Introduction
Lung cancer results in the death of ~1.59 million
individuals worldwide each year, and is a leading cause of cancer
mortality (1,2). According to the pathological
diagnostic classification, non-small cell lung cancer (NSCLC)
accounts for >85% of cases of lung cancer (3). The molecular pathogenesis of NSCLC
has received increasing attention in recent years (4–7).
Thus, non-coding RNAs, which serve crucial functions in numerous
biological processes have become of interest. Based on their size,
sequence and function, non-coding RNAs can be classified into
various subclasses, the two most notable of which are
long-noncoding RNAs (lncRNAs) and microRNAs (miRNAs).
LncRNAs are parts of non-coding RNAs with >200
nucleotides and do not encode proteins (8). Previous studies have indicated that
lncRNAs may have an important involvement in cancer biology and
cellular processes (9,10). LncRNAs may contribute to these
processes via various mechanisms at the post-transcriptional level,
for example, by serving as precursors to miRNA and regulating mRNA
by interacting with miRNAs (11,12). Unlike lncRNAs, miRNAs are
generally 18–25 nucleotides in length, and are small non-coding
protein RNAs that are able to bind to the 3′ untranslated region of
target mRNAs and result in the degradation and translational
downregulation of target mRNAs (13,14). In addition, miRNAs have the
ability to regulate the coding and noncoding transcriptome.
It is worthy of note that lncRNAs and miRNAs serve
pivotal roles in numerous similar processes, including cell
proliferation, apoptosis and carcinogenesis. Due to the similarity
of their functions, it may be hypothesized that an interaction
between lncRNAs and miRNAs occurs during the tumorigenic process.
For instance, the competing endogenous RNA (ceRNA) hypothesis that
lncRNAs serve as molecular sponges for miRNAs has been proposed
(15,16). Thus, the discovery of
lncRNA-miRNA-mRNA networks may lead to a more comprehensive
understanding of the etiology and metastasis mechanism of cancer
and to potential therapeutic targets, and such networks been
studied for several types of cancer (17,18).
In a previous study, the present authors identified
47 lncRNAs, including LINC00968, that were differentially expressed
between normal lung tissues and tumor samples by bioinformatic
analysis (19). Furthermore, a
significant downregulation of LINC00968 in lung adenocarcinoma
(LUAD) tissues and A549 cells was discovered (data not shown).
These experiments suggest that LINC00968 may serve important roles
in lung cancer progression. In the present study, using miRNA
profiling by microarray following the overexpression of LINC00968
in A549 cells, Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analyses of the differentially expressed
miRNAs (DEMs) were performed. Furthermore, a prospective
lncRNA-miRNA-mRNA regulatory network of LINC00968 was constructed.
These results may assist in the exploration of the underlying
regulatory mechanisms in the progression of lung cancer.
Materials and methods
Overexpression of LINC00968 in A549
cells
A549 cells were obtained from the Cell Bank of the
Chinese Academy of Sciences (Shanghai, China). Following culture in
6-well plates overnight, the cells (2×106) were
transfected with a solution of lentiviruses in F-12 culture medium
containing 10% serum (Gibco, Grand Island, NY, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (Hyclone, Logan, UT,
USA) for 24 h. The solution was then removed, fresh F-12 culture
medium was added and the A549 cells were incubated for another
24–72 h. The conventional culture environment was 5% CO2
air in a humidified incubator at 37°C. Lentiviruses encoding
LINC00968 were designed and synthesized by Shanghai GenePharma Co.,
Ltd. (Shanghai, China). An inverted fluorescence microscope was
applied to examine the transfection efficiency (data not
shown).
RNA isolation and miRNA microarray
detection
The sample analysis and microarray hybridization
were conducted by KangChen Bio-tech (Shanghai, China). Total RNA
was isolated using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and purified using an RNeasy mini kit
(Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's
protocol. The quality and quantity of DNA was monitored using a
NanoDrop spectrophotometer (ND-2000; Thermo Fisher Scientific,
Inc.) followed by gel electrophoresis to determine the integrity of
the RNA. The miRNA was labeled using a miRCURY™ Hy3™/Hy5™ Power
labeling kit and hybridized with the miRCURY™ LNA array (version
18.0) (both from Exiqon A/S, Vedbaek, Denmark). The slides were
then scanned with the Axon GenePix 4000B microarray scanner and the
scanned images were analyzed using GenePix Pro 6.0 software (both
from Molecular Devices, LLC, Sunnyvale, CA, USA). DEMs were
identified according to fold changes and P-value at the threshold
set of |log2 FC| >0.58 and P<0.05.
Target gene prediction of DEMs
In order to determine the potential association
between mRNAs and miRNAs, the potential transcriptional targets of
the DEMs were predicted using five miRNA target prediction
algorithms: miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), miRDB (http://www.mirdb.org/), GeneCards (http://www.genecards.org), TargetScan (http://www.Targetscan.org/) and RNA22 (https://cm.Jefferson.edu/rna22/). Target genes
that were commonly predicted by at least three of these algorithms
were considered predicted target genes. Validated target genes
verified by experiments were obtained from miRecords (http://c1.accurascience.com/miRecords/),
miRTarBase and TarBase (http://diana.imis.athenainnovation.gr/DianaTools/index.php?r=tarbase/index).
Target genes from any one of these three algorithms were listed as
validated target genes.
GO and KEGG pathway analysis
Functional enrichment analysis is essential to
uncover biological functions of miRNA target genes. To gain an
understanding of the biological functions of miRNA target genes, GO
classification and KEGG pathway enrichment analysis were performed
using the bioinformatics software DAVID 6.7 (https://david-ncifcrf.gov/). GO classifies genes
according to three categories, biological process, molecular
function and cellular component, and presents a common descriptive
framework of gene annotation and classification for the analysis of
gene-set data. In addition, the potential involvement in biological
pathways of the DEMs were detected based on analysis using the KEGG
pathway database, which is a credible and informative database that
includes almost all biological signal pathways (20). The false discovery rate (FDR)
<0.05 was set as the cut-off for selecting significantly
enriched KEGG pathways.
Construction of a network of
lncRNA-miRNA-mRNA and protein-protein interaction (PPI)
Following the integration of the results of the
predicted targets and validated targets, prospective targets of the
DEMs were obtained. In order to demonstrate the association among
lncRNAs, miRNAs and mRNAs, an lncRNA-miRNA-mRNA interaction network
was created. The network was visualized using Cytoscape (version
3.4.0) (21). The online tool
STRING (http://string-db.org/) was then used to
draw a PPI network of the prospective targets. The highest
confidence of 0.9 was selected as the minimum required interaction
score.
Extraction of gene and miRNA expression
profiles from the Cancer Genome Atlas (TCGA)
A download of RNA-seq data and miRNA-seq data of
lung cancer from the TCGA (https://cancergenome.nih.gov/) data portal was
conducted, in which the extracted expression data included
LINC00968, DEMs and mRNAs associated with LINC00968.
Statistical analysis
Statistical analyses were performed with SPSS
software version 22.0 (IBM Corp., Armonk, NY, USA). The mean ±
standard deviation was used to present the experimental results. A
two-tailed t-test was used to assess the differences between the
different groups. The correlation among lncRNAs, miRNAs and mRNAs
was analyzed by Pearson's correlation analysis. A statistically
significant threshold was defined as P<0.05.
Results
miRNA profiling following the
overexpression of LINC00968
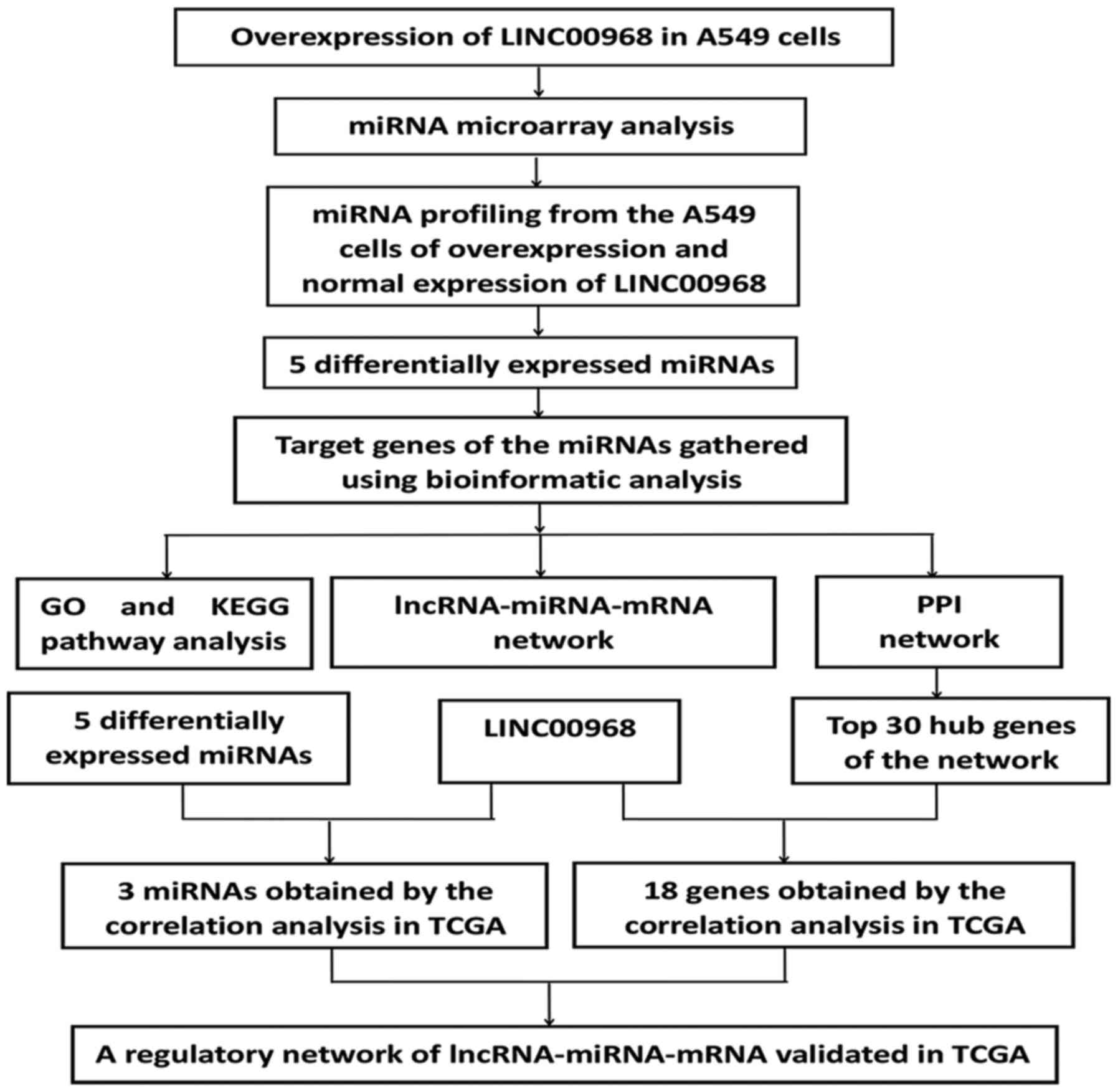
An experimental flow chart of the study is shown in
Fig. 1. To uncover the
association between LINC00968 and miRNAs in lung cancer, microarray
expression profiling of miRNAs in A549 cells transfected with
LINC00968 was performed. In the microarray analysis, 166 miRNAs
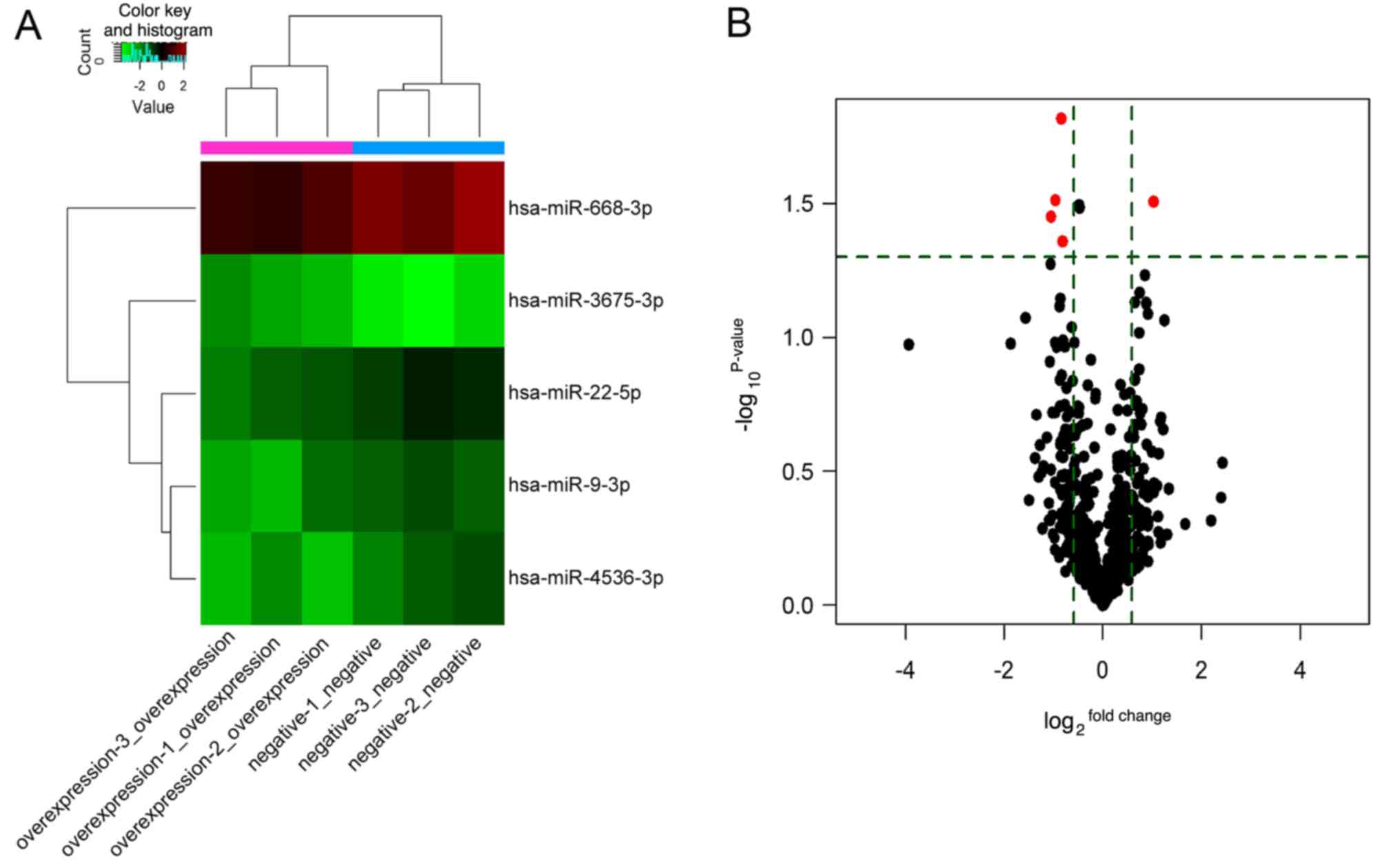
were detected, of which 75 were upregulated and 91 downregulated.
Among these, five DEMs were identified, of which miR-3675 was
highly expressed, and four miRNAs (miR-9, miR-22, miR-668 and
miR-4536) exhibited low expression in A549 cells (Table I), suggesting that these DEMs were
associated with LINC00968 in lung cancer cells. Additionally, the
expression profile of the five DEMs is shown in a heatmap and
volcano plot (Fig. 2). The DEMs
were then subjected to further analysis.
 | Table ISummary of differentially expressed
miRNAs detected by microarray analysis. |
Table I
Summary of differentially expressed
miRNAs detected by microarray analysis.
| ID | Name | Fold change | P-value |
|---|
| Upregulated
miRNA | | | |
| 148214 | miR-3675-3p | 2.034 | 0.031 |
| Downregulated
miRNA | | | |
| 145701 | miR-668-3p | 0.513 | 0.031 |
| 168616 | miR-4536-3p | 0.483 | 0.035 |
| 42532 | miR-22-5p | 0.559 | 0.015 |
| 29852 | miR-9-3p | 0.569 | 0.044 |
Collection of prospective target genes of
DEMs via bioinformatic approaches
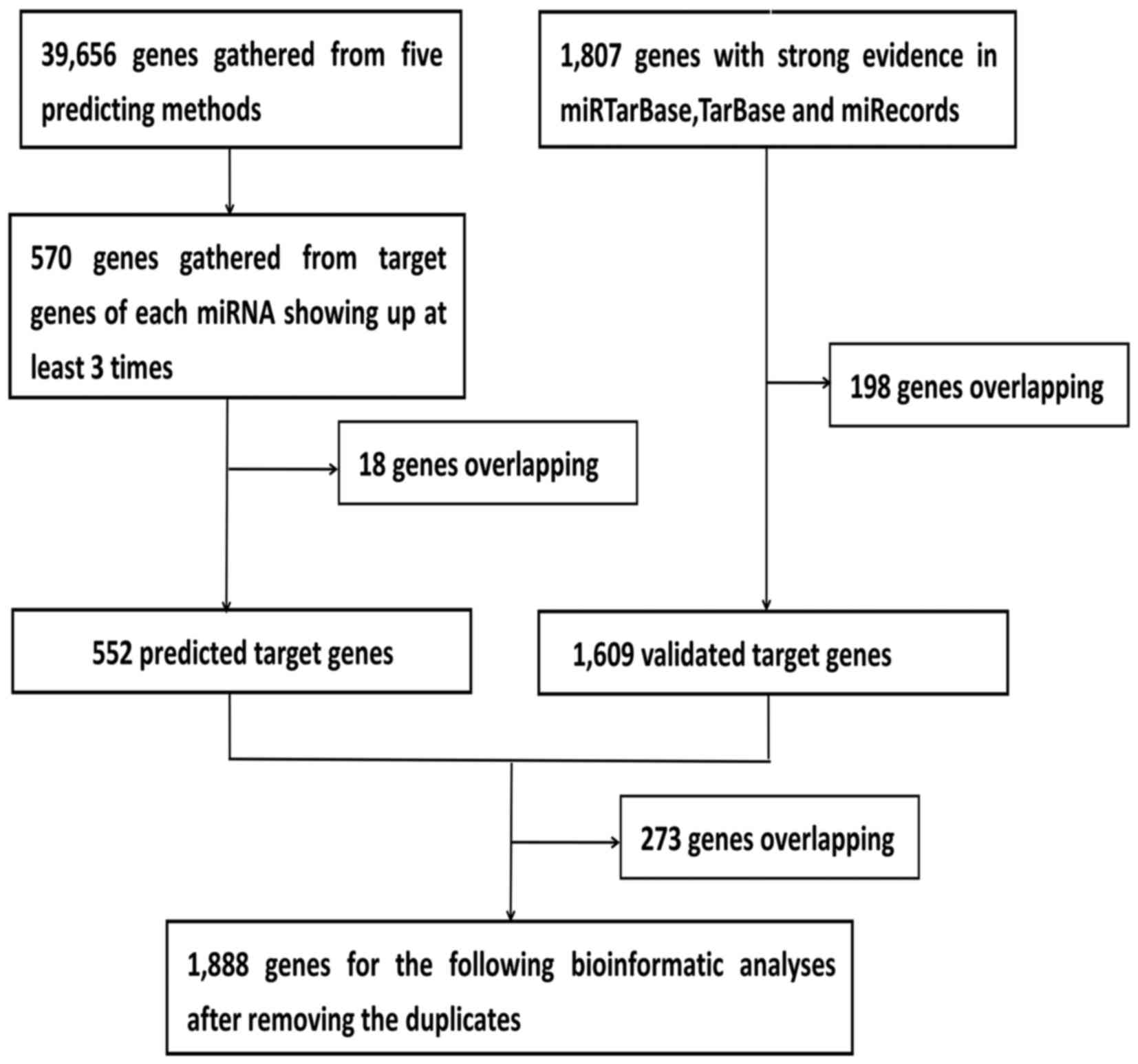
Given that the biological significance of DEMs
depends on their effect upon their targets, the predicted target
genes of the five DEMs were identified. Using this approach, 552
predicted target genes were obtained three times in the five
software programs. In addition, 1,609 validated target genes with
experimental validation were collected from the miRTarBase, TarBase
and miRecords databases. Lastly, 1,888 prospective target genes
were identified by integrating the results of the predicted target
genes and validated target genes (Fig. 3).
GO classification and KEGG pathways of
miRNA prospective target genes
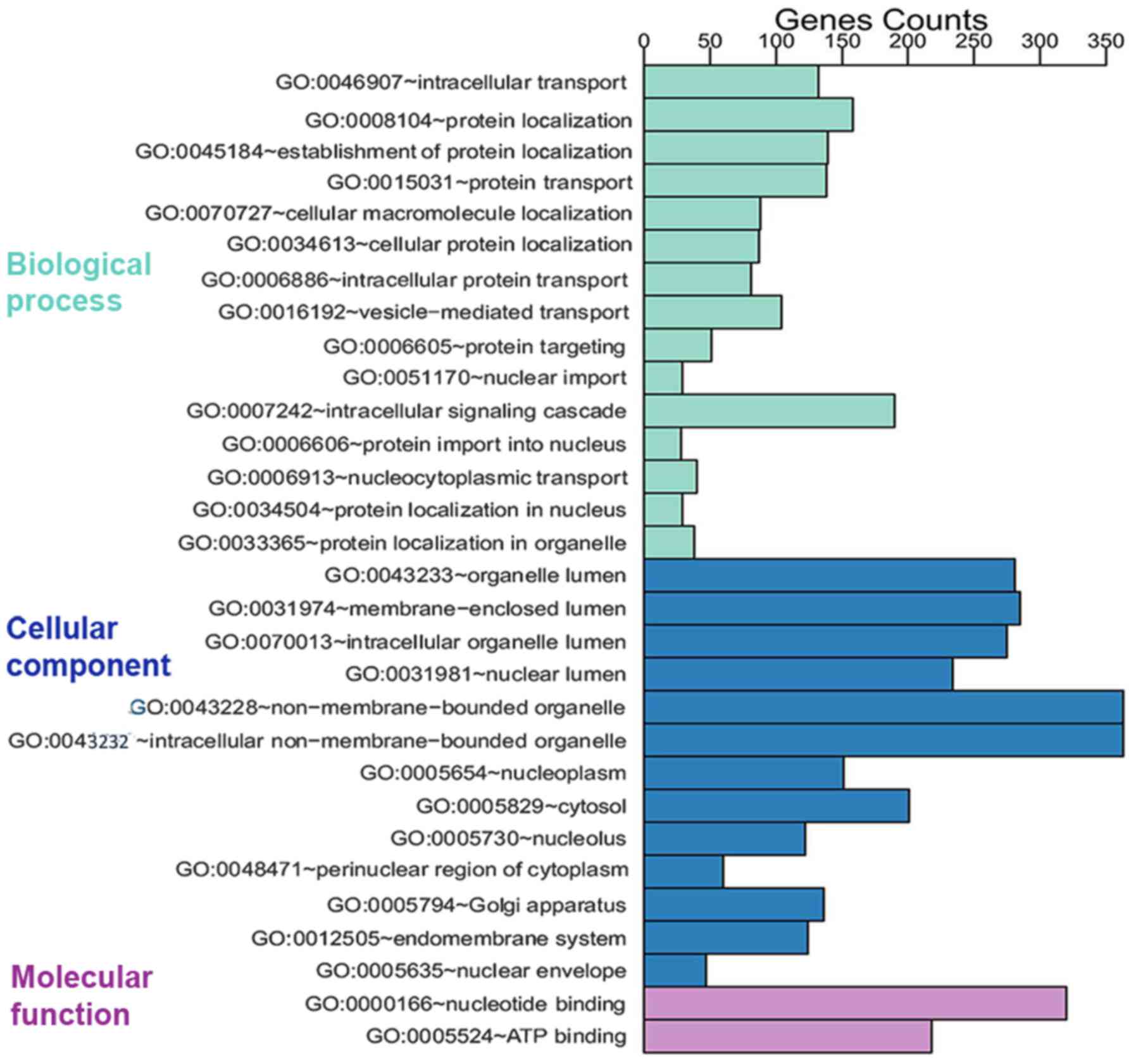
The 1,888 prospective target genes of five DEMs were
then subjected to GO analysis. Through GO annotation and enrichment
analysis, the roles of gene products from biological process,
cellular component and molecular function were identified (Table II). The most significant term in
biological process was intracellular transport (GO:0046907,
P=9.75×10−14), that in cellular component was organelle
lumen (GO:0043233, P=3.96×10−16) and that in molecular
function was nucleotide binding (GO:0000166,
P=2.76×10−9). To better represent gene enrichment in the
three major categories, a pathway schematic was constructed
consisting of the top 30 enriched GO terms (Fig. 4). In the KEGG analysis, four
significantly enriched pathways (FDR <0.05) were obtained
(Table III), including adherens
junction, focal adhesion, long-term potentiation and renal cell
carcinoma, among which the two most significant pathways were
adherens junction (P=3.13×10−7) and focal adhesion
(P=6.90×10−7).
 | Table IITop five enriched GO terms of
prospective target genes from three GO categories. |
Table II
Top five enriched GO terms of
prospective target genes from three GO categories.
| GO ID | GO Term | P-value | FDR | Count | Genes |
|---|
| Biological
process | | | | | |
| GO:0046907 | Intracellular
transport |
9.75×10−14 |
1.80×10−10 | 132 | NCBP2, GRPEL2,
XPO1, GRPEL1, SEC31A, LTBP2, AP1G2, TGFB2, HOOK1, CRY2 |
| GO:0008104 | Protein
localization |
5.59×10−12 |
1.03×10−8 | 158 | GRPEL2, XPO1,
GRPEL1, SEC31A, LTBP2, AP1G2, CHMP4B, MXI1, VPS33A, CTNNB1 |
| GO:0045184 | Establishment of
protein localization |
6.89×10−11 |
1.27×10−7 | 139 | GRPEL2, XPO1,
GRPEL1, SEC31A, LTBP2, AP1G2, CHMP4B, VPS33A, TGFB2, HOOK1 |
| GO:0015031 | Protein
transport |
7.01×10−11 |
1.30×10−7 | 138 | GRPEL2, XPO1,
GRPEL1, SEC31A, LTBP2, AP1G2, CHMP4B, VPS33A, TGFB2, HOOK1 |
| GO:0070727 | Cellular
macromolecule localization |
1.05×10−10 |
1.94×10−7 | 88 | GRPEL2, COPA, XPO1,
GRPEL1, SEC24A, LTBP2, AP1G2, AP1B1, FGF9, CLTC |
| Cellular
component | | | | | |
| GO:0043233 | Organelle
lumen |
3.96×10−16 |
6.66×10−13 | 281 | XRCC5, PDP1, MEF2C,
MRPL40, PNMA3, PDP2, NAA15, SYNCRIP, INTS2, CDC16 |
| GO:0031974 | Membrane-enclosed
lumen |
4.28×10−16 |
6.66×10−13 | 285 | XRCC5, PDP1, MEF2C,
MRPL40, PNMA3, PDP2, NAA15, SYNCRIP, INTS2, CDC16 |
| GO:0070013 | Intracellular
organelle lumen |
7.94×10−16 |
1.15×10−12 | 275 | XRCC5, MEF2C, PDP1,
MRPL40, PNMA3, PDP2, NAA15, SYNCRIP, INTS2, CDC16 |
| GO:0031981 | Nuclear lumen |
1.71×10−15 |
2.48×10−12 | 234 | MEF2C, XRCC5,
PNMA3, NAA15, SYNCRIP, INTS2, CDC16, SART3, WTAP, CD2AP |
| GO:0043228 |
Non-membrane-bounded organelle |
2.50×10−14 |
3.72×10−11 | 363 | XRCC5, MRPL40,
KIFC1, PNMA3, UTRN, CDC16, CCT3, MYLIP, REST, WTAP |
| Molecular
function | | | | | |
| GO:0000166 | Nucleotide
binding |
2.76×10−9 |
4.45×10−6 | 320 | XRCC5, LDHB, KIFC1,
ADCY7, U2AF2, SYNCRIP, LEMD3, CCT3, SART3, RNF213 |
| GO:0005524 | ATP binding |
1.45×10−7 |
2.34×10−4 | 218 | XRCC5, KIFC1,
ADCY7, CCT3, ACTG1, KIF13A, MAP3K7, WNK4, DHX33, DHX36 |
| GO:0032559 |
Adenyl-ribonucleotide binding |
3.94×10−7 |
6.37×10−4 | 218 | XRCC5, KIFC1,
ADCY7, CCT3, ACTG1, KIF13A, MAP3K7, WNK4, DHX33, DHX36 |
| GO:0032555 | Purine
ribonucleotide binding |
5.89×10−7 |
9.51×10−4 | 258 | XRCC5, KIFC1,
ADCY7, CCT3, MAP3K7, ACTG1, KIF13A, WNK4, RAB23, DHX33 |
| GO:0032553 | Ribonucleotide
binding |
5.89×10−7 |
9.51×10−4 | 258 | XRCC5, KIFC1,
ADCY7, CCT3, MAP3K7, ACTG1, KIF13A, WNK4, RAB23, DHX33 |
 | Table IIITop four enriched KEGG pathways
obtained after KEGG pathway analysis. |
Table III
Top four enriched KEGG pathways
obtained after KEGG pathway analysis.
| KEGG ID | KEGG term | P-value | FDR | Count | Genes |
|---|
| hsa04520 | Adherens
junction |
3.13×10−7 |
3.84×10−4 | 26 | PARD3, ERBB2,
CTNND1, CDH1, SRC, IQGAP1, CTNNB1, VCL, ACTG1, MAP3K7, CSNK2A2,
IGF1R, CDC42, SSX2IP, EGFR, PTPRM, PTPRF, TGFBR1, TGFBR2, CREBBP,
SMAD2, MAPK1, TJP1, EP300, FYN, MAPK3 |
| hsa04510 | Focal adhesion |
6.90×10−7 |
8.47×10−4 | 47 | CAV1, TLN1, GRB2,
ERBB2, PIP5K1C, ELK1, ITGB1, PTEN, SRC, VCL, CTNNB1, AKT1, ACTG1,
CDC42, IGF1R, PAK4, PPP1R12A, LAMB1, THBS1, THBS2, PIK3R1, SHC4,
FN1, PRKCA, EGFR, COL4A2, COL4A1, FLT1, MAP2K1, BRAF, ROCK2, MYLK3,
MYL12B, MAPK10, FLNC, FLNB, MAPK1, FYN, ITGA5, JUN, MAPK3, RAP1A,
MAPK9, RAP1B, COL1A1, CRK, MYLK |
| hsa04720 | Long-term
potentiation |
2.54×10−5 |
3.12×10−2 | 21 | PRKCA, MAP2K1,
BRAF, CREBBP, PPP3R1, ITPR3, MAPK1, NRAS, RPS6KA3, EP300, RPS6KA2,
MAPK3, PPP3CB, PPP1R12A, RAP1A, RAP1B, PRKACA, PPP3CA, PRKACB,
PLCB1, PLCB2 |
| hsa05211 | Renal cell
carcinoma |
4.05×10−5 |
4.97×10−2 | 21 | MAP2K1, EPAS1,
BRAF, GRB2, CREBBP, EGLN1, ARNT, TGFB2, AKT1, CDC42, MAPK1, NRAS,
EP300, HIF1A, JUN, PAK4, MAPK3, RAP1A, RAP1B, CRK, PIK3R1 |
Regulatory network of
lncRNA-miRNA-mRNA
To identify the potential links among LINC00968,
DEMs and mRNAs, miRNA microarray and bioinformatic analyses were
performed. Consequently, five DEMs were detected via the
overexpression of LINC00968, which provided the five lncRNA-miRNA
interactions. In addition, to obtain the prospective target genes,
the results of predicted targets and validated targets were
integrated, and the miRNA-mRNA interactions were subsequently
obtained. By combining the lncRNA-miRNA and miRNA-mRNA
interactions, an lncRNA-miRNA-mRNA network was constructed and
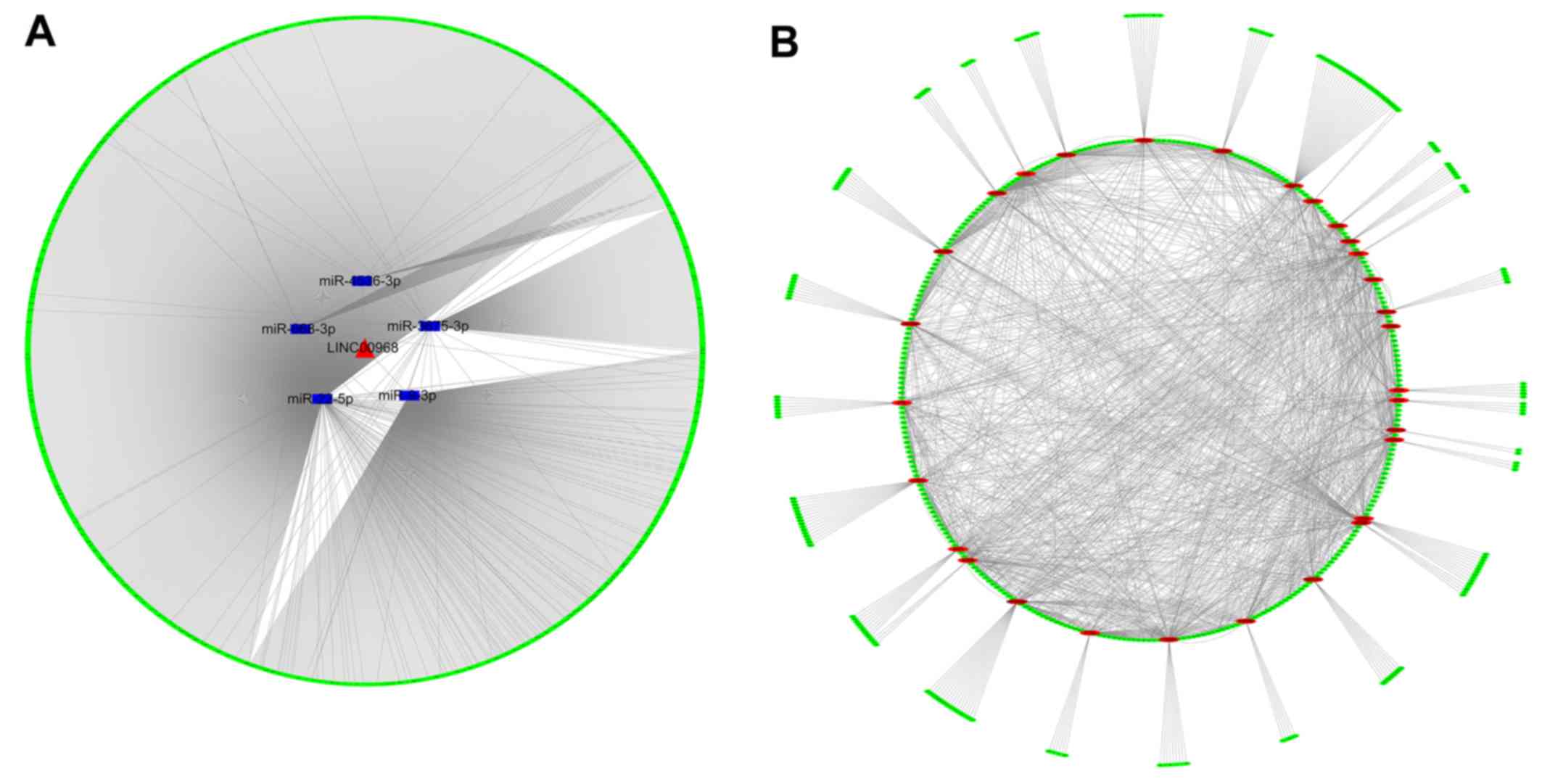
visualized with Cytoscape (Fig.
5A). The network revealed a preliminary connection between
LINC00968, the five DEMs miR-9-3p, miR-22-5p, miR-668-3p,
miR-3675-3p and miR-4536-3p, and 1,888 prospective target genes,
which required further verification.
PPI network of 1,888 prospective target
genes
To identify the association between target genes, a
PPI network was generated using STRING that presented the strength
of the links of different genes. In the PPI network analysis of the
prospective targets, the top 30 genes among all hub genes were
identified, on the basis of the connectivity between genes
(Fig. 5B). These top 30 genes may
have tight connections to the lncRNA-miRNA-mRNA network.
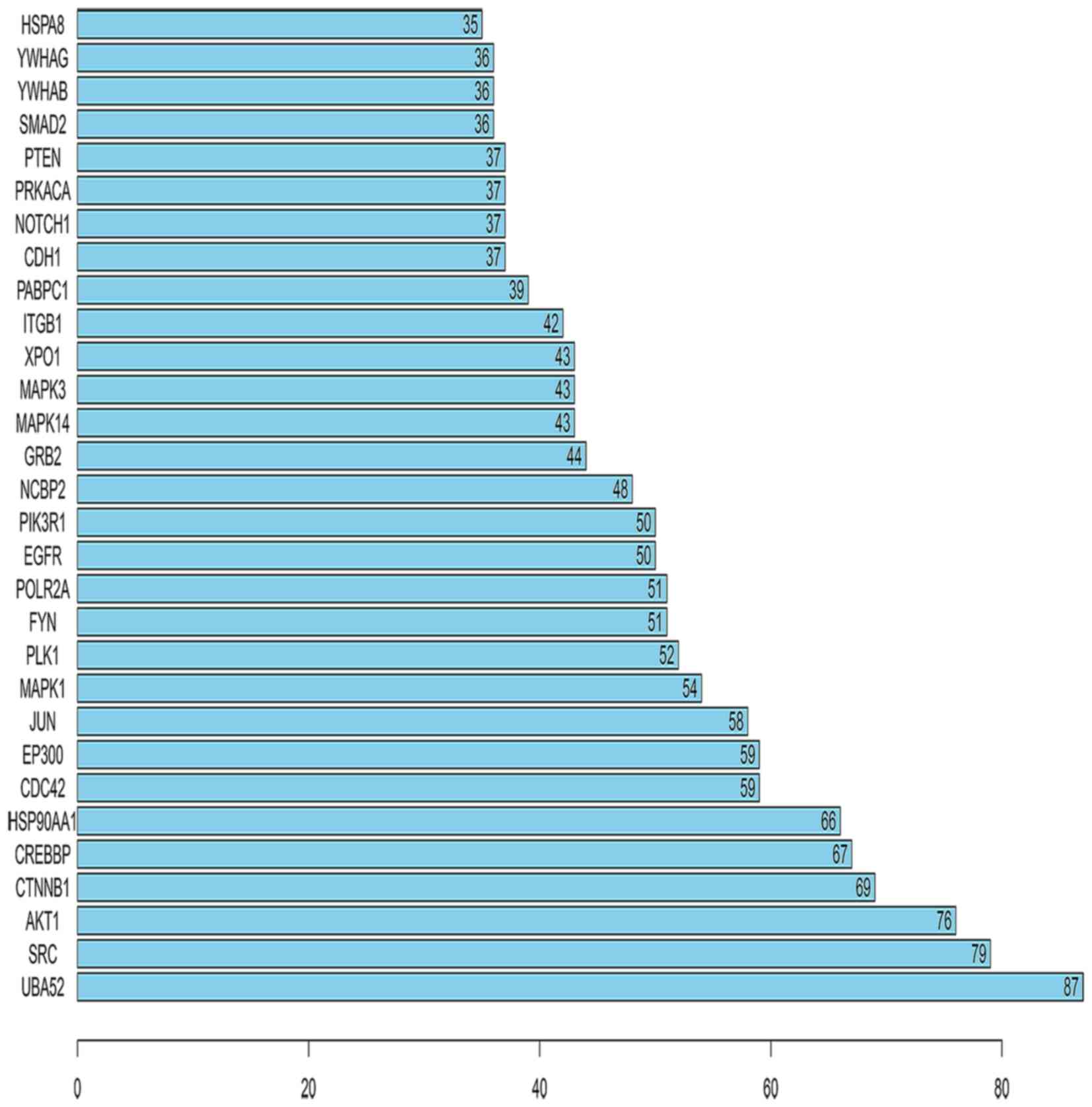
Furthermore, the number of links to each of the top 30 genes was
calculated, of which UBA52 was the gene with the highest
connectivity in the top 30 genes (Fig. 6).
Validation of the correlation among
LINC00968, DEMs and hub genes in the TCGA database
The correlation among LINC00968, DEMs and hub genes
was evaluated by bivariate correlation analysis from the
aforementioned lncRNA-miRNA-mRNA network in LUAD patients based
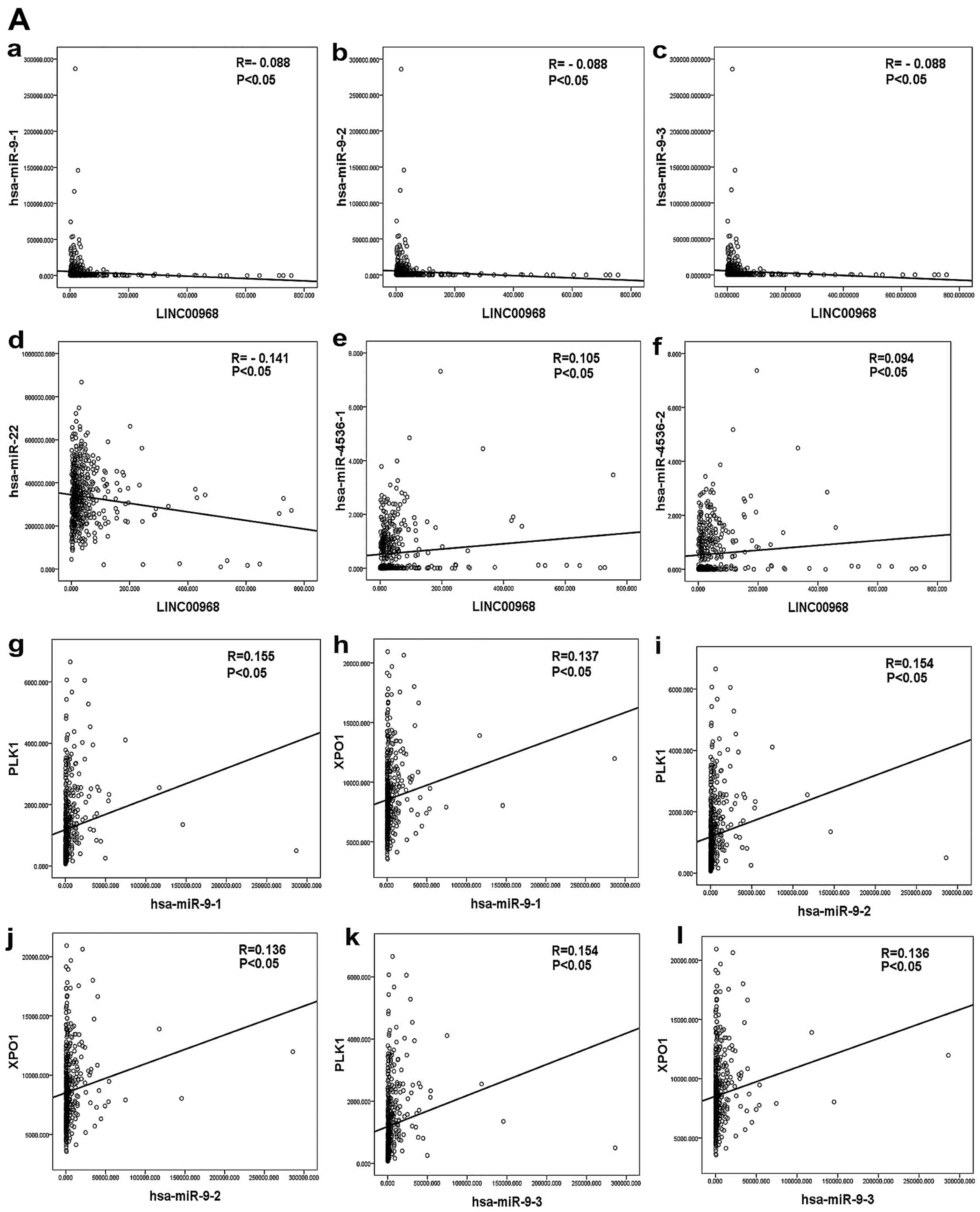
upon TCGA data. Three DEMs were significantly correlated with
LINC00968 in TCGA, and the results suggested that miR-9 and miR-22
were negatively correlated with LINC00968 and miR-4536 was
positively correlated with LINC00968 in LUAD patients (Fig. 7A-a-f). In addition, 18 hub genes
of the top 30 genes were identified as being significantly
correlated with LINC00968 in LUAD based upon TCGA (P<0.05). The
correlation between the three miRNAs and the 18 hub genes was then
analyzed (Table IV). Polo-like
kinase 1 (PLK1) and exportin-1 (XPO1) were found to
be co-associated with miR-9, miR-22 and miR-4536 (Fig. 7A-g-l and B-a-f). Furthermore, an
lncRNA-miRNA-mRNA network of LINC00968 was validated in LUAD
patients based on the TCGA dataset by combining correlations
between LINC00968 and the co-associated genes (Fig. 7B-g-h), which included the three
miRNAs (miR-9, miR-22 and miR-4536) and two hub genes (PLK1
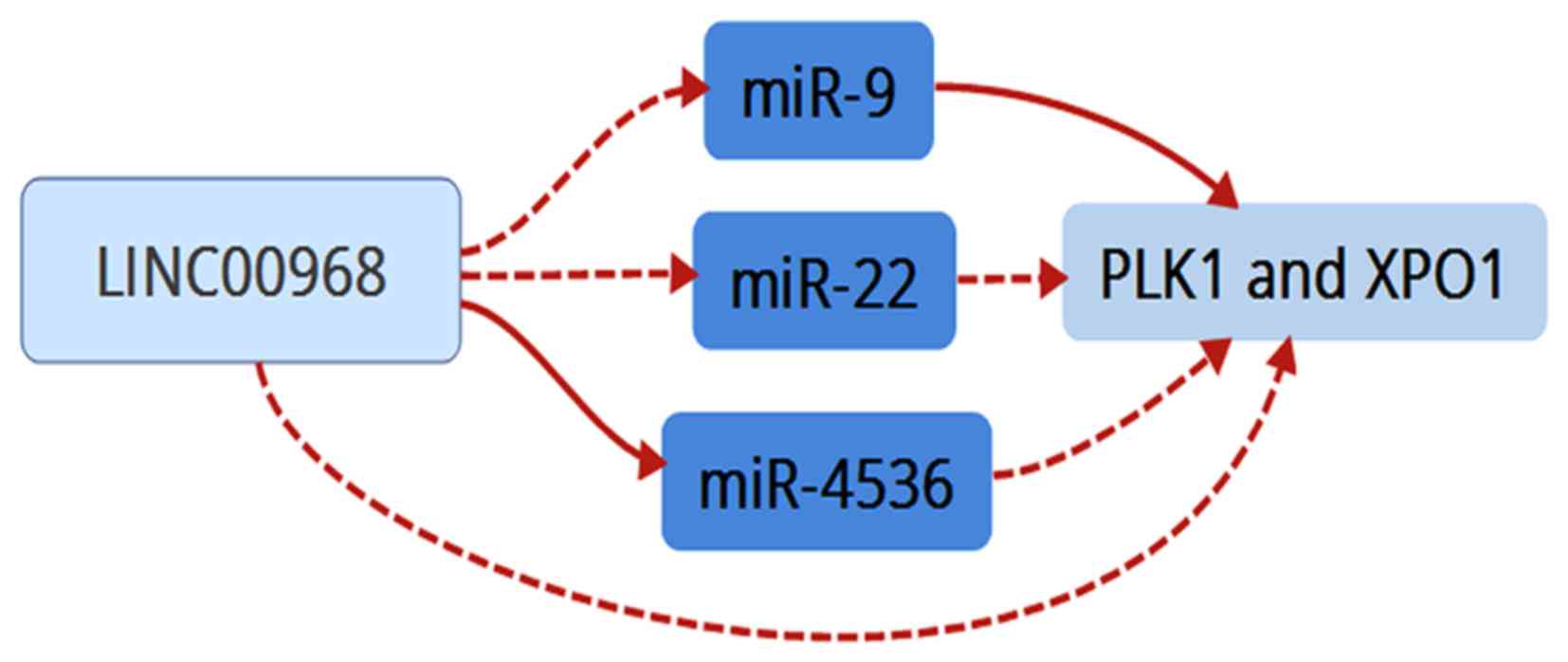
and XPO1). Finally, these data suggested that LINC00968
implemented its biofunction in LUAD patients by directly or
indirectly targeting PLK1 and XPO1 (Fig. 8).
 | Table IVGenes associated with the three
differentially expressed miRNAs that have a significant association
with LINC00968 in the 18 hub genes based on the TCGA database. |
Table IV
Genes associated with the three
differentially expressed miRNAs that have a significant association
with LINC00968 in the 18 hub genes based on the TCGA database.
| miRNA | Associated
genes |
|---|
| miR-9-1 | JUN, PLK1, FYN,
PIK3R1, NCBP2, XPO1, PABPC1, NOTCH1 |
| miR-9-2 | JUN, PLK1, FYN,
PIK3R1, NCBP2, XPO1, PABPC1, NOTCH1 |
| miR-9-3 | JUN, PLK1, FYN,
PIK3R1, NCBP2, XPO1, PABPC1, NOTCH1 |
| miR-22 | SRC, HSP90AA1,
CDC42, PLK1, FYN, NCBP2, XPO1, PABPC1, YWHAG |
| miR-4536-1 | SRC, HSP90AA1,
PLK1, PIK3R1, XPO1, NOTCH1, YWHAG |
| miR-4536-2 | SRC, PLK1, PIK3R1,
NCBP2, XPO1, NOTCH1, PTEN, YWHAG |
Discussion
As described in a previous study by the present
research team, 47 differentially expressed lncRNAs were discovered
in tumor tissues and normal lung tissues based on gene expression
files from five GEO datasets, one of which was LINC00968 (19). In addition, the downregulation of
LINC00968 expression was found in LUAD tissues and A549 cells,
suggesting its potential tumor suppressor role in LUAD. Therefore,
the biological mechanisms of LINC00968 in LUAD patients were
explored by lncRNA-miRNA-mRNA network and bioinformatic analysis in
the present study.
To date, lncRNAs and miRNAs are known to function as
key regulators in the biological processes of numerous cancers and
may have good diagnostic and prognostic values for a variety of
cancers, including lung cancer (22). Moreover, accumulating evidence
indicates that lncRNAs are capable of binding specific miRNAs and
regulating their function. For instance, the interaction data from
the Starbase database and miRanda algorithm have been used to
generate a global triple network based on the ceRNA theory that the
lncRNA and mRNA shared the same miRNA (23). Another study has shown that
C032469 is able to regulate human telomerase reverse transcriptase
expression by binding to miR-1207-5p in gastric cancer (24). These studies provide an approach
for understanding the connection between lncRNAs, miRNAs and mRNAs
in the progression and development of lung cancer. Therefore, it
was necessary to construct an lncRNA-miRNA-mRNA regulatory network
of LINC00968 in the present study.
The overexpression of LINC00968 in A549 cells
induced a variation of the miRNA profile that was detected using an
miRNA microarray. The results of the miRNA microarray analysis
indicated that certain interactions exist between LINC00968 and
five DEMs, namely miR-9-3p, miR-22-5p, miR-668-3p, miR-3675-3p and
miR-4536-3p. Furthermore, by integrating predicted target genes and
validated target genes of the five DEMs, 1,888 prospective target
genes were obtained for further analysis.
GO analysis subsequently revealed that the five DEMs
participated in a variety of biological processes potentially
important to lung cancer progression, in the categories of
biological process, cellular component and molecular function.
Intracellular transport, organelle lumen and nucleotide binding
were selected as representatives of the three major categories,
respectively. Intracellular transport is of critical importance for
a variety of life functions and is tightly associated with certain
serious diseases, such as myocardial and neurodegenerative diseases
and renal cancer (25–27). Studies have indicated that
intracellular transport not only regulates lung cancer cell growth
(28) but also regulates the
intracellular levels of anticancer drugs (29). Organelles mainly include
mitochondria, endoplasmic reticulum, centrosome and ribosomes,
which maintain the normal structure and functions of cells. Results
of previous studies suggest that mitochondria and ribosome act as
key regulators in A549 lung cancer cells (30,31). With regard to nucleotide binding,
lncRNAs and miRNAs may bind to specific molecules: the lncRNA HOX
transcript antisense intergenic RNA has been indicated to regulate
the cell biological function of NSCLC through binding to
hypoxia-inducible factor-1α (32), and miR-486-5p may downregulate
cyclin-dependent kinase 4 expression to inhibit the development of
NSCLC (33). These results
indicate that nucleotide binding may serve vital roles in the
progression of lung cancer.
KEGG pathway analysis determined that these five
DEMs participate in adherens junction and focal adhesion as these
were the two most significantly enriched pathways. Adherens
junctions maintain cell-cell adhesion and ensure the normal
transmission of cellular signals. β-catenin and E-cadherin as core
structural components of adherens junctions participate in the
regulation of lung cancer (34,35). Focal adhesions, highly regulated
multi-protein complexes, are critical in the regulation of a number
of pathological processes, such as the progression of lung cancer
(36,37). Under normal circumstances,
adherens junction and focal adhesion are necessary for the
maintenance of homeostasis (38–40). Therefore, their dysregulation in
cancer cells may be closely associated with the development and
progression of lung cancer.
The results of the GO and KEGG analyses suggested
that LINC00968 may function as a regulator in lung cancer. Thus, an
original network of LINC00968, 5 DEMs and 1,888 target genes was
constructed. In addition, a PPI network of the 1,888 target genes
was generated. Based on the connectivity of genes in the PPI
network, the top 30 hub genes were obtained. Additionally, by
correlation analysis of 18 of the top 30 hub genes with miR-9,
miR-22 and miR-4536 in TCGA, two genes, namely PLK1 and
XPO1, were found to be co-associated with miR-9, miR-22 and
miR-4536. Thus, the three miRNAs and the two genes were chosen for
continued investigation via in-depth studies, and an
lncRNA-miRNA-mRNA network of LINC00968 validated in TCGA was
constructed.
The three independent miRNAs miR-9-1, miR-9-2 and
miR-9-3 are transcription products of miR-9 involved in tumor
growth (41). miR-9 has shown the
ability to enhance the effect of anticancer drugs in NSCLC
(42) and acts as an important
regulator in the evolution and progression of NSCLC (43). Similarly, studies have shown that
miR-22 not only acts as a novel biomarker for NSCLC (44), but also inhibits tumor growth and
metastasis in lung cancer (45).
Unfortunately, the regulatory mechanism of miR-4536 in lung cancer
has not been reported; however, the common downstream target genes
PLK1 and XPO1 of these three miRNAs serve important
roles in carcinogenesis.
PLK1 regulates the development of numerous cancers
by participating in the mitotic process. Previous evidence suggests
that tumor progression is inhibited by the targeting of PLK1
(46,47). In addition, XPO1 is a nuclear
exporter that mediates the nuclear export of multiple tumor
suppressors (48). Kim et
al (49) discovered that the
inhibition of XPO1 was a promising therapeutic strategy for
a cohort of patients with lung cancer.
These studies suggest that the three miRNAs and the
two genes exert important influences on the regulation of lung
cancer. Therefore, the network comprising LINC00968, the three
miRNAs (miR-9, miR-22 and miR-4536) and the two hub genes (PLK1 and
XPO1) validated in the TCGA database may be a potential regulatory
mechanism with vital roles in the progression and prognosis of lung
cancer. However, LINC00968 is a novel lncRNA that has not been
reported previously, the precise functions of which will be the
subject of subsequent studies by the present research team. The
regulatory mechanisms of LINC00968 will also be analyzed.
Acknowledgments
The present study was supported by Guangxi Key
Project of Science and Technology (grant no. 1598012-30), the Fund
of the Guangxi Provincial Health Bureau Scientific Research Project
(grant no. Z2013201), the Fund of the National Natural Science
Foundation of China (grant nos. NSFC81660488, NSFC81360327 and
NSFC81560469), and the Natural Science Foundation of Guangxi, China
(grant no. 2015GXNSFCA139009).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Siegel RL and Jemal A: Lung
Cancer Statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar
|
|
3
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou M, Li J, Li C, Guo L, Wang X, He Q,
Fu Y and Zhang Z: Tertiary amine mediated targeted therapy against
metastatic lung cancer. J Control Release. 241:81–93. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khandelwal A, Bacolla A, Vasquez KM and
Jain A: Long non-coding RNA: A new paradigm for lung cancer. Mol
Carcinog. 54:1235–1251. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Castellano JJ, Navarro A, Viñolas N,
Marrades RM, Moises J, Cordeiro A, Saco A, Muñoz C, Fuster D,
Molins L, Ramirez J and Monzo M: LincRNA-p21 impacts prognosis in
resected non-small cell lung cancer patients through angiogenesis
regulation. J Thorac Oncol. 11:2173–2182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chang TH, Tsai MF, Gow CH, Wu SG, Liu YN,
Chang YL, Yu SL, Tsai HC, Lin SW, Chen YW, et al: Upregulation of
microRNA-137 expression by Slug promotes tumor invasion and
metastasis of non-small cell lung cancer cells through suppression
of TFAP2C. Cancer Lett. 402:190–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duval M, Cossart P and Lebreton A:
Mammalian microRNAs and long noncoding RNAs in the host-bacterial
pathogen crosstalk. Semin Cell Dev Biol. 65:11–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi X, Sun M, Wu Y, Yao Y, Liu H, Wu G,
Yuan D and Song Y: Post-transcriptional regulation of long
noncoding RNAs in cancer. Tumour Biol. 36:503–513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H and Zhu JK: Emerging roles of RNA
processing factors in regulating long non-coding RNAs. RNA Biol.
11:793–797. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guz M, Rivero-Müller A, Okoń E,
Stenzel-Bembenek A, Polberg K, Słomka M and Stepulak A:
MicroRNAs-role in lung cancer. Dis Markers. 2014:2181692014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valinezhad Orang A, Safaralizadeh R and
Kazemzadeh-Bavili M: Mechanisms of miRNA-Mediated Gene Regulation
from Common Downregulation to mRNA-Specific Upregulation. Int J
Genomics. 2014:9706072014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H19
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar
|
|
17
|
Ye S, Yang L, Zhao X, Song W, Wang W and
Zheng S: Bioinformatics method to predict two regulation mechanism:
TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell
Biochem Biophys. 70:1849–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Fan D, Jian Z, Chen GG and Lai
PB: Cancer specific long noncoding RNAs show differential
expression patterns and competing endogenous RNA potential in
hepatocellular carcinoma. PLoS One. 10:e01410422015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang J, Lin J, Liu T, Chen T, Pan S, Huang
W and Li S: Analysis of lncRNA expression profiles in non-small
cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer.
85:110–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016. View Article : Google Scholar :
|
|
21
|
Czerwinska U, Calzone L, Barillot E and
Zinovyev A: DeDaL: Cytoscape 3 app for producing and morphing
data-driven and structure-driven network layouts. BMC Syst Biol.
9:462015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vencken SF, Greene CM and McKiernan PJ:
Non-coding RNA as lung disease biomarkers. Thorax. 70:501–503.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song C, Zhang J, Liu Y, Pan H, Qi HP, Cao
YG, Zhao JM, Li S, Guo J, Sun HL, et al: Construction and analysis
of cardiac hypertrophy-associated lncRNA-mRNA network based on
competitive endogenous RNA reveal functional lncRNAs in cardiac
hypertrophy. Oncotarget. 7:10827–10840. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lü MH, Tang B, Zeng S, Hu CJ, Xie R, Wu
YY, Wang SM, He FT and Yang SM: Long noncoding RNA BC032469, a
novel competing endogenous RNA, upregulates hTERT expression by
sponging miR-1207-5p and promotes proliferation in gastric cancer.
Oncogene. 35:3524–3534. 2016. View Article : Google Scholar
|
|
25
|
Frismantiene A, Kyriakakis E, Dasen B,
Erne P, Resink TJ and Philippova M: Actin cytoskeleton regulates
functional anchorage-migration switch during T-cadherin-induced
phenotype modulation of vascular smooth muscle cells. Cell Adh
Migr. 1–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pluskota E, Bledzka KM, Bialkowska K,
Szpak D, Soloviev DA, Jones SV, Verbovetskiy D and Plow EF:
Kindlin-2 interacts with endothelial adherens junctions to support
vascular barrier integrity. J Physiol. Aug 11–2017.Epub ahead of
print. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kleinschmidt EG and Schlaepfer DD: Focal
adhesion kinase signaling in unexpected places. Curr Opin Cell
Biol. 45:24–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song S, Jacobson KN, McDermott KM, Reddy
SP, Cress AE, Tang H, Dudek SM, Black SM, Garcia JG, Makino A, et
al: ATP promotes cell survival via regulation of cytosolic
[Ca2+] and Bcl-2/Bax ratio in lung cancer cells. Am J
Physiol Cell Physiol. 310:C99–C114. 2016.
|
|
29
|
Galetti M, Petronini PG, Fumarola C,
Cretella D, La Monica S, Bonelli M, Cavazzoni A, Saccani F,
Caffarra C, Andreoli R, et al: Effect of ABCG2/BCRP expression on
efflux and uptake of gefitinib in NSCLC cell lines. PLoS One.
10:e01417952015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Holandino C, Teixeira CA, de Oliveira FA,
Barbosa GM, Siqueira CM, Messeder DJ, de Aguiar FS, da Veiga VF,
Girard-Dias W, Miranda K, et al: Direct electric current treatment
modifies mitochondrial function and lipid body content in the A549
cancer cell line. Bioelectrochemistry. 111:83–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim BG, Kwon HY, Sohn EJ, Hwang S, Kwon OS
and Kim SH: Activation of caspases and inhibition of ribosome
biogenesis mediate antitumor activity of Chijongdan in A549
non-small lung cancer cells. BMC Complement Altern Med. 14:4202014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou C, Ye L, Jiang C, Bai J, Chi Y and
Zhang H: Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α
activated driver of malignancy, enhances hypoxic cancer cell
proliferation, migration, and invasion in non-small cell lung
cancer. Tumour Biol. 36:9179–9188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ,
Lu SD, He YY, Wang P, Sun QL, Jin YX, et al: Direct repression of
the oncogene CDK4 by the tumor suppressor miR-486-5p in non-small
cell lung cancer. Oncotarget. 7:34011–34021. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dong S, Khoo A, Wei J, Bowser RK,
Weathington NM, Xiao S, Zhang L, Ma H, Zhao Y and Zhao J: Serum
starvation regulates E-cadherin upregulation via activation of
c-Src in non-small-cell lung cancer A549 cells. Am J Physiol Cell
Physiol. 307:C893–C899. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu X, Kim JE, Sun PL, Yoo SB, Kim H, Jin Y
and Chung JH: Immunohistochemical demonstration of alteration of
β-catenin during tumor metastasis by different mechanisms according
to histology in lung cancer. Exp Ther Med. 9:311–318. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bennett DT, Reece TB, Foley LS, Sjoberg A,
Meng X, Fullerton DA and Weyant MJ: C-terminal tensin-like protein
mediates invasion of human lung cancer cells and is regulated by
signal transducer and activator of transcription 3. J Thorac
Cardiovasc Surg. 149:369–375. 2015. View Article : Google Scholar
|
|
37
|
Lin TY and Hsu HY: Ling Zhi-8 reduces lung
cancer mobility and metastasis through disruption of focal adhesion
and induction of MDM2-mediated Slug degradation. Cancer Lett.
375:340–348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Colman MA, Pinali C, Trafford AW, Zhang H
and Kitmitto A: A computational model of spatiotemporal cardiac
intracellular calcium handling with realistic structure and spatial
flux distribution from sarcoplasmic reticulum and t-tubule
reconstructions. PLoS Comput Biol. 13:e10057142017. View Article : Google Scholar
|
|
39
|
Mukherjee R, Majumder P and Chakrabarti O:
MGRN1 mediated ubiquitination of α-tubulin regulates microtubule
dynamics and intracellular transport. Traffic. Sep 13–2017.Epub
ahead of print. View Article : Google Scholar
|
|
40
|
Yang Q, Wang Y, Yang Q, Gao Y, Duan X, Fu
Q, Chu C, Pan X, Cui X and Sun Y: Cuprous oxide nanoparticles
trigger ER stress-induced apoptosis by regulating copper
trafficking and overcoming resistance to sunitinib therapy in renal
cancer. Biomaterials. 146:72–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yuva-Aydemir Y, Simkin A, Gascon E and Gao
FB: MicroRNA-9: Functional evolution of a conserved small
regulatory RNA. RNA Biol. 8:557–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen X, Zhu L, Ma Z, Sun G, Luo X, Li M,
Zhai S, Li P and Wang X: Oncogenic miR-9 is a target of erlotinib
in NSCLCs. Sci Rep. 5:170312015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mitra R, Edmonds MD, Sun J, Zhao M, Yu H,
Eischen CM and Zhao Z: Reproducible combinatorial regulatory
networks elucidate novel oncogenic microRNAs in non-small cell lung
cancer. RNA. 20:1356–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu C, Zheng Y, Lian D, Ye S, Yang J and
Zeng Z: Analysis of microRNA expression profile identifies novel
biomarkers for non-small cell lung cancer. Tumori. 101:104–110.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xin M, Qiao Z, Li J, Liu J, Song S, Zhao
X, Miao P, Tang T, Wang L, Liu W, et al: miR-22 inhibits tumor
growth and metastasis by targeting ATP citrate lyase: Evidence in
osteosarcoma, prostate cancer, cervical cancer and lung cancer.
Oncotarget. 7:44252–44265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang XH, Lu Y, Liang JJ, Cao JX, Jin YQ,
An GS, Ni JH, Jia HT and Li SY: MiR-509-3-5p causes aberrant
mitosis and anti-proliferative effect by suppression of PLK1 in
human lung cancer A549 cells. Biochem Biophys Res Commun.
478:676–682. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu C, Li S, Chen T, Hu H, Ding C, Xu Z,
Chen J, Liu Z, Lei Z, Zhang HT, et al: miR-296-5p suppresses cell
viability by directly targeting PLK1 in non-small cell lung cancer.
Oncol Rep. 35:497–503. 2016. View Article : Google Scholar
|
|
48
|
Chen Y, Camacho C, Silvers TR, Razak AR,
Gabrail NY, Gerecitano JF, Kalir E, Pereira E, Evans BR, Ramus SJ,
et al: Inhibition of the nuclear export receptor XPO1 as a
therapeutic target for platinum resistant ovarian cancer. Clin
Cancer Res. 23:1552–1563. 2016. View Article : Google Scholar
|
|
49
|
Kim J, McMillan E, Kim HS, Venkateswaran
N, Makkar G, Rodriguez-Canales J, Villalobos P, Neggers JE,
Mendiratta S, Wei S, et al: XPO1-dependent nuclear export is a
druggable vulnerability in KRAS-mutant lung cancer. Nature.
538:114–117. 2016. View Article : Google Scholar : PubMed/NCBI
|