Introduction
Rheumatoid arthritis (RA) is an autoimmune disease
that is characterized by chronic progressive arthropathy (1,2).
Apoptosis is considered to serve an important role in the
pathogenesis of RA; in particular, there is a lack of apoptosis in
synovial cells and an excess of apoptosis in cartilage cells
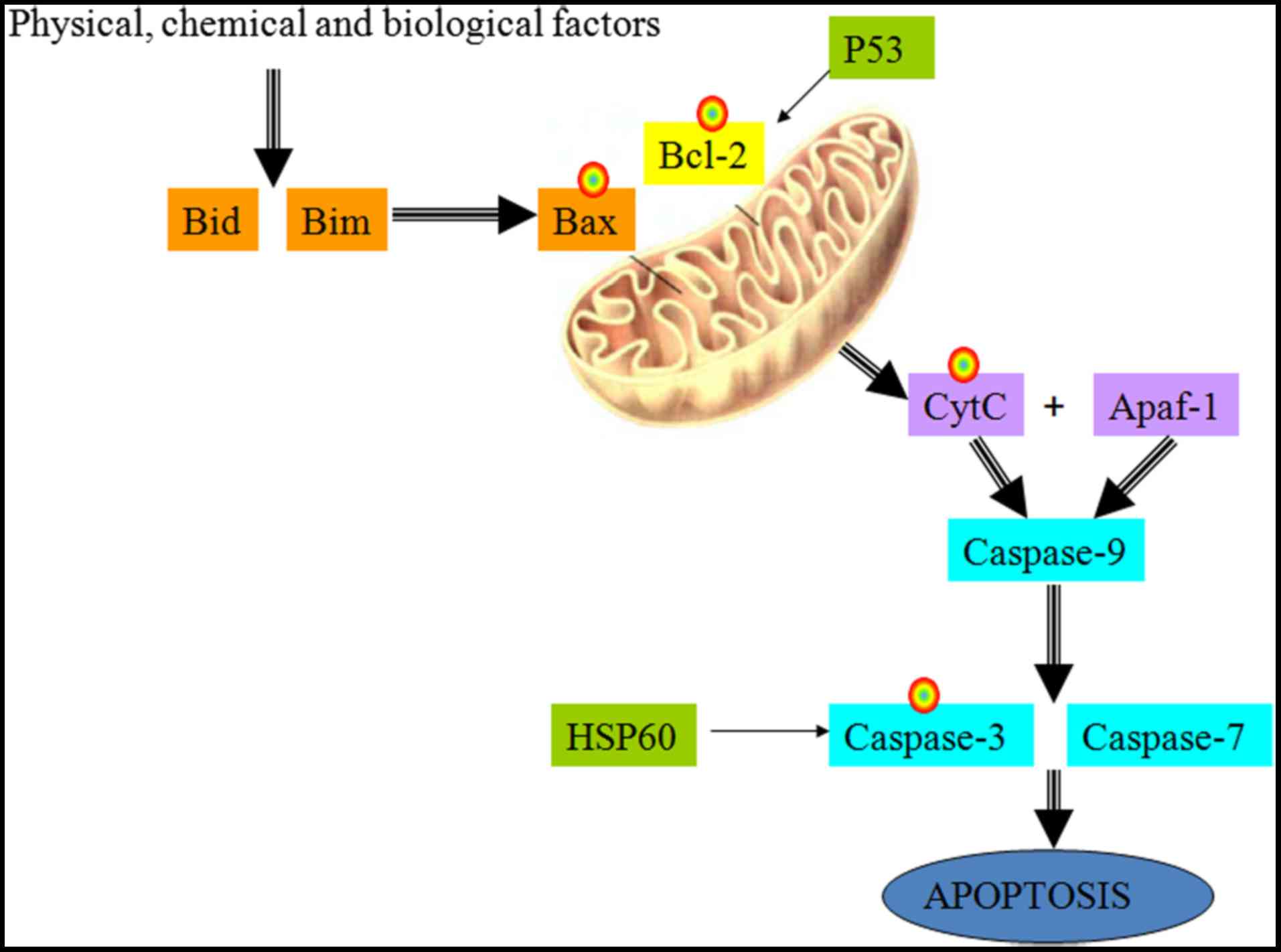
(3,4). The mitochondrial signaling pathway
is a common apoptotic pathway, in which the Bcl-2 family members
function as pro-apoptotic and anti-apoptotic signal transduction
factors. When apoptotic activation signals are received, Bax
oligomerizes, escaping inhibition by Bcl-2, and is inserted into
the mitochondrial membrane; the subsequent changes facilitate the
release of CytC into the cytosol, where it interacts with the
activating factor Apaf-1 to form a multimeric complex. Caspase-9
recruitment initiates the caspase cascade, which involves the
activation of downstream Caspase-3 and eventually results in
apoptosis (Fig. 1) (5–7).
As Bcl-2 is the initiating factor of the
mitochondrial pathway, and its transcripts have been found to be
highly expressed in the synovial tissues and cells of patients with
RA (8), the present study aimed
to design and synthesize human Bcl-2-short hairpin (sh)RNA
expression vectors and assess their effects. The vectors were
transformed into competent DH5α Escherichia coli (a
genetically engineered Escherichia coli) cells, and then
transfected into the human synovial sarcoma cell line SW982 for
screening of an effective interference sequence. The expression
levels of molecules associated with the mitochondrial pathway were
then detected. The present study provides a theoretical and
experimental basis for a potential molecular targeting treatment
for RA.
Materials and methods
Materials
Type I collagenase, Dulbecco's modified Eagle's
medium (DMEM)/F12 (glucose-free) and trypsin were purchased from
Corning Inc. (Corning, NY, USA). DNA endonuclease enzymes
(XhoI and MluI) were purchased from Shanghai Yu Bo
Biological Technology Co., Ltd. (Shanghai, China). Lipofectamine
2000, First Strand cDNA Synthesis kit, Plasmid Extraction kit, Cell
Total RNA Extraction kit, 2X SG Fast qPCR Master Mix and fetal
bovine serum (FBS) were purchased from Bio Basic Inc. (Amherst, NY,
USA). Human synovial sarcoma SW982 cells were purchased from Bohu
Biotechnology Co. (Shanghai, China). Cells between passages 3 and 5
(P3–P5) were used in experiments.
Experiments were carried out in the Laboratory of
Molecular Biology at the Bioengineering Biotechnology Company of
Shanghai (Shanghai, China) between September and December 2016.
Design and synthesis of Bcl-2-shRNA
Using bioinformatics methods (9,10),
the complete sequence of Bcl-2 mRNA (serial number: NM_000633.2)
was acquired from GenBank, three sequences were designed for the
target gene, and the corresponding sense and antisense
oligonucleotides were designed and synthesized (Table I).
 | Table IThe sequences of Bcl-2 pre-short
hairpin RNA. |
Table I
The sequences of Bcl-2 pre-short
hairpin RNA.
| Oligo name | Single stranded
oligonucleotide sequence (5′-3′) |
|---|
| Bcl-2 I-F |
CACCCGGGAGATAGTGATGAAGTTTCAAGAGAACTTCATCACTATCTCCCGTTTTTTG |
| Bcl-2 I-R |
AGCTCAAAAAACGGGAGATAGTGATGAAGTTCTCTTGAAACTTCATCACTATCTCCCG |
| Bcl-2 II-F |
CACCTGGATGTTCTGTGCCTGTA
TTCAAGAGATACAGGCACAGAACATCCATTTTTTG |
| Bcl-2 II-R |
AGCTCAAAAAATGGATGTTCTGTGCCTGTATCTCTTGAATACAGGCACAGAACATCCA |
| Bcl-2 III-F |
CACCTGTCTTTTGTTGTTGTTCATTCAAGAGATGAACAACAACAAAAGACATTTTTTG |
| Bcl-2 III-R |
AGCTCAAAAAATGTCTTTTGTTGTTGTTCATCTCTTGAATGAACAACAACAAAAGACA |
Construction of Bcl-2-shRNA interference
vector
According to the instructions of the plasmid
construction kit (DNA Blunting kit; Takara Bio, Inc., Shiga, Japan;
cat. no. 6025), three pairs of shRNAs and a negative control
oligonucleotide strand (each 5 µl) were respectively heated
(95°C) for 5 min in the annealing buffer, and cooled for 20 min at
room temperature. Following the formation of double chains, the
oligonucleotides were ligated into the pHAV3.1-shRNA-tGFP vector by
T4 DNA ligase. Subsequently, this mixture (10 µl) was added
to 200 µl competent DH5α E. coli cells (Beijing World
Gold Biotech Co., Ltd., Beijing, China) for the transformation
step, in which the system was incubated on ice for 30 min, then
heat-shocked at 42°C for 45 sec. Subsequently, lysogeny broth (LB)
plates with ampicillin were coated with the E. coli, and the
cells were cultured overnight at 37°C. Finally, positive clones
were selected for the extraction of the DNA plasmids. The positive
strains were cultured overnight in LB liquid culture medium, and
the plasmids were extracted using the Plasmid Extraction kit, and
then, using a double DNA endonuclease (XhoI and MluI)
digestion for identification, the plasmids were sent to Invitrogen
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) to be
sequenced.
Culture and passage of SW982 cells
The SW982 cells were cultured in DMEM-F12 containing
2.5% FBS and 5% horse serum, and were placed in an incubator at a
temperature of 37°C with saturated humidity and 5% CO2.
After the cells had grown to 80% confluence, they were transferred
into a maintenance culture medium (DMEM/F12, 100 nmol/l
dexa-methasone and 100 nmol/l insulin) for 8 days. The day after
this, the medium was replaced and the cells were stained with Oil
Red O.
Transfection of SW982 cells with the
shRNA expression plasmid
In serum-free DMEM-F12 medium, SW982 cells were
transfected with the Bcl-2 shRNAs and negative control plasmids
using Lipofectamine 2000. SW982 cells were grouped into a control
group (transfected with a negative control shRNA), and Bcl-2-sh1,
Bcl-2-sh2 and Bcl-2-sh3 groups (transfected with Bcl-2-sh1, 2 and
3, respectively). Following transfection for 6 h, the culture
medium was replaced by DMEM-F12 medium containing 10% FBS and the
cells were cultured for a further 48–72 h. The number, intensity
and distribution of successfully transfected cells, identified by
their expression of green fluorescent protein (GFP), were observed
by fluorescence microscopy at different time-points. If the
fluorescence intensity was uniform and bright, the transfection
efficiency was deemed to be high and the total RNA of the cells was
extracted.
Screening for effective interference
sequence by reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cultured cells
using the UNlQ-10 Column TRIzol Total RNA Isolation kit (Sangon
Biotech Co., Ltd., Shanghai, China; cat. no. B511321) and was
quantified by UV spectrophotometry (260 nm; NanoDrop ND-100; Thermo
Fisher Scientific, Inc., Wilmington, DE, USA). RT was performed
with the First Strand cDNA Synthesis kit (Sangon Biotech) and the
resultant cDNA was used as the template for qPCR using a Prism 9700
StepOne™ Real-Time PCR system (Eastwin Life Sciences, Inc.,
Beijing, China). The primers were synthesized by Sangon Biotech.
The thermal cycling conditions were as follows: 1 cycle of 95°C for
10 min; and 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The
2−ΔΔCt method was used to calculate the transcript
expression levels relative to those of β-actin, which served as an
internal control (11). The
sequence with the highest interference efficiency was selected
according to the results of the quantitative detection of Bcl-2
mRNA.
Quantitative measurement of mitochondrial
pathway gene expression levels
shRNA transfection, total RNA extraction and RT-qPCR
were performed as described above. The primers used for
gene-specific amplification are listed in Table II.
 | Table IIGene-specific primers used. |
Table II
Gene-specific primers used.
| Gene | GenBank ID | Primer sequence
(5′-3′)
|
|---|
| Forward | Reverse |
|---|
| Bcl-2 | NM_000633.2 |
TTGCCAGCCGGAACCTATG |
CGAAGGCGACCAGCAATGATA |
| Bax | NM_001291428.1 |
CCCGAGAGGTCTTTTTCCGAG |
CCAGCCCATGATGGTTCTGAT |
| CytC | NM_018947.5 |
TTTGGTTGCACTTACACCGG |
GGACGTCCCCACTCTCTAAG |
| Caspase-3 | NM_004346.3 |
CATGGAAGCGAATCAATGGACT |
CTGTACCAGACCGAGATGTCA |
| β-actin | NM_001101.3 |
CATCCGCAAAGACCTGTACG |
CCTGCTTGCTGATCCACATC |
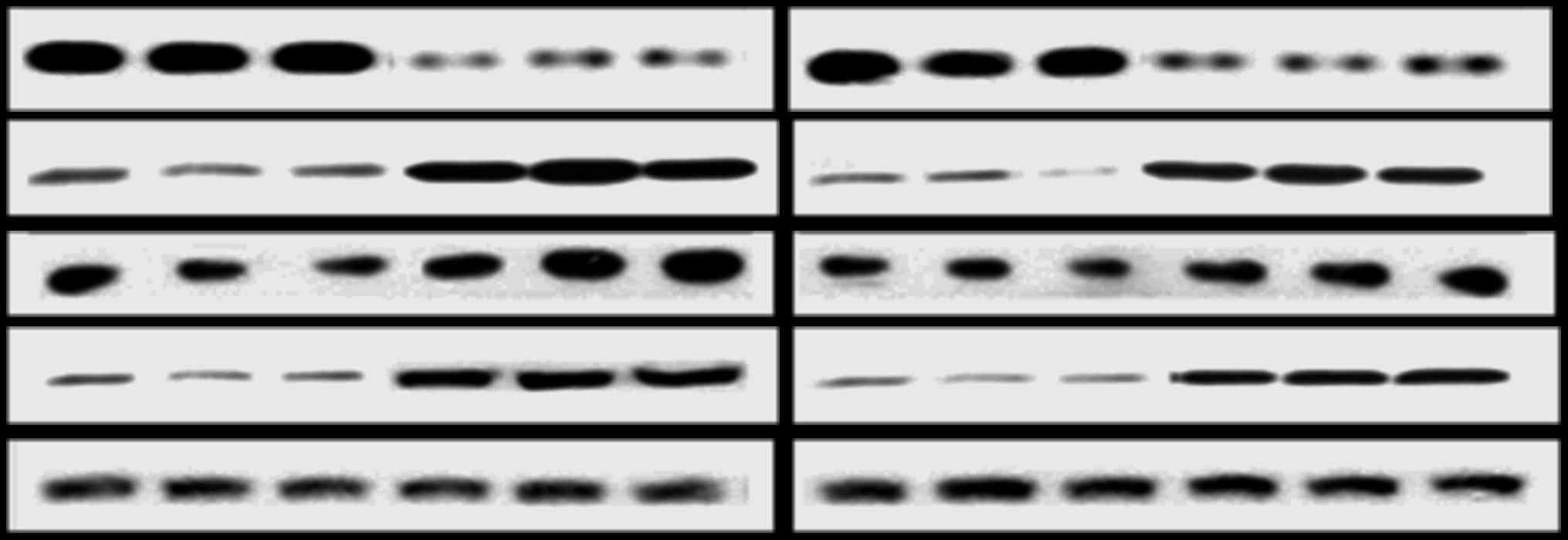
Quantitative measurement of mitochondrial
pathway protein expression levels using western blotting
Total protein was isolated from the Bcl-2-sh1- and
negative control-transfected cells using RIPA lysis buffer
(Beyotime Biotech Co., Ltd., Shanghai, China) and subjected to
western blot analysis. The protein concentration was determined by
BCA assay (BCA assay kit; Beyotime Biotech). The total protein
samples (40 µg) were resolved by 15% SDS-PAGE and
electro-transferred onto nitrocellulose membranes. Non-specific
binding sites were blocked by incubating the membranes at room
temperature with 5X TBS with 10% BSA. Subsequently, the membranes
were probed for mitochondrial pathway proteins through incubation
with primary antibodies (all diluted 1:1,000; anti-Bcl-2, ab47489;
anti-Bax, ab54829; anti-CytC, ab90529; anti-caspase-3, ab59388; and
anti-tubulin, ab6046; all from Abcam, Cambridge, UK) for 60 min at
37°C, followed by incubation with the appropriate secondary
antibodies (horseradish peroxidase-labeled goat anti-rabbit IgG;
Sangon Biotech) for 60 min at 37°C. Immunoreactivity was detected
by the enhanced chemiluminescence method using an ECL kit (Beyotime
Institute of Biotechnology, Haimen, China). Data were obtained from
at least three individual experiments performed in triplicate, and
the expression levels of the mitochondrial pathway proteins were
normalized to those of tubulin.
Statistical analysis
Statistical analyses were performed with the SPSS
software (version 18.0; SPSS, Inc., Chicago, IL, USA). Data are
expressed as the mean ± standard deviation, and inter-group
differences were evaluated with a Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Construction and identification of Bcl-2
shRNA expression plasmid
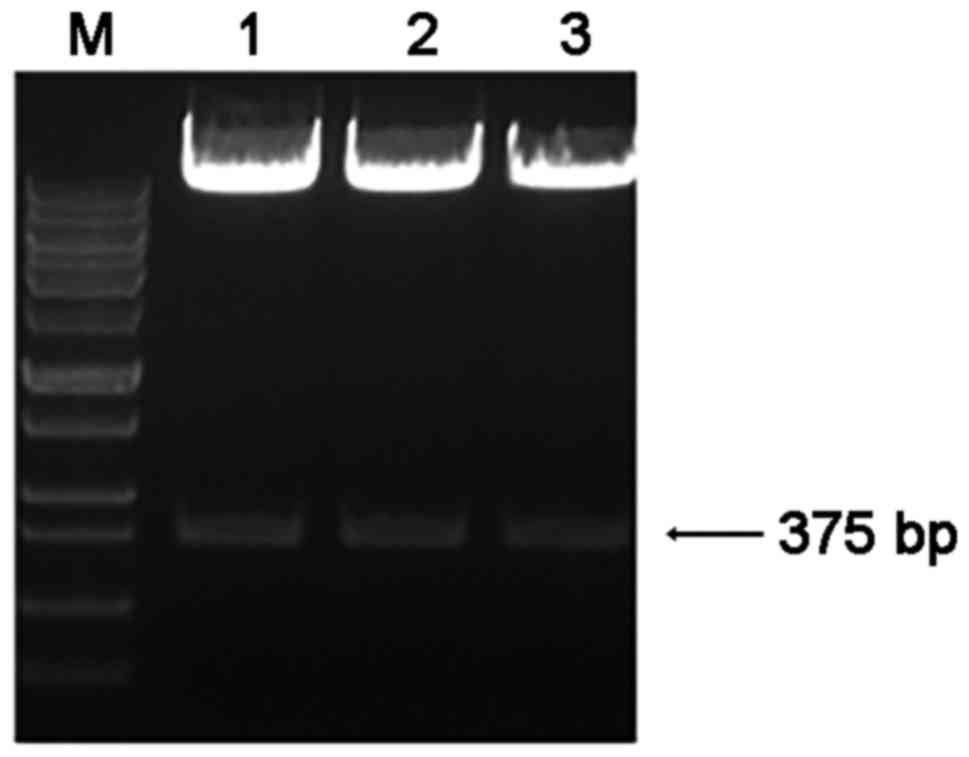
Bcl-2-shRNA expression plasmids 1, 2 and 3, which
were identified by double enzyme digestion and 1% agarose gel
electrophoresis, were synthesized successfully (Fig. 2). The shRNA expression plasmids
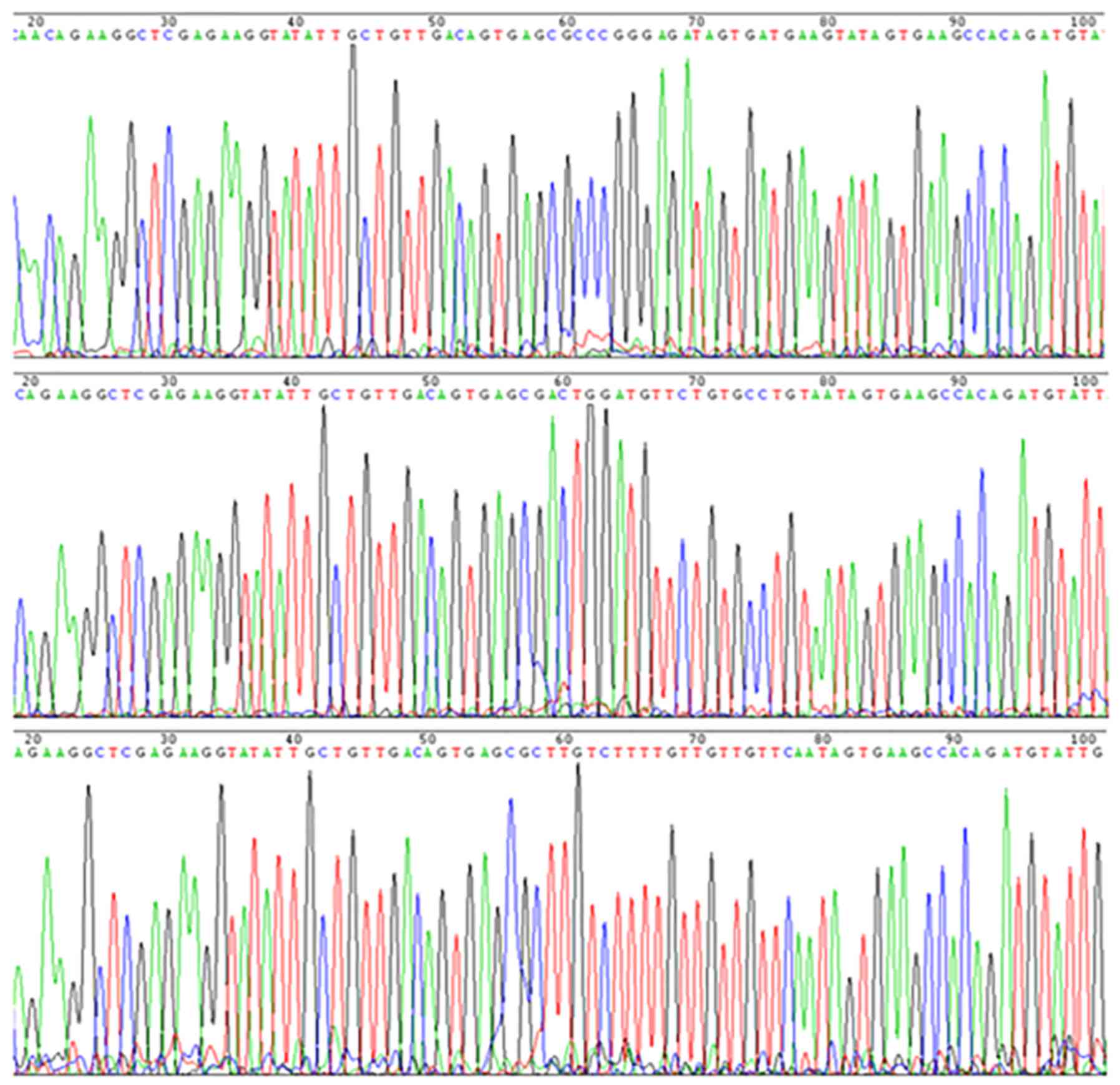
were confirmed by sequencing analysis; the recombinant plasmids
contained shRNA fragments, and the nucleotide sequences of the
inserted fragments were complete, and were consistent with the
designed sequences (Fig. 3).
Observation of Bcl-2 shRNA expression
plasmid-transfected SW982 cells by fluorescence microscopy
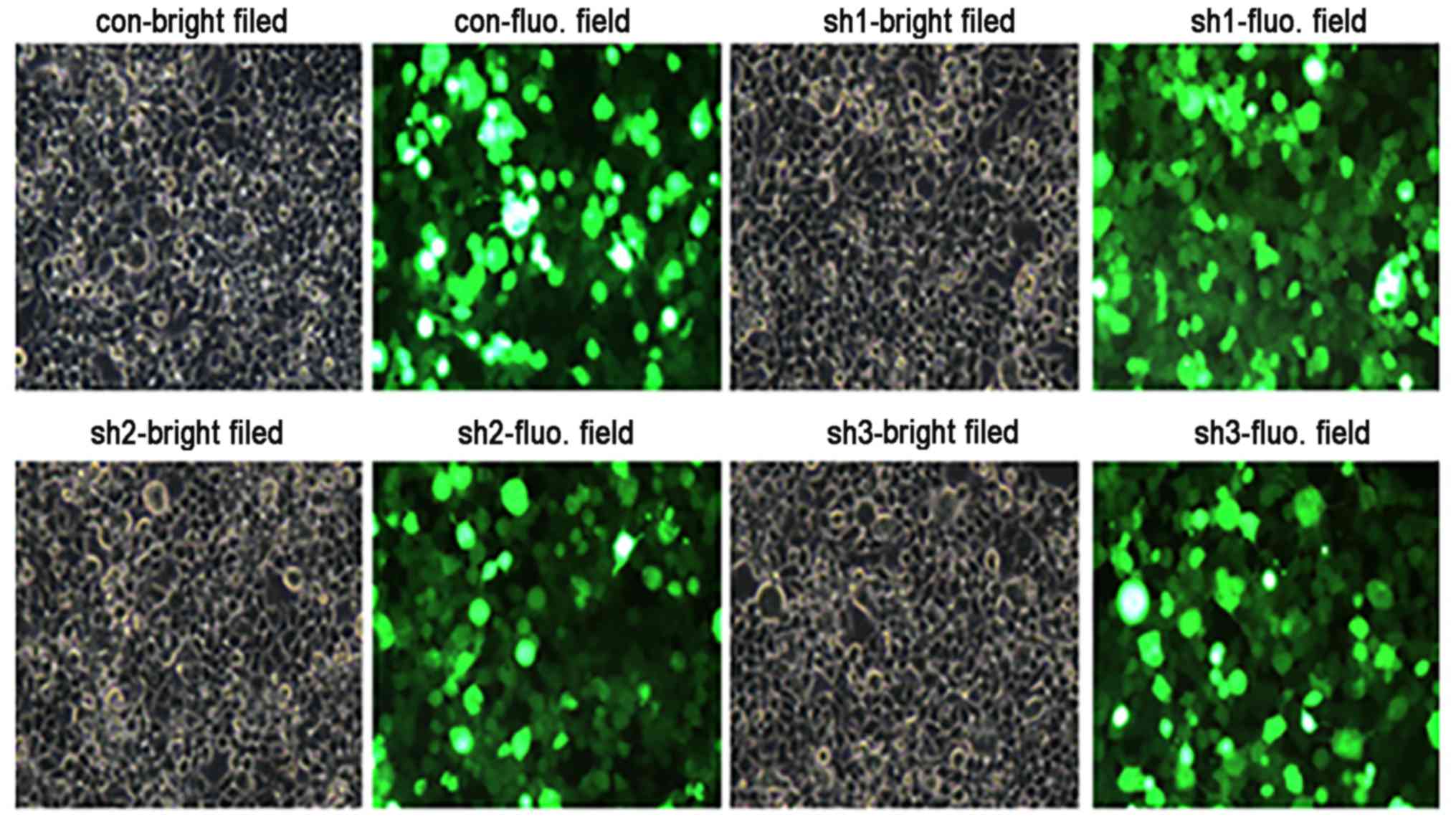
Following the transfection of Bcl-2 shRNA expression
plasmids into SW982 cells (P3–P5), GFP expression was observed by
fluorescence microscopy. GFP expression peaked at 48 h, and the
fluorescence intensity in the cytoplasm was uniform and bright,
which indicated that the transfection was successful (Fig. 4).
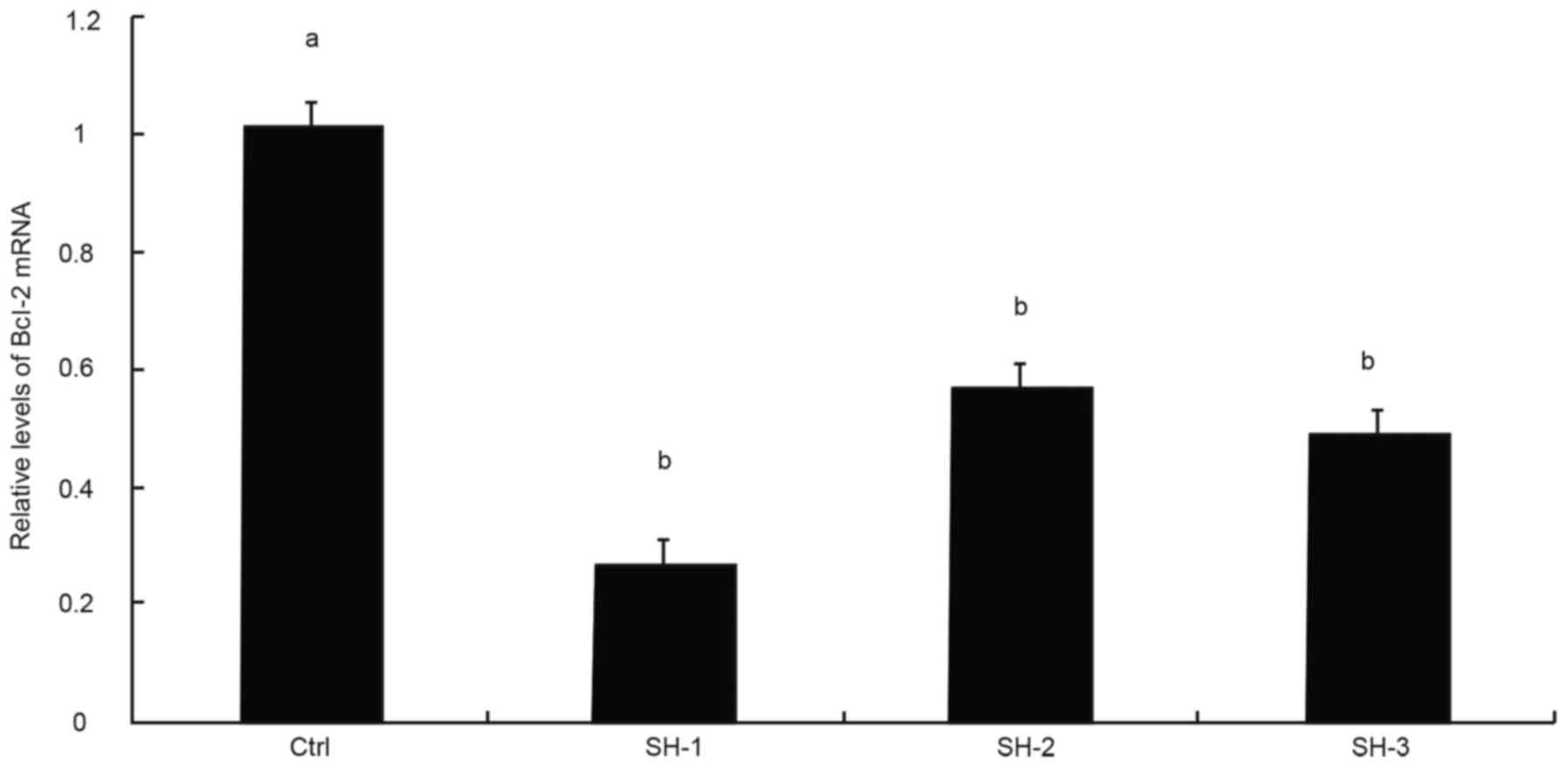
Expression of target gene Bcl-2 and
screening for an effective interference sequence
RT-qPCR was used to detect the efficacy of Bcl-2
inhibition in transfected SW982 cells, which revealed that the
Bcl-2-sh1, Bcl-2-sh2 and Bcl-2-sh3 plasmids significantly reduced
Bcl-2 mRNA levels compared with the negative control group
(P<0.05). The effect of Bcl-2-sh1 was the most pronounced
(inhibition rate >75%). Therefore, Bcl-2-sh1 was selected as the
most effective interference sequence and was used for subsequent
experiments (Fig. 5).
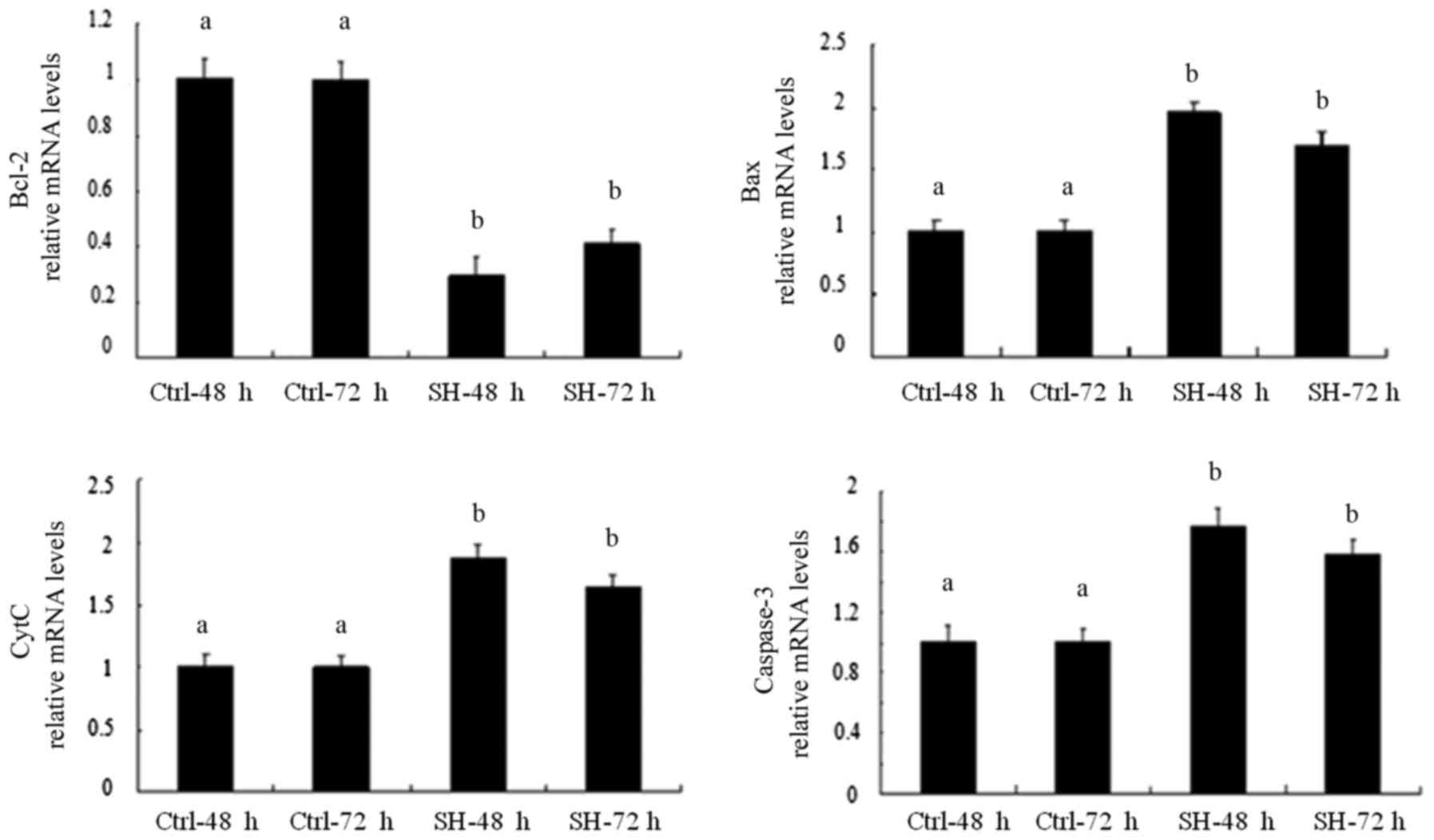
Effect of Bcl-2-sh1 on the expression of
mitochondrial pathway molecules
At 48 and 72 h following the transfection of
Bcl-2-sh1 into SW982 cells, the mRNA and protein expression levels
of Bcl-2, Bax, caspase-3, CytC were assessed. The results indicated
that, compared with the control group, the expression of Bcl-2 was
significantly decreased, while on the contrary, Bax, CytC and
Caspase-3 were significantly increased at the mRNA and protein
levels; all differences were statistically significant (P<0.05).
The effects of the Bcl-2 shRNA on the levels of Bcl-2, Bax, CytC
and Caspase-3 were more pronounced at 48 h than at 72 h
post-transfection (Figs. 6 and
7).
Discussion
Previous studies have demonstrated that the
biological characteristics of synovial cells in patients with RA
are markedly altered. Notably, significant enhancements in the
proliferative rate and migratory ability of the cells have been
observed. In addition, the Bcl-2 gene has been shown to be
overexpressed in the synovial tissue and fibroblast-like synovial
cells of RA patients, resulting in a deficiency in the apoptosis of
inflammatory cells and an imbalance in immune homeostasis (12,13). The human synovial sarcoma cell
line SW982 possesses the characteristic of abnormal proliferation,
similar to RA synovial tissue; therefore, its use for the study of
RA has been recognized (14–16).
shRNA is a highly efficient gene-silencing molecule;
compared with the traditional gene-silencing technologies,
including gene knockout, negative mutation and antisense RNA, it
has several advantages (17–19). Therefore, in the present study,
the authors designed and synthesized Bcl-2 shRNA and transfected it
into the SW982 cell line to observe its effects on the expression
of mitochondrial apoptosis pathway genes, including Bcl-2, Bax,
CytC and Caspase-3, in order to further study the relevance of this
pathway to RA.
The reason for the selection of the Bcl-2 gene
fragment for shRNA interference in the present study was the
position and function of this gene in the mitochondrial pathway.
Bcl-2 is an inhibitor of apoptosis and, as it is located upstream
of the mitochondrial pathway, it is critical to the inhibition of
this pathway. The Bcl-2 transmembrane protein contains two types of
Bcl-2 homology (BH) domains: BH1 and BH2. At these regions, Bcl-2
and the pro-apoptotic gene Bax can form heterodimers or homodimers;
this interaction is the basic mechanism by which Bcl-2 suppresses
apoptosis (20,21). The abnormal proliferation of
synovial cells in patients with RA is associated with high
expression of Bcl-2. Therefore, the authors hypothesized that the
construction of a Bcl-2-shRNA could directly activate the
mitochondrial pathway at the source, enabling abnormal synovial
cells to undergo apoptosis.
In the present study, bioinformatics methods were
used in the design process, and the Bcl-2 mRNA sequences were
obtained from GenBank. Sequences that were highly homologous with
other genes were removed, and the GC content of the sequence was
strictly limited to 35–55%. For the loop structure in the shRNA
template, in order to avoid the formation of a termination signal,
the TTCAAGAGA sequence was selected. Three target sequences were
designed, and the expression-vector method was used to prepare the
shRNA (22–24). This method uses a plasmid with a
resistance marker as a vector for transfection of the shRNA into
cells, so as to achieve sustained suppression of target gene
expression (25,26). The results demonstrated that all
three Bcl-2 shRNAs could inhibit the expression of Bcl-2 in SW982
cells, and the effect of Bcl-2-sh1 was the most obvious.
At 48 and 72 h following the transfection of
Bcl-2-sh1 into SW982 cells, the results showed that the expression
of Bcl-2 was significantly decreased, while the expression levels
of Bax, CytC and Caspase-3 were significantly increased compared
with those in the control group. Thus, the effectiveness of
shRNA-mediated interference of Bcl-2 in human SW982 cells was
confirmed, and it was demonstrated that this could indirectly
promote the expression of other pro-apoptotic genes in the
mitochondrial pathway. Since Bcl-2 and Bax exist in the form of
homodimers or heterodimers, once the level of Bcl-2 is greater,
Bcl-2/Bcl-2 homodimers are formed, and apoptosis is inhibited.
Bcl-2-shRNA inhibited the expression of the Bcl-2 gene and thereby
changed the proportion of Bcl-2/Bax, thus enhancing the expression
of the pro-apoptotic gene Bax. Bax is a promoter of the
mitochondrial pathway that promotes the release of CytC, activates
Caspase-3 and induces apoptosis of synoviocytes (27).
In summary, the interference effect of Bcl-2-sh1 on
BCL-2 was more pronounced than that of the other two sequences,
which demonstrated that, although many shRNA sequences may be
designed for the same target gene, the interference effect might
differ due to the different target sequences. With regard to the
time-points, the interference effect of Bcl-2-sh1 was greater at 48
h than at 72 h post-transfection, indicating that the inhibitory
effect of the shRNA was decreased over time; therefore, it is
necessary to investigate ways of prolonging the silencing
effect.
Acknowledgments
The present study was supported by grants from the
Development Fund and Innovation Fund of Science and Technology
(Medical and Health Projects) in Pudong New District of Shanghai,
China (grant nos. PKJ2015-Y24 and 2015/05-2018/05).
References
|
1
|
Macintyre NJ, Muller ME, Webber CE and
Adachi JD: The relationship between radial bone properties and
disease activity and physical function in individuals with
rheumatoid arthritis. Physiother Can. 64:284–291. 2012. View Article : Google Scholar :
|
|
2
|
Navalho M, Resende C, Rodrigues AM,
Pereira da Silva JA, Fonseca JE, Campos J and Canhão H: Bilateral
evaluation of the hand and wrist in untreated early inflammatory
arthritis: A comparative study of ultrasonography and magnetic
resonance imaging. J Rheumatol. 40:1282–1292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krabben A, Abhishek A, Britsemmer K, Filer
A, Huizinga TW, Raza K, van Schaardenburg DJ and van der Helm-van
Mil AH: Risk of rheumatoid arthritis development in patients with
unclassified arthritis according to the 2010 ACR/EULAR criteria for
rheumatoid arthritis. Rheumatology (Oxford). 52:1265–1270. 2013.
View Article : Google Scholar
|
|
4
|
Shin SY, Katz P, Wallhagen M and Julian L:
Cognitive impairment in persons with rheumatoid arthritis.
Arthritis Care Res (Hoboken). 64:1144–1150. 2012.
|
|
5
|
Li R, Yan G, Li Q, Sun H, Hu Y, Sun J and
Xu B: MicroRNA-145 protects cardiomyocytes against hydrogen
peroxide (H2O2)-induced apoptosis through
targeting the mitochondria apoptotic pathway. PLoS One.
7:449072012. View Article : Google Scholar
|
|
6
|
Mane SD, Thoh M, Sharma D, Sandur SK and
Naidu KA: Ascorbyl stearate promotes apoptosis through intrinsic
mitochondrial pathway in HeLa cancer cells. Anticancer Res.
36:6409–6417. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang WD, Zhang Z, Zhang H, et al: Oxygen
free radicals and mitochondrial signaling in oligospermia and
asthenospermia. Mol Med Rep. 10:1875–1880. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yan C, Kong D, Ge D, Zhang Y, Zhang X, Su
C and Cao X: Mitomycin C induces apoptosis in rheumatoid arthritis
fibroblast-like synoviocytes via a mitochondrial-mediated pathway.
Cell Physiol Biochem. 35:1125–1136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu L, Liu Y, Zhang X, Chen M, Wu H, Lin
M, Zhan Y, Zhuang C, Lin J, Li J, et al: Inhibiting cell migration
and cell invasion by silencing the transcription factor ETS-1 in
human bladder cancer. Oncotarget. 7:25125–25134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li G, Zhang L, Liu J, Xiao T, Liu G, Wang
J and Hou M: shRNA-mediated RPS15A silencing inhibits U937 acute
myeloid leukemia cell proliferation and enhances apoptosis. Mol Med
Rep. 13:4400–4406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
12
|
Liu H, Yang Y, Cai X, Gao Y, Du J and Chen
S: The effects of arctigenin on human rheumatoid arthritis
fibroblast-like synoviocytes. Pharm Biol. 53:1118–1123. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu QS, Luo XY, Jiang H, Xing Y, Yang MH,
Yuan GH, Tang Z and Wang H: Salvia miltiorrhiza injection restores
apoptosis of fibroblast-like synoviocytes cultured with serum from
patients with rheumatoid arthritis. Mol Med Rep. 11:1476–1482.
2015. View Article : Google Scholar
|
|
14
|
Sugiyama R, Agematsu K, Migita K, Nakayama
J, Mokuda S, Ogura F, Haraikawa K, Okumura C, Suehiro S, Morikawa
S, et al: Defect of suppression of inflammasome-independent
interleukin-8 secretion from SW982 synovial sarcoma cells by
familial Mediterranean fever-derived pyrin mutations. Mol Biol Rep.
41:545–553. 2014. View Article : Google Scholar
|
|
15
|
Beaulieu E, Green L, Elsby L, Alourfi Z,
Morand EF, Ray DW and Donn R: Identification of a novel cell
type-specific intronic enhancer of macrophage migration inhibitory
factor (MIF) and its regulation by mithramycin. Clin Exp Immunol.
163:178–188. 2011. View Article : Google Scholar :
|
|
16
|
Gupta A, Niger C, Buo AM, Eidelman ER,
Chen RJ and Stains JP: Connexin43 enhances the expression of
osteoarthritis-associated genes in synovial fibroblasts in culture.
BMC Musculoskelet Disord. 15:4252014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Snead NM and Rossi JJ: RNA interference
trigger variants: Getting the most out of RNA for RNA
interference-based therapeutics. Nucleic Acid Ther. 22:139–146.
2012.PubMed/NCBI
|
|
18
|
Nishioka N, Matsuoka T, Yashiro M,
Hirakawa K, Olden K and Roberts JD: Plasminogen activator inhibitor
1 RNAi suppresses gastric cancer metastasis in vivo. Cancer Sci.
103:228–232. 2012. View Article : Google Scholar
|
|
19
|
Jung HS and Shin YK: The potential
RNAi-based combination therapeutics. Arch Pharm Res. 34:1–2. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stornaiuolo M, La Regina G, Passacantilli
S, Grassia G, Coluccia A, La Pietra V, Giustiniano M, Cassese H, Di
Maro S, Brancaccio D, et al: Structure-based lead optimization and
biological evaluation of BAX direct activators as novel potential
anticancer agents. J Med Chem. 58:2135–2148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siddiqui WA, Ahad A and Ahsan H: The
mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update.
Arch Toxicol. 89:289–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Afonin KA, Grabow WW, Walker FM, Bindewald
E, Dobrovolskaia MA, Shapiro BA and Jaeger L: Design and
self-assembly of siRNA-functionalized RNA nanoparticles for use in
automated nanomedicine. Nat Protoc. 6:2022–2034. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mysara M, Garibaldi JM and Elhefnawi M:
MysiRNA-designer: A workflow for efficient siRNA design. PLoS One.
6:e256422011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hefferon KL: Innovations in siRNA
research: A technology comes of age. Recent Pat Antiinfect Drug
Discov. 5:226–239. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Li Z, Han Y, Liang LH and Ji A:
Nanoparticle-based delivery system for application of siRNA in
vivo. Curr Drug Metab. 11:182–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sarret P, Doré-Savard L and Beaudet N:
Direct application of siRNA for in vivo pain research. Methods Mol
Biol. 623:383–395. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yuan S, Wu B, Yu Z, Fang J, Liang N, Zhou
M, Huang C and Peng X: The mitochondrial and endoplasmic reticulum
pathways involved in the apoptosis of bursa of Fabricius cells in
broilers exposed to dietary aflatoxin B1. Oncotarget.
7:65295–65306. 2016.PubMed/NCBI
|
|
28
|
Zhang W: Research of the pathogensis of
oxygen free radicals-mitochondrion pathway in oligospermia and
asthenospermia. Chinese doctoral dissertation full text database.
May;2015.
|