Introduction
Antimicrobial peptides (AMPs) in the cathelicidin
family are multifunctional molecules found in all mammalian species
(1). Human cathelicidin
hCAP18/LL-37 was reported to prevent infections via the innate
immune system (2). LL-37 is
widely expressed in several types of cells and organs, including
neutrophils, skin, epithelial cells of the airways and colon
epithelium (3-6), and is considered to suppress
cancers, such as colon cancer (7).
Colon cancer is one of the most commonly diagnosed
malignant tumors in men and women. A total of 1.4 million colon
cancer cases were estimated and ~700,000 deaths were reported in
2012 worldwide (8). There are six
types of standard treatment for colon cancer, including surgery and
chemotherapy (9). However, the
effectiveness of chemotherapeutic agents differs among patients.
Therefore, novel biological agents and treatment approaches for
colon cancer are needed. The present study focused on AMPs as one
of the possible biological therapeutic agents.
Our group previously reported that a 27-residue
analog of the LL-37 peptide, FF/CAP18, induced apoptosis in SAS-H1
oral squamous cell carcinoma cells (10) and the colon carcinoma-derived cell
line HCT116 (11). FF/CAP18
upregulates miR-663a in HCT116 cells. MicroRNAs (miRNAs) regulate
the growth of HCT116 cells by targeting the CXCR4-p21 pathway
(12).
Exosomes are small extracellular membrane vesicles
(30-100 nm) secreted by numerous types of cells, including cancer
cells. Exosomes contain protein and nucleic acids, such as miRNAs,
and act as important mediators of intercellular communication
(13,14). The association between exosomes
and cancer cells has been widely investigated. Specific miRNAs
transported in cancer-secreted exosomes have been found to promote
metastasis (15) and are
potential biomarkers of cancer (16).
Exosomal miRNAs play critical roles in regulating
cancer development. miRNAs are small non-coding RNAs that control
gene expression by binding to the 3′-untranslated region of their
target mRNA (17). Alterations of
miRNAs are involved in the initiation and progression of cancer
(18), whereas miRNAs may act as
tumor suppressors as well as oncogenes (19). miRNAs are present in cancer cells
and extracellularly in exosomes (13,20). However, cell-to-cell communication
in colon cancer cells following treatment with AMPs has not yet
been elucidated. Therefore, the aim of the present study was to
focus on the release of exosomes from HCT116 cells treated with
FF/CAP18 and cell-to-cell communication following treatment with
AMPs. The quality and quantity of exosomes secreted by colon cancer
HCT116 cells and upregulated miRNAs in both the cells and exosomes
were analyzed following treatment with FF/CAP18.
Materials and methods
Cell culture
Cells from the HCT116 human colon carcinoma-derived
cell line were provided by Dr Bert Vogelstein (The Johns Hopkins
University, Baltimore, MD, USA). The cells were cultured in
Dulbecco’s modified Eagle’s medium (DMEM; Nacalai Tesque, Kyoto,
Japan) containing 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc., Carlsbad, CA, USA) and 5% antibiotic/antimycotic
mixed stock solution (Nacalai Tesque) at 37°C and 5%
CO2. Cell passage and maintenance were performed as
previously described (12).
Peptides
The primary amino acid structure of the original
LL-37 is LLGD FFR KSK EKI GKE FKR IVQ RIK DFL RNL VPR TES. hCAP is
a 27-mer peptide that lacks the first and last 5 amino acids of
LL-37. FF/CAP18 (FRK SKE KIG KFF KRI VQR IFD FLR NLV) was designed
by replacing a glutamic acid residue and lysine residue with
phenylalanine of hCAP18 to enhance its antimicrobial and anticancer
properties (11).
Exosome isolation
HCT116 cells were seeded at a density of
1.2×106 cells/ml in a 100-mm dish and incubated for 2
days. The cells were washed with phosphate-buffered saline and
cultivated in DMEM containing 10% exosome-depleted fetal bovine
serum Media Supplement (System Biosciences, Palo Alto, CA, USA) and
5% antibiotic/antimycotic mixed stock solution (Nacalai Tesque).
After 48 h, the medium was harvested to collect exosomes. Exosomes
were isolated by ultrafiltration using a Vivaspin 20-100K (GE
Healthcare, Little Chalfont, UK) and miRCURY™ Exosome Isolation
Kit-Cells, Urine and CSF (Exiqon, Vedbaek, Denmark). The culture
medium was centrifuged at 300 x g for 5 min and at 1,200 x g for 20
min to remove cells and debris. The supernatant (20 ml) was
ultrafiltered and condensed to 1 ml using the Vivaspin 20-100K to
concentrate the exosomes. Exosome isolation was performed using a
miRCURY™ Exosome Isolation Kit according to the manufacturer’s
instructions (Exiqon). The protein levels of exosomes resuspended
in 100 µl of resuspension buffer (Exiqon) were quantified
using the DC protein assay (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). After confirmation by transmission electron microscopy
(TEM), a combination method was used to isolate exosomes from
FF/CAP18-treated cells.
TEM
To examine the quality of isolated exosomes, the
size, size distribution and morphology of non-treated HCT116 cells
were evaluated. A total of 5 µl of each exosome sample (n=3)
was applied to a 400-mesh copper grid for 10 min prior to negative
staining with 10 µl of 2% phosphotungstic acid for 10 min.
The grid was washed with 10 µl of distilled water, dried,
and viewed by TEM using a model H-7650 microscope (Hitachi, Tokyo,
Japan).
Western blot analysis
Cells and exosomes were homogenized in lysis buffer
[1 M Tris-HCl (pH 7.4), 3 M NaCl, 1% Triton X-100, 6 mM sodium
deoxycholate, and 0.5% protease inhibitor; Nacalai Tesque] by
ultrasonic fragmentation as previously described (12). Protein concentration was measured
using the DC protein assay kit. Cell lysates (20 µg) and
exosome lysates (10 µg) were electrophoresed on 5-20%
polyacrylamide gels (ATTO, Tokyo, Japan) and transferred to
Immobilon-P membranes (Merck Millipore, Darmstadt, Germany). The
membranes were blocked with phosphate-buffered saline containing 3%
skimmed milk and 0.05% Tween-20, and then incubated with primary
mouse monoclonal antibodies against CD63 (1:1,000; 10628D;
Invitrogen; Thermo Fisher Scientific, Inc.), CD81 (1:1,000; 10630D;
Invitrogen; Thermo Fisher Scientific, Inc.) and α-tubulin (1:1,000;
sc-32293; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). After
washing, the membranes were incubated with sheep anti-mouse IgG
conjugated to horseradish peroxidase (1:2,000; GE Healthcare) as
the secondary antibody. Signals were detected using a SuperSignal™
West Femto Maximum Sensitivity Substrate Trial Kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and images were obtained with
the LumiCube device (Liponics, Inc., Tokyo, Japan) and analyzed
with Just TLC software (Sweday, Sodra Sandby, Sweden).
Localization of FAM-FF/CAP18
Synthetic FF/CAP18 was conjugated with
5-carboxyfluorescein (FAM; Scrum Co. Ltd., Tokyo, Japan). HCT116
cells were examined at 1 and 6 h after treatment with FAM-FF/CAP18
(40 µg/ml). The nuclei were visualized by staining with 1
µg/ml 4′,6-diamidino-2-phenyl-indole (Dojindo Laboratories,
Kumamoto, Japan) 6 h after treatment with FAM-FF/CAP18. The
mitochondria were also visualized by staining with 200 nM MitoRed
(Dojindo Laboratories). The staining was visualized using a BZ-8100
fluorescence microscope (Keyence, Tokyo, Japan). Each experiment
was conducted in triplicate.
Detection of apoptosis using Annexin
V-7-amino-actinomycin D (7-AAD)
The rate of cell apoptosis following treatment with
FF/CAP18 was measured using the Muse® Annexin V and Dead
Cell assay kit (Merck Millipore). This assay is based on the
appearance of phosphatidylserine on the surface of cells early
during apoptosis, which can bind Annexin V. In the late phase of
apoptosis or necrosis, phosphatidylserine on the cell surface binds
to Annexin V and DNA binds to 7-AAD, which is a fluorescent
intercalator of GC regions. The combination of Annexin V and 7-AAD
is used to detect apoptotic cells and distinguish between apoptotic
phases. HCT116 cells were treated with FF/CAP18 (20, 30 or 40
µg/ml). After 48 h, the cells were treated with trypsin and
collected in 1.5-ml micro-tubes. These tubes were centrifuged at
800 x g for 5 min. The supernatant was aspirated and the cell
pellets were individually resuspended in 100 µl fresh medium
and added to 100 µl of Muse Annexin V and Dead Cell assay
kit reagent. The cellular mixture was incubated for 20 min at room
temperature and the cells were examined using a Muse Cell Analyzer
(Merck Millipore). These experiments were repeated three times
(n=3).
Uptake of exosomes and cell viability
analysis
Cell viability was measured by the WST-8 assay
(Dojindo Laboratories). HCT116 cells were seeded (2×103
cells/well) in a 96-well plate with complete culture medium and
incubated at 37°C in 5% CO2. After 24 h, exosomes
released by FF/CAP18-treated or untreated HCT116 cells were added
to the corresponding cells and co-incubated for another 48 h. WST-8
reagent was added to each well and the samples were incubated for 4
h. Absorbance was measured at a wavelength of 450 nm using a
Synergy™ HT (BioTek, Winooski, VT, USA). These experiments were
repeated three times (n=3). Cell viabilities (%) were calculated as
relative values based on the absorbance of non-treated cells
(100%).
RNA extraction and miRNA microarray
Exosomes were collected according to the exosome
isolation protocol. Total RNA, including miRNAs, was extracted
using the miRCURY™ RNA Isolation Kit-Cell and Plant (Exiqon)
according to the appendix protocol of the miRCURY™ Exosome
Isolation Kit-Cells, Urine and CSF (Exiqon). RNA quality
[concentration, optical density (OD) 260/280, 260/230 nm] was
determined using a BioSpecnano device (Shimadzu, Kyoto, Japan). The
3D-Gene™ human microRNA chips for miRNA expression analysis (Toray
Industries, Inc., Tokyo, Japan) were used to analyze the effect of
FF/CAP18 treatment on miRNA expression in exosomes. Total RNAs
labeled with 3D-Gene miRNA labeling kit (Toray Industries, Inc.)
were hybridized onto the 3D-Gene Human miRNA Oligo chip (Toray
Industries, Inc.). Fluorescent signals were scanned with the
3D-Gene Scanner (Toray Industries, Inc.) and analyzed using 3D-Gene
Extraction software (Toray Industries, Inc.). The relative
expression level of a given miRNA was calculated by comparing the
signal intensities of the valid spots throughout the microarray
experiments. The data were globally normal-ized per array, such
that the median of the signal intensity was adjusted to 25.
Cellular RNA extraction and miRNA microarray were performed as
previously described (12).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical significance between two groups was determined by the
Student’s t-test of variance with least difference post hoc test.
P<0.05 was considered to indicate statistically significant
differences.
Results
Characterization of exosomes secreted
from HCT116 cells
To ensure successful isolation of exosomes, the
collected exosomes from non-treated HCT116 cells were observed as
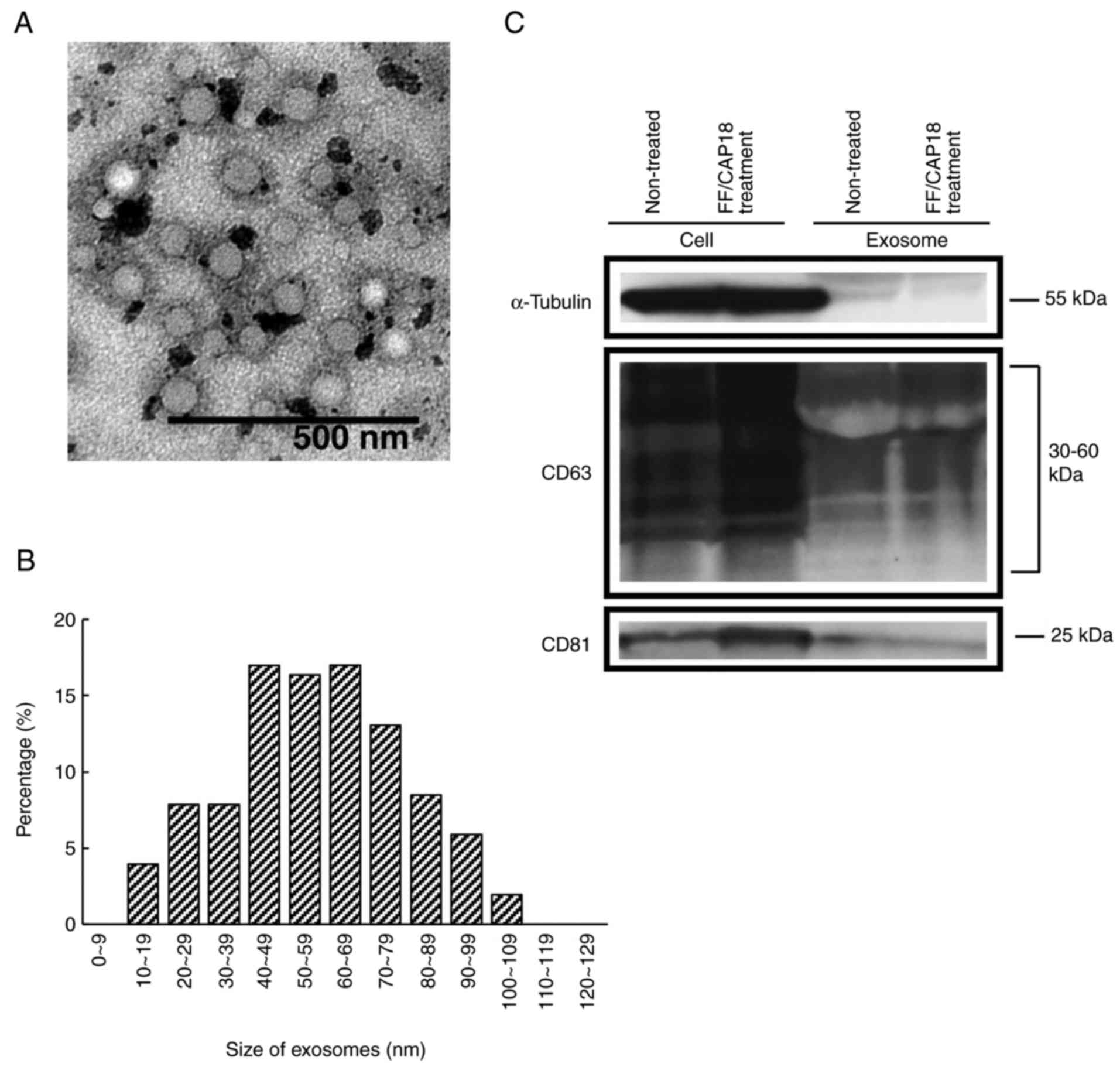
microvesicle clusters (Fig. 1A).
For size distribution, 78% of exosomes were 40-100 nm (Fig. 1B). The amount of exosomes was
3.8×10−8 and 4.9×10−8 mg/cell in untreated
controls and FF/CAP18-treated cells, respectively.
The characterization of exosomes, as determined by
western blotting, is shown in Fig.
1C. The investigated CD63 and CD81 proteins were previously
reported as being exosome-specific markers (21,22). CD63 was detected at 30-60 kDa, as
expected from the datasheet for the CD63 antibody. CD81 was
detected at 25 kDa. These two proteins were detected in HCT116
cells and exosomes before and after treatment with FF/CAP18. In
HCT116 cells, the levels of CD63 and CD81 were increased after
FF/CAP18 treatment. The expression levels of exosomes from
FF/CAP18-treated cells were similar to those in controls, although
both proteins were detected.
Localization of FF/CAP18
At 1 h after treatment with FAM-FF/CAP18, FF/CAP18
interacted with the cell membranes (Fig. 2A). Some FAM-FF/CAP18 was observed
in the cells. At 6 h after treatment, FF/CAP18 was incorporated in
the cytoplasm (Fig. 2B). No
FF/CAP18 was detected on the cell membrane. As shown in Fig. 2B, the peptide localized in the
cytoplasm, but not in the mitochondria or nucleus.
Apoptosis detection using the Annexin
V-7-AAD assay
Cells were classified into four groups based on the
reactivity of Annexin V and 7-AAD combined, as previously described
(23). An increased concentration
of FF/CAP18 was accompanied by an increased affinity for Annexin V
(Fig. 2C). Treatment with
FF/CAP18 at 20 and 30 µg/ml decreased the number of live
cells (Annexin V- and 7-AAD−) (Fig. 2C, upper right panel and lower left
panel). Treatment with FF/CAP18 at 40 µg/ml increased the
number of late apoptotic cells and apoptotic cell death (Annexin
V+ and 7-AAD+). To determine the density of
FF/CAP18 treatment, HCT116 cells were exposed to FF/CAP18 at a
concentration of 33.8 µg/ml; this concentration was selected
for the experiment as this dose caused the death of ~50% of HCT116
cells (Fig. 2D).
Viability of HCT116 cells after treatment
with exosomes
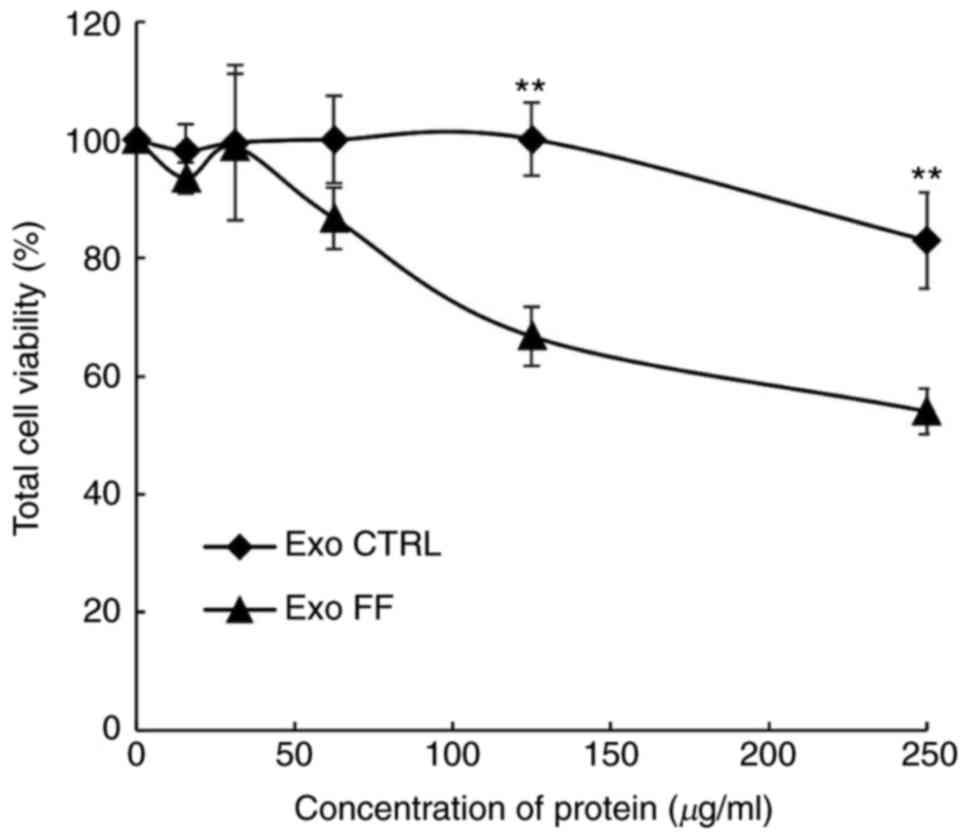
Exosomes were quantified with the DC protein assay
to evaluate the effect of FF/CAP18 on the exosome secretion volume.
The amount of pooled exosomes released from cells during FF/CAP18
exposure was 5.0×10-8 mg/cell. The amount of exosomes
released from non-treated control cells was 3.8×10−8 mg
per cell. To determine whether exosomes affect the viability of
HCT116 cells, exosomes released from control HCT116 cells (Exo
CTRL) and from cells treated with FF/CAP18 (Exo FF) were harvested
and added to HCT116 cells in the WST-8 assay. The effect on cell
viability was evaluated at exosome protein concentrations of 15.63,
31.25, 62.5, 125 and 250 µg/ml. The cell viability (Exo FF
group) was significantly decreased at 125 and 250 µg/ml
(Fig. 3, P<0.01). The effect
of Exo CTRL was less prominent compared with that of Exo FF.
miRNA expression profiles of exosomes
released by HCT116 cells treated with FF/CAP18
An attempt was made to analyze the alterations in
the miRNAs in exosomes to reveal the cause of cell viability
suppression. A miRNA microarray revealed that 137 miRNAs were
increased by 2-fold or more in exosomes released by
FF/CAP18-treated HCT116 cells compared with exosomes released by
non-treated cells (Table I). By
contrast, 17 miRNAs were increased by 2-fold or more in HCT116
cells following FF/CAP18 treatment compared with non-treated cells
(Table II). Three miRNAs
(miR-584-5p, miR-1202 and miR-3162-5p) were upregulated in both
HCT116 cells and exosomes following treatment with FF/CAP18.
 | Table IUpregulated miRNAs in exosomes
released from HCT116 cells treated with FF/CAP18. |
Table I
Upregulated miRNAs in exosomes
released from HCT116 cells treated with FF/CAP18.
| Fold change | Upregulated miRNAs
(miRs) in exosomes |
|---|
| 2-3 | 194-3p, 6795-5p,
4485-3p, 378i, 125b-5p, 491-5p, 151b, 31-5p, 500a-3p, 6753-5p,
1273a, 4747-3p, 4689, 765, 1247-5p, 361-5p, 1184, 6861-5p, 493-3p,
1180-3p, 3199, 365a-5p, 3132, 4476, 193b-3p, 502-3p, 378a-3p, 378b,
1910-3p, 769-3p, 6781-3p, 3160-5p, 4291, 375, 1307-5p, 27a-3p,
19a-3p, 4650-3p, 6761-5p, 1202, 93-3p, 4732-5p, 4756-5p, 423-3p,
6722-5p, 320c, 1273e, 92a-3p, 5585-3p, 208a-5p, 4787-3p, 18a-5p,
4804-3p, 4641, 487b-3p, 128-3p, 100-5p, 345-5p, 320a, 3945, 92b-3p,
6774-3p, 151a-3p, 711, 769-5p, 4749-5p, 378g, 7d-3p, 6767-5p, 4450,
4520-5p, 6833-5p, 652-5p, 323a-5p, 584-5p, 6893-5p, 4725-3p,
584-3p, 4472, 18b-5p, 6129, 619-5p, 320b, 3678-3p, 185-5p,
4433b-5p, 8073, 3685, 5088-5p, 151a-5p, 885-3p, 8082, 7854-3p,
28-5p, 130b-3p, 99b-3p, 4669, 634, 106b-5p, 21-3p, 593-5p, 6805-3p,
4306, 320e, 4267 |
| 3-4 | 378d, 4531,
378a-5p, 3162-5p, 6790-3p, 6790-3p, 3619-3p, 659-5p, 6776-5p,
508-5p, 378c, 19b-3p, 4324, 191-5p, 378e, 3616-3p, 3649, 378f,
8078 |
| 4-5 | 4447, 6782-5p,
3126-5p |
| 5-6 | 4482-3p, 371a-5p,
hsa-let-7b-5p, 921 |
| 6-7 | has-let-7c-5p,
6887-5p |
| 7-8 | 3687, 15b-5p,
hsa-let-7a-5p |
| 8< | 4261,
hsa-let-7d-5p |
 | Table IIUpregulated miRNAs in HCT116 cells
after treatment with FF/CAP18. |
Table II
Upregulated miRNAs in HCT116 cells
after treatment with FF/CAP18.
| miRNAs (miRs) | Fold change in
HCT116 after treatment with FF/CAP18 | More than 2‑fold
upregulation in exosomes |
|---|
| 4271 | 359 | No |
| 575 | 316 | No |
| 483-5p | 287 | No |
| 663a | 277 | No |
| 4298 | 237 | No |
| 630 | 195 | No |
| 601 | 109 | No |
| 939-5p | 84 | No |
| 572 | 71 | No |
| 3663-3p | 58 | No |
| 513a-5p | 8 | No |
| 4257 | 3 | No |
| 1202 | 3 | Yes |
| 513b | 3 | No |
| 584-5p | 3 | Yes |
| 3679-5p | 3 | No |
| 3162-5p | 2 | Yes |
Discussion
AMPs play a key role in cancer regulation (24,25). Previous studies revealed the
detailed mechanism of the suppression of colon cancer cell growth
by the human cathelicidin AMP, LL-37, and its analog, FF/CAP18.
This suppression involved the CXCR4-p21 pathway, which is
independent of p53, in an miR-663a-targeted manner with an altered
metabolic profile (11,12,23). The present study provides insight
into the mechanism of cancer growth suppression by AMPs, which
occurs via cell-to-cell communication (Fig. 3 and Tables I and II). To the best of our knowledge, the
impact of AMPs on tumor cell-to-cell communication has not been
previously investigated. In the present study, treatment with
FF/CAP18, an analog of LL-37, caused HCT116 cells to secrete more
exosomes compared with untreated cells, and these exosomes
suppressed HCT116 cell growth. Several miRNAs exhibiting increased
expression in cells as well as exosomes following FF/CAP18
treatment were considered to suppress cancer cell growth.
FF/CAP18 became localized on the HCT116 cell
membrane, was incorporated into the cells and induced apoptosis
(Fig. 2A-C). LL-37 is considered
to act by rapidly disrupting the bacterial membrane (26,27). Recently, Shahmiri et al
reported that cathelicidin LL-37 exhibits membrane-disrupting
antimicrobial activity and two distinct interaction pathways: Pore
formation in bilayers of unsaturated phospholipids, and membrane
modulation with saturated phospholipids (28). In the present study, membrane
disruption was not observed in HCT116 cells following treatment
with FF/CAP18. FF/CAP18 was detected on the membrane of HCT116
cells 1 h after treatment and in the cytoplasm of the cells 6 h
after treatment. These results suggest that, in cancer cells,
FF/CAP18 exerted its effects without disrupting the membrane.
Additionally, FF/CAP18 treatment of HCT116 cells caused the cells
to secrete more exosomes than in the absence of treatment. The
secretion of exosomes is regulated by cellular factors, such as
intracellular calcium levels, and extracellular factors, such as
chemical treatment (29,30). The enhanced exosomal export may be
a stress response of HCT116 cells to FF/CAP18. The exosomes
released by HCT116 cells during exposure to FF/CAP18 suppressed the
viability of HCT116 cells (Fig.
3). This result indicates that exosomes released in the
presence of FF/CAP18 contain a tumor suppression factor, such as
miR-584-5p, miR-1202 and miR-3162-5p.
The contents of protein and nucleic acids, including
miRNAs, of exosomes were previously determined (13,14). miRNAs are crucial for cancer
regulation (18,19). FF/CAP18 treatment changed the
expression levels of miRNAs in exosomes released by HCT116 cells
(Table I). FF/CAP18 treatment
induced an increase of 2-fold or more in the expression of three
miRNAs (miR-584-5p, miR-1202 and miR-3162-5p) in both HCT116 cells
and exosomes, among which miR-584-5p and miR-1202 reportedly act as
cancer suppressors. miR-584-5p was reported to inhibit
proliferation and induce apoptosis of colon and gastric cancer
cells (31,32). miR-584-5p induces apoptotic death
by inhibiting the interaction between hnRNP A1 and CDK6 mRNA
(32). miR-1202 is downregulated
in ovarian cancer and clear cell papillary renal cell carcinoma
(33,34). Additionally, miR-1202 suppresses
glioma cell proliferation and induces apoptosis by targeting and
inhibiting Rab1A (35).
miR-584-5p and miR-1202 may suppress the proliferation of HCT116
cells via exosomes. By contrast, miR-3162-5p may be a new
regulatory factor in colon cancer. Our group reported that AMPs
upregulate the expression of miR-663a in HCT116 cells, and that
this miRNA regulates the proliferation of colon cancer HCT116 cells
via the CXCR4-p21 pathway (12).
However, the expression of miR-663a in exosomes was upregulated by
<2-fold following FF/CAP18 treatment. Therefore, these three
miRNAs may exert anticancer effects via exosomes to a greater
extent compared with miR-663a. More studies are required to
elucidate the mechanism by which these three miRNAs suppress cancer
in order to support the use of AMPs as anti-cancer agents.
The present study focused on the role of FF/CAP18.
LL-37 may act in regulating cancer via exosomes. The results of
this study indicate that exosomes released by cancer cells in the
presence of AMPs suppress tumor growth. Several studies have
suggested that exosomes secreted by cancer cells assist in cancer
growth and angiogenesis, leading to cancer metastasis (15,36). By contrast, specific exosomes were
found to suppress cancer growth. The secretion of miRNAs was
mediated through exosomes and their quality and quantity were
altered after treatment with FF/CAP18. Alterations in the miRNAs in
exosomes released by cells may suppress cancer.
Additionally, LL-37 and FF/CAP18, the analog of the
LL-37 peptide, were previously described as anticancer agents
(7,10,11). In the present study, FF/CAP18 was
also found to inhibit cancer growth through exosomes. Therefore,
AMPs act directly on target cancer cells and indirectly via
exosomes. The effect of exosomes from HCT116 cells on other type of
cancer cells remains unclear. If the exosomes secreted by
FF/CAP18-treated HCT116 cells regulate the viability of other types
of cancer cells, AMPs may exert anticancer effects against
metastatic cancer in other organs.
In recent years, the potential of using exosomes as
therapeutic agents, such as exosomes released by mesenchymal stem
cells, has attracted attention (37). Studies on exosomes released by
cancer cells during treatment with AMPs are important for
understanding the therapeutic effects of AMPs against colon
cancer.
Funding
This study was supported by a Grand-in-Aid for
Scientific Research from the Japan Society for the Promotion of
Science (2016-2018: Grant number 16H05036).
Availability of data and materials
All the data generated and analyzed in the present
study are available from the corresponding author upon reasonable
request.
Authors’ contributions
MH, KK, KI, TI and EI conceived and designed the
experiments. MH and KK performed the experiments. MH, KK and EI
analyzed the date. MH and EI wrote paper. All the authors have read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests to disclose.
Acknowledgments
Not applicable.
References
|
1
|
Zanetti M, Gennaro R, Scocchi M and
Skerlavaj B: Structure and biology of cathelicidins. Adv Exp Med
Biol. 479:203–218. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zanetti M: The role of cathelicidins in
the innate host defenses of mammals. Curr Issues Mol Biol.
7:179–196. 2005.PubMed/NCBI
|
|
3
|
Cowland JB, Johnsen AH and Borregaard N:
hCAP-18, a cathelin/probactenecin-like protein of human neutrophil
specific granules. FEBS Lett. 368:173–176. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Frohm M, Agerberth B, Ahangari G,
Ståhle-Bäckdahl M, Lidén S, Wigzell H and Gudmundsson GH: The
expression of the gene coding for the antibacterial peptide LL-37
is induced in human keratinocytes during inflammatory disorders. J
Biol Chem. 272:15258–15263. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bals R, Wang X, Zasloff M and Wilson JM:
The peptide anti-biotic LL-37/hCAP-18 is expressed in epithelia of
the human lung where it has broad antimicrobial activity at the
airway surface. Proc Natl Acad Sci USA. 95:9541–9546. 1998.
View Article : Google Scholar
|
|
6
|
Hase K, Eckmann L, Leopard JD, Varki N and
Kagnoff MF: Cell differentiation is a key determinant of
cathelicidin LL-37/human cationic antimicrobial protein 18
expression by human colon epithelium. Infect Immun. 70:953–963.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren SX, Cheng AS, To KF, Tong JH, Li MS,
Shen J, Wong CC, Zhang L, Chan RL, Wang XJ, et al: Host immune
defense peptide LL-37 activates caspase-independent apoptosis and
suppresses colon cancer. Cancer Res. 72:6512–6523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
PDQ Adult Treatment and Editorial Board:
Colon Cancer Treatment (PDQ®): Patient Version PDQ Cancer
Information Summaries edn. Bethesda (MD): National Cancer Institute
(US); 2002
|
|
10
|
Okumura K, Itoh A, Isogai E, Hirose K,
Hosokawa Y, Abiko Y, Shibata T, Hirata M and Isogai H: C-terminal
domain of human CAP18 antimicrobial peptide induces apoptosis in
oral squamous cell carcinoma SAS-H1 cells. Cancer Lett.
212:185–194. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuroda K, Fukuda T, Yoneyama H, Katayama
M, Isogai H, Okumura K and Isogai E: Anti-proliferative effect of
an analogue of the LL-37 peptide in the colon cancer derived cell
line HCT 116 p53+/+ and p53−/−. Oncol Rep. 28:829–834. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuroda K, Fukuda T, Krstic-Demonacos M,
Demonacos C, Okumura K, Isogai H, Hayashi M, Saito K and Isogai E:
miR-663a regulates growth of colon cancer cells, after
administration of antimicrobial peptides, by targeting CXCR4-p21
pathway. BMC Cancer. 17:332017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L,
Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, et al:
Cancer-secreted miR-105 destroys vascular endothelial barriers to
promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. PLoS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Redis RS, Calin S, Yang Y, You MJ and
Calin GA: Cell-to-cell miRNA transfer: From body homeostasis to
therapy. Pharmacol Ther. 136:169–174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun Y, Zheng W, Guo Z, Ju Q, Zhu L, Gao J,
Zhou L, Liu F, Xu Y, Zhan Q, et al: A novel TP53 pathway influences
the HGS-mediated exosome formation in colorectal cancer. Sci Rep.
6:280832016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zaharie F, Muresan MS, Petrushev B, Berce
C, Gafencu GA, Selicean S, Jurj A, Cojocneanu-Petric R, Lisencu CI,
Pop LA, et al: Exosome-carried microRNA-375 inhibits cell
progression and dissemination via Bcl-2 blocking in colon cancer. J
Gastrointest Liver Dis. 24:435–443. 2015.
|
|
23
|
Kuroda K, Fukuda T, Isogai H, Okumura K,
Krstic-Demonacos M and Isogai E: Antimicrobial peptide FF/CAP18
induces apoptotic cell death in HCT116 colon cancer cells via
changes in the metabolic profile. Int J Oncol. 46:1516–1526. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu WK, Wang G, Coffelt SB, Betancourt AM,
Lee CW, Fan D, Wu K, Yu J, Sung JJ and Cho CH: Emerging roles of
the host defense peptide LL-37 in human cancer and its potential
therapeutic applications. Int J Cancer. 127:1741–1747. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuroda K, Okumura K, Isogai H and Isogai
E: The human cathelicidin antimicrobial peptide LL-37 and mimics
are potential anticancer drugs. Front Oncol. 5:1442015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Neville F, Cahuzac M, Konovalov O,
Ishitsuka Y, Lee KYC, Kuzmenko I, Kale GM and Gidalevitz D: Lipid
headgroup discrimination by antimicrobial peptide LL-37: Insight
into mechanism of action. Biophys J. 90:1275–1287. 2006. View Article : Google Scholar
|
|
27
|
Turner J, Cho Y, Dinh NN, Waring AJ and
Lehrer RI: Activities of LL-37, a cathelin-associated antimicrobial
peptide of human neutrophils. Antimicrob Agents Chemother.
42:2206–2214. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shahmiri M, Enciso M, Adda CG, Smith BJ,
Perugini MA and Mechler A: Membrane core-specific antimicrobial
action of cathelicidin LL-37 peptide switches between pore and
nanofibre formation. Sci Rep. 6:381842016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Savina A, Furlán M, Vidal M and Colombo
MI: Exosome release is regulated by a calcium-dependent mechanism
in K562 cells. J Biol Chem. 278:20083–20090. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Safaei R, Larson BJ, Cheng TC, Gibson MA,
Otani S, Naerdemann W and Howell SB: Abnormal lysosomal trafficking
and enhanced exosomal export of cisplatin in drug-resistant human
ovarian carcinoma cells. Mol Cancer Ther. 4:1595–1604. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Li, Wei ZQ, Wang S, Chen W, Zhang Z,
Chen L, Li L, Sun B, Xu GJ, et al: Overexpression of miR-584 5p
inhibits proliferation and induces apoptosis by targeting WW
domain-containing E3 ubiquitin protein ligase 1 in gastric cancer.
J Exp Clin Cancer Res. 36:592017. View Article : Google Scholar
|
|
32
|
Konishi H, Fujiya M, Ueno N, Moriichi K,
Sasajima J, Ikuta K, Tanabe H, Tanaka H and Kohgo Y: microRNA-26a
and -584 inhibit the colorectal cancer progression through
inhibition of the binding of hnRNP A1-CDK6 mRNA. Biochem Biophys
Res Commun. 467:847–852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim
SW and Kim YT: MicroRNA profiling of a CD133+
spheroid-forming subpopulation of the OVCAR3 human ovarian cancer
cell line. BMC Med Genomics. 5:182012. View Article : Google Scholar
|
|
34
|
Munari E, Marchionni L, Chitre A, Hayashi
M, Martignoni G, Brunelli M, Gobbo S, Argani P, Allaf M, Hoque MO,
et al: Clear cell papillary renal cell carcinoma: micro-RNA
expression profiling and comparison with clear cell renal cell
carcinoma and papillary renal cell carcinoma. Hum Pathol.
45:1130–1138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Quan Y, Song Q, Wang J, Zhao L, Lv J and
Gong S: MiR-1202 functions as a tumor suppressor in glioma cells by
targeting Rab1A. Tumor Biol. 39:1010428317697562017. View Article : Google Scholar
|
|
36
|
Meehan K and Vella LJ: The contribution of
tumour-derived exosomes to the hallmarks of cancer. Crit Rev Clin
Lab Sci. 53:121–131. 2016. View Article : Google Scholar
|
|
37
|
Lai RC, Chen TS and Lim SK: Mesenchymal
stem cell exosome: A novel stem cell-based therapy for
cardiovascular disease. Regen Med. 6:481–492. 2011. View Article : Google Scholar : PubMed/NCBI
|