholar :
|
|
29
|
Healy AM, Schwartz JJ, Zhu X, Herrick BE,
Varnum B and Farber HW: Gas 6 promotes Axl-mediated survival in
pulmonary endothelial cells. Am J Physiol Lung Cell Mol Physiol.
280:L1273–L1281. 2001.PubMed/NCBI
|
|
30
|
Frausto RF, Wang C and Aldave AJ:
Transcriptome analysis of the human corneal endothelium. Invest
Ophthalmol Vis Sci. 55:7821–7830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sörensen I, Adams RH and Gossler A:
DLL1-mediated Notch activation regulates endothelial identity in
mouse fetal arteries. Blood. 113:5680–5688. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Johnson KD, Hsu AP, Ryu MJ, Wang J, Gao X,
Boyer ME, Liu Y, Lee Y, Calvo KR, Keles S, et al: Cis-element
mutated in GATA2-dependent immunodeficiency governs hematopoiesis
and vascular integrity. J Clin Invest. 122:3692–3704. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lim KC, Hosoya T, Brandt W, Ku CJ,
Hosoya-Ohmura S, Camper SA, Yamamoto M and Engel JD: Conditional
Gata2 inactivation results in HSC loss and lymphatic mispatterning.
J Clin Invest. 122:3705–3717. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hamik A, Lin Z, Kumar A, Balcells M, Sinha
S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER and Jain MK:
Kruppel-like factor 4 regulates endothelial inflammation. J Biol
Chem. 282:13769–13779. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Roche F, Sipilä K, Honjo S, Johansson S,
Tugues S, Heino J and Claesson-Welsh L: Histidine-rich glycoprotein
blocks collagen-binding integrins and adhesion of endothelial cells
through low-affinity interaction with α2 integrin. Matrix Biol.
48:89–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peters MA, Wendholt D, Strietholt S, Frank
S, Pundt N, Korb-Pap A, Joosten LA, van den Berg WB, Kollias G,
Eckes B, et al: The loss of α2β1 integrin suppresses joint
inflammation and cartilage destruction in mouse models of
rheumatoid arthritis. Arthritis Rheum. 64:1359–1368. 2012.
View Article : Google Scholar
|
|
37
|
Sandrim VC, Dias MC, Bovolato AL,
Tanus-Santos JE, Deffune E and Cavalli RC: Plasma from
pre-eclamptic patients induces the expression of the
anti-angiogenic miR-195-5p in endothelial cells. J Cell Mol Med.
20:1198–1200. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu S, Hong Q, Wang Y, Hou K, Wang L, Zhang
Y, Fu B, Zhou Y, Zheng W, Chen X, et al: High concentrations of
uric acid inhibit angiogenesis via regulation of the Kruppel-like
factor 2-vascular endothelial growth factor-A axis by miR-92a. Circ
J. 79:2487–2498. 2015. View Article : Google Scholar
|
|
39
|
Xue Y, Wei Z, Ding H, Wang Q, Zhou Z,
Zheng S, Zhang Y, Hou D, Liu Y, Zen K, et al: MicroRNA-19b/221/222
induces endothelial cell dysfunction via suppression of PGC-1α in
the progression of atherosclerosis. Atherosclerosis. 241:671–681.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zheng X, Li A, Zhao L, Zhou T, Shen Q, Cui
Q and Qin X: Key role of microRNA-15a in the KLF4 suppressions of
proliferation and angiogenesis in endothelial and vascular smooth
muscle cells. Biochem Biophys Res Commun. 437:625–631. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Plummer PN, Freeman R, Taft RJ, Vider J,
Sax M, Umer BA, Gao D, Johns C, Mattick JS, Wilton SD, et al:
MicroRNAs regulate tumor angiogenesis modulated by endothelial
progenitor cells. Cancer Res. 73:341–352. 2013. View Article : Google Scholar
|
|
42
|
Lüscher TF and Barton M: Biology of the
endothelium. Clin Cardiol. 20(Suppl 2): II-3-II-101997.
|
|
43
|
Axtell AL, Gomari FA and Cooke JP:
Assessing endothelial vasodilator function with the Endo-PAT 2000.
J Vis Exp. Oct 15–2010.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hartge MM, Kintscher U and Unger T:
Endothelial dysfunction and its role in diabetic vascular disease.
Endocrinol Metab Clin North Am. 35:551–560. viii–ix. 2006.
View Article : Google Scholar : PubMed/NCBI
|