Introduction
Spinal cord injury (SCI) leads to severe motor and
sensory dysfunction, and severely affects patients both mentally
and physically (1). SCI is one of
the major causes of disability (2), the clinical treatment of which is
complex. At present, there is no available interventional treatment
that is completely effective for SCI (3). The pathophysiological process of SCI
includes primary and secondary injuries (4). Secondary injury generally occurs
within several hours, days or weeks after the primary injury
(4). It comprises post-traumatic
inflammatory response, neuronal apoptosis and necrosis, local
tissue microcirculatory ischemia and hypoxia, oxidative stress
response, and the excessive production of peroxide and reactive
oxygen species (2). Secondary
injury further damages the spinal cord and can lead to akinesthesia
(5); however, the mechanism
underlying secondary injury is currently unclear. It is known that
secondary injury induces recruitment of a large number of
inflammatory cells at the early stage, which is accompanied by a
strong inflammatory response, and neuronal necrosis and apoptosis
(5).
Silent information regulator 2 (Sir2)-associated
enzymes are highly conserved NAD+-dependent histone
deacetylases (6). There are seven
Sir2 homologous genes in the human body, namely, sirtuin (SIRT)1-7
(6). SIRT1 has been the focus of
much research in recent years. The SIRT1 protein has been revealed
to exert anti-inflammatory and antioxidative effects, and can
alleviate cell injury. In addition, SIRT1 is able to markedly
reduce the amyloid-β protein-induced nuclear factor (NF)-κB pathway
and decrease the production of inflammatory factors (7). It can also suppress inflammatory
response-induced injury to protocortical neurons. A possible
underlying mechanism for these effects is that SIRT1 acts on the
NF-κB subunit, RelA/p65, through deacetylation; therefore, SIRT1
can participate in regulating NF-κB active genes. In particular,
SIRT1 can reduce the binding of NF-κB with intranuclear
inflammatory genes, and can thus reduce the production of
inflammatory factors, such as TNF-α and IL-1β (8).
MicroRNAs (miRNAs/miRs) are gene expression
regulatory factors, which were discovered >20 years ago
(3). Expression of numerous
miRNAs is altered in response to SCI. It has previously been
reported that miRNAs participate in the regulation of post-SCI gene
expression; such gene expression alterations are closely associated
with certain pathological processes (3), including ischemia and swelling of
the spinal cord, the inflammatory response and neuronal necrosis.
Molecular biology has developed rapidly; as a result, research
regarding miRNA has attracted increasing attention. It has
previously been indicated that miRNAs serve vital regulatory roles
in spinal development, spinal plasticity, and post-SCI pathogenesis
and developmental processes (9).
Therefore, some miRNAs may be considered effective targets for
post-SCI therapeutic intervention, and investigating the function
and role of miRNAs in the pathological development of SCI may aid
understanding of the pathogenesis of secondary SCI. Furthermore,
research into miRNAs may provide novel therapeutic targets and
intervention strategies for SCI treatment and rehabilitation
(1). The present study aimed to
investigate the function and mechanism of SIRT1 in SCI. In the
present study, an in vivo model was used to study primary
SCI injury, and an in vitro model was used to study
secondary SCI injury
Materials and methods
Experimental animals and establishment of
an animal model
The present study was approved by the Scientific
Review Committee and the Institutional Review board of Dalian
University (Dalian, China). Healthy male Sprague-Dawley rats
(weight, 200-250 g; age, 9-11 weeks, n=12) were obtained from the
Animal Center of Dalian University. All rats were maintained under
a 12-h dark/light cycle at 22-24°C (relative humidity 55-60%), and
received ad libitum access to a standard laboratory diet and
water. The rats were randomly assigned into the following two
groups: Control group (n=6) and SCI model group (n=6). All rats in
the SCI model group were anesthetized with an intraperitoneal
(i.p.) injection of 30 mg/kg pentobarbital sodium, after which
T8-T9 spinous processes and lamina were uncovered, and paraspinal
muscles were stripped. Subsequently, the T8 and T9 spinous
processes were clamped with forceps and lamina was removed to
expose the dura mater. The underlying cord was exposed to contusion
injury without disrupting the dura (10). All rats in the control group were
anesthetized with an i.p. injection of 30 mg/kg pentobarbital
sodium; however, they did not undergo SCI. A total of 1 day after
induction of the SCI model, rats were anesthetized with an i.p.
injection of 30 mg/kg pentobarbital sodium and were sacrificed by
decapitation. Subsequently, spinal cord samples were collected and
stored at −80°C.
Basso, Beattie, Bresnahan (BBB) score and
water content analysis
The BBB score of the rats was determined after 4 min
in an open field (1), and was
scored between 0 (no observable hind-limb movements) and 21 (normal
locomotion). For water content analysis of the spinal cord, spinal
cord samples were weighed, dried at 80°C for 24 h and were then
weighed again. The following formula was used to calculate the
spinal cord water content: [(Wet weight-dry weight)/wet weight]
×100%.
Hematoxylin and eosin staining
Spinal cord samples were fixed with 4%
paraformaldehyde for 48 h at room temperature, embedded in paraffin
and cut into 10-µm sections. The tissue samples were then
stained with hematoxylin and eosin for 10 min, and examined under a
fluorescence microscope (BX53; Olympus Corporation, Tokyo,
Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from spinal cord tissues and
transfected cells using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer′s protocol. cDNA was obtained using the ImProm-II
Reverse Transcription system (Promega Corporation, Madison, WI,
USA), according to manufacturer′s protocol. RT-qPCR was performed
with SYBR Green PCR Mater Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) on an ABI 7900HT fast real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primer
sequences were as follows: SIRT1, forward 5′-TCA GTG TCA TGG TTC
CTT TGC-3′, reverse 5′-AAT CTG CTC CTT TGC CAC TCT-3′; GAPDH,
forward 5′-CCC CTG GCC AAG GTC ATC CA-3′, reverse 5′-CGG AAG CCA
TGC CAG TGA G-3′; miR-494, forward 5′-CAT AGC CCG TGA AAC ATA CAC
G-3′, reverse 5′-GTG CAG GGT CCG AGG T-3′; and U6 forward 5′-CGC
TTC GGC AGC ACA TAT ACT A-3′ and reverse 5′-GCG AGC ACA GAA TTA ATA
CGA C-3′. The qPCR thermo-cycling conditions were as follows: 95°C
for 10 min; followed by 40 cycles at 95°C for 25 sec, 60°C for 25
sec and 72°C for 25 sec, and a final extension step at 4°C for 10
min. Relative expression levels were calculated using the
2-ΔΔCq method (11).
Gene microarray hybridization
Total RNA (500 ng) was labeled with Cyanine-5-CTP
(cat. no. NEL474001EA; PerkinElmer, Inc., Waltham, MA, USA) and
hybridized using the SurePrint G3 Mouse Whole Genome GE Microarray
G4852A platform (cat. no. G4851A; Agilent Technologies, Inc., Santa
Clara, CA, USA) with an equimolar concentration of
cyanine-3-cTP-labelled universal rat reference (Stratagene; Agilent
Technologies, Inc.). Images were analyzed using Agilent Feature
Extraction 10.7.3.1 software (Agilent Technologies, Inc.).
Cell line, and cell culture and
transfection
PC12 cells were used to study SCI in vitro,
as previously reported (12-14). PC12 cells were purchased from the
American Type Culture Collection (Manassas, VA, USA) and were
maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in a humidified
atmosphere containing 5% CO2 at 37°C. miR-494 mimics
(5′-UGA AAC AUA CAC GGG AAA CCU C-3′), anti-miR-494 mimics (5′-GGU
UUC CCG UGU AUG UUU CAU U-3′) and a negative control (5′-UUC UCC
GAA CGU GUC ACG U-3′) were obtained from Shanghai GenePharma Co.,
Ltd. (Shanghai, China). Cell transfection (1×106
cells/ml) was performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C with 100 ng
miR-494 mimics, anti-miR-494 or negative control mimics. A total of
4 h post-transfection, DMEM was removed and replaced with FBS-free
DMEM; the cells were then treated with 100 ng/ml lipopolysaccharide
(Beyotime Institute of Biotechnology, Haimen, China) to induce an
in vitro SCI model, as reported in the literature (15), or with 10 µM CAY10602 at
37°C for 48 h. After 4 h of transfection, cells were treated with
the p53 agonist, 20 nM Nutlin 3 (MCE China, Shanghai, China), for
44 h at 37°C. In addition, to confirm that LPS induced an in
vitro SCI model, PC12 cells were separated into the following
two groups: The control group, in which cells were not treated with
LPS; and the LPS group, in which cells were treated with 100 ng/ml
LPS (Beyotime Institute of Biotechnology).
ELISA kit
Cell supernatants were collected at 1,000 x g for 10
min at 4°C and were used to measure tumor necrosis factor (TNF)-α
(cat. no. H052), interleukin (IL)-1β (cat. no. H002), IL-6 (cat.
no. H007) and IL-18 (cat. no. H015) levels using ELISA kits
(Nanjing Jiancheng Bioengineering Institute, Nanjing, China),
according to the manufacturer′s protocol. Absorbance was detected
using an automatic multi-well spectrophotometer (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) at 450 nm.
MTT assay and LDH activity assay
At different time points (24 and 48 h)
post-transfection, 20 µl MTT solution (5 mg/ml) was added to
each well and incubated at 37°C for 4 h. The culture medium was
then removed and dimethyl sulfoxide was added to dissolve formazan
for 20 min at 37°C. The absorbance was detected using an automatic
multi-well spectrophotometer (Bio-Rad Laboratories, Inc.) at 492
nm.
A total of 48 h post-transfection, LDH activity was
analyzed using LDH activity kits (cat. no. C0016, Beyotime
Institute of Biotechnology), according to manufacturer’s protocol,
and absorbance was detected using an automatic multi-well
spectrophotometer (Bio-Rad Laboratories, Inc.) at 450 nm.
miR-494 target prediction and
dual-luciferase reporter assay
miR-494 target prediction was conducted using
TargetScan (http://www.targetscan.org/index.html) and miRanda
(http://www.microrna.org). SIRT1 luciferase
reporter plasmids containing the 3′ untranslated region targeting
miR-494 (100 ng) were co-transfected into the cells
(1×106 cell/ml) with 100 ng of miRNA-494 mimics using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. Cells were plated in 24-well plates and
were transfected for 48 h. The Dual Luciferase Assay system
(Promega Corporation) was used to measure luciferase reporter
activities.
Apoptosis assay
Cells were washed with PBS for 5 min and fixed with
4% paraformaldehyde for 15 min at room temperature. Subsequently,
cells were stained with 5 µl Annexin V-phycoerythrin
conjugate and 5 µl propidium iodide (BD Biosciences,
Franklin Lakes, NJ, USA) for 15 min in the dark at room
temperature. Flow cytometry was performed using BD AccuriC6 (BD
Biosciences) and data were analyzed by FlowJo 7.6.1 (FlowJo, LLC,
Ashland, OR, USA).
Caspase-3/9 activity
Total protein was extracted using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology) and protein concentration was evaluated using the
bicinchoninic acid assay (Beyotime Institute of Biotechnology),
according to the manufacturer’s protocols. Protein (10 µg)
was then used to measure caspase-3/9 activity using the Caspase-3/9
activity kits (cat. nos. C1115 and C1158; Beyotime Institute of
Biotechnology), according to the manufacturer’s protocol.
Western blot analysis
Total protein was extracted using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology) and protein concentration was evaluated using the
bicinchoninic acid assay (Beyotime Institute of Biotechnology),
according to the manufacturer’s protocols. Proteins (50 µg)
were separated by 10% SDS-PAGE and were transferred to
polyvinylidene difluoride membranes. The membranes were then
blocked with 5% non-fat dry milk in Tris-buffered saline-0.1% Tween
for 1 h at 37°C, and were incubated with primary antibodies against
SIRT1 (cat. no. sc-135792, 1:1,000), cyclin E (cat. no. sc-48420,
1:1,000), p53 (cat. no. sc-47698, 1:1,000), Bax (cat. no. sc-20067,
1:1,000), p21 (cat. no. sc-817, 1:1,000) and GAPDH (cat. no.
sc-51631, 1:5,000) (all Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) at 4°C overnight. After washing with PBST for 15 min,
membranes were incubated with a horseradish peroxidase-conjugated
anti-mouse secondary antibody (cat. no. sc-2005, 1:5,000; Santa
Cruz Biotechnology, Inc.) for 1 h at 37°C. Proteins were visualized
using an enhanced chemiluminescence kit (Pierce; Thermo Fisher
Scientific, Inc.) and were analyzed using Image Lab 3.0 (Bio-Rad
Laboratories, Inc.).
Immunofluorescence staining
Cells were washed with PBS for 5 min and fixed with
4% paraformaldehyde for 15 min at room temperature. Subsequently,
cells were penetrated with 0.25% Triton X-100 in PBS for 15 min at
room temperature, washed with PBS for 15 min, and blocked with 5%
bovine serum albumin (Beyotime Institute of Biotechnology) in PBS
for 1 h at 37°C. The cells were then incubated with anti-SIRT1
(cat. no. sc-135792, 1:100; Santa Cruz Biotechnology, Inc.) at 4°C
overnight, washed with PBS for 15 min, and incubated with goat
anti-mouse immunoglobulin G-CruzFluor™ 555 (cat. no. sc-362267,
1:5,000; Santa Cruz Biotechnology, Inc.) for 1 h at room
temperature. Finally, cells were washed for 15 min and incubated
with DAPI in the dark. Cells were observed using confocal
microscopy (Leica SP5, Argon laser; Leica Microsystems, Inc.,
Buffalo Grove, IL, USA).
Statistical analysis
All data are presented as the means ± standard
deviation (n=3), and were analyzed using SPSS 17.0 (SPSS, Inc.,
Chicago, IL, USA). Data were compared using two-tailed Student’s
t-test or one-way analysis of variance and Tukey’s post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
SIRT1 and miR-494 expression in the SCI
rat model
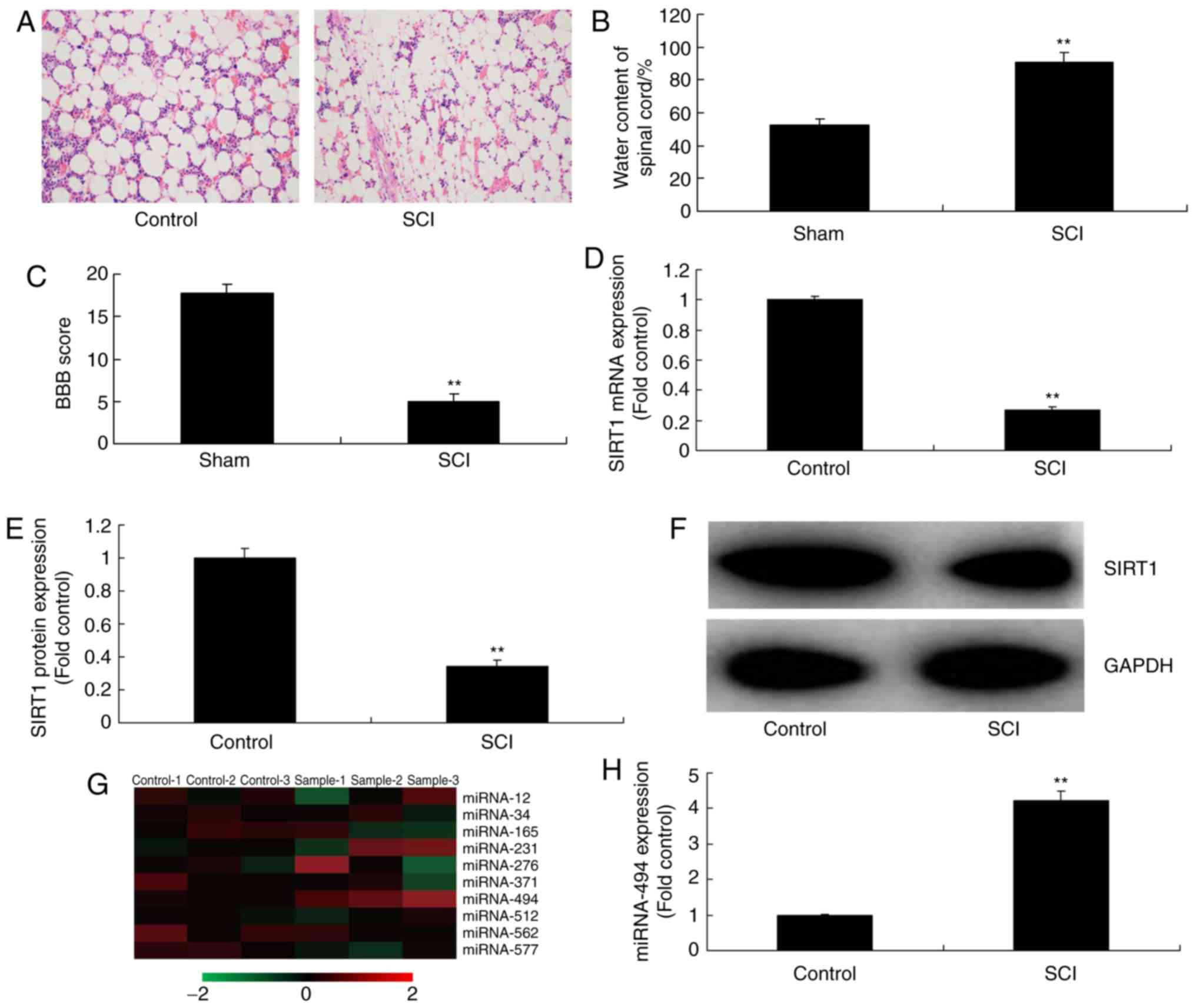
The present study evaluated the expression levels of
SIRT1 in SCI tissues using qPCR. Hematoxylin and eosin staining
indicated that inflammation was markedly increased in SCI tissues
compared with in the control group (Fig. 1A). Furthermore, water content in
the spinal cord was increased, and the BBB score was reduced in SCI
tissues, compared with in the control group (Fig. 1B and C). As shown in Fig. 1D-F, the mRNA and protein
expression levels of SIRT1 in SCI tissues were significantly
inhibited compared with in the control group. Subsequently, a gene
chip was used to analyze the alterations in miRNA expression in SCI
tissues; it was revealed that miR-494 expression was increased in
rats in the SCI group compared with in the control group (Fig. 1G and H). These results indicated
that an association may exist between miR-494 and SIRT1 in SCI.
miR-494 regulates SIRT1 expression in an
SCI model
Initially, it was revealed that LPS induced an
increase in tumor necrosis factor (TNF)‑α, interleukin (IL)‑1β,
IL‑6 and IL‑18 levels in PC12 cells compared with in the control
group (Fig. 2). Subsequently, in
the in vitro model, SIRT1 was identified as a direct target
of miR‑494 and inhibited luciferase activity (Fig. 3A and B). miR‑494 mimics and
anti‑miR‑494 mimics were separately transfected into cells, and
increased or decreased miR‑494 expression compared with in the
control group, respectively (Fig.
3C and D). Following miR‑494 overexpression, a gene chip
analysis was conducted, which revealed that SIRT1 mRNA expression
was suppressed in the in vitro model compared with in the
control group (Fig. 3E). Western
blotting and immunofluorescence revealed that miR-494
overexpression suppressed SIRT1 protein expression in vitro,
in comparison with the control group (Fig. 3F-H). However, miR-494
downregulation induced SIRT1 mRNA and protein expression in
vitro compared with in the control group (Fig. 3I-L).
miR-494 regulates cell apoptosis in an in
vitro model of SCI
To determine the effects of miR-494 on the SCI
model, cell apoptosis and cell growth were analyzed. As shown in
Fig. 4A-D, overexpression of
miR-494 promoted cell apoptosis and LDH activity, and inhibited
cell growth compared with in the negative control group.
Conversely, miR-494 knockdown promoted cell growth, and inhibited
cell apoptosis and LDH activity in the SCI model compared with in
the negative control group (Fig.
4E-H).
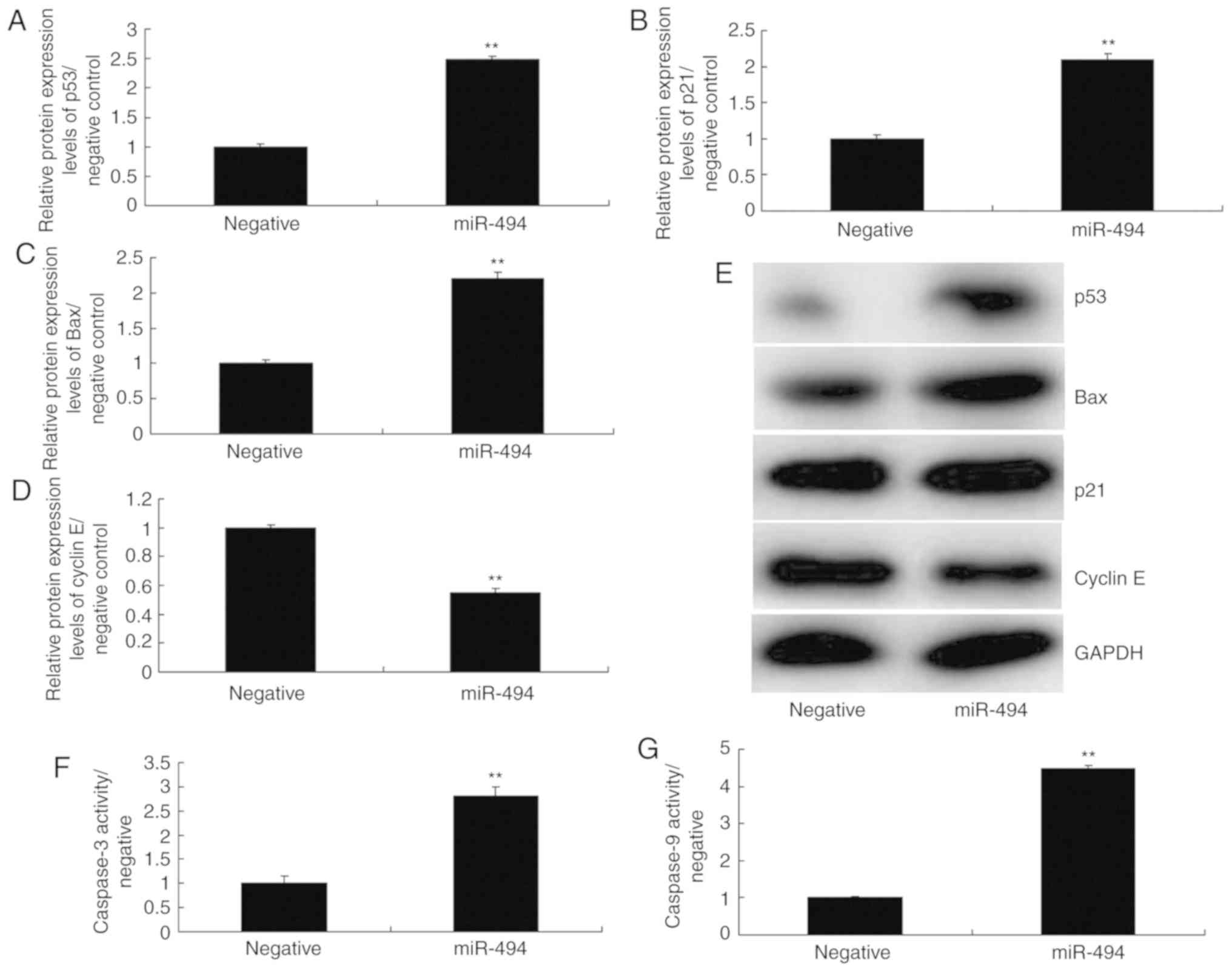
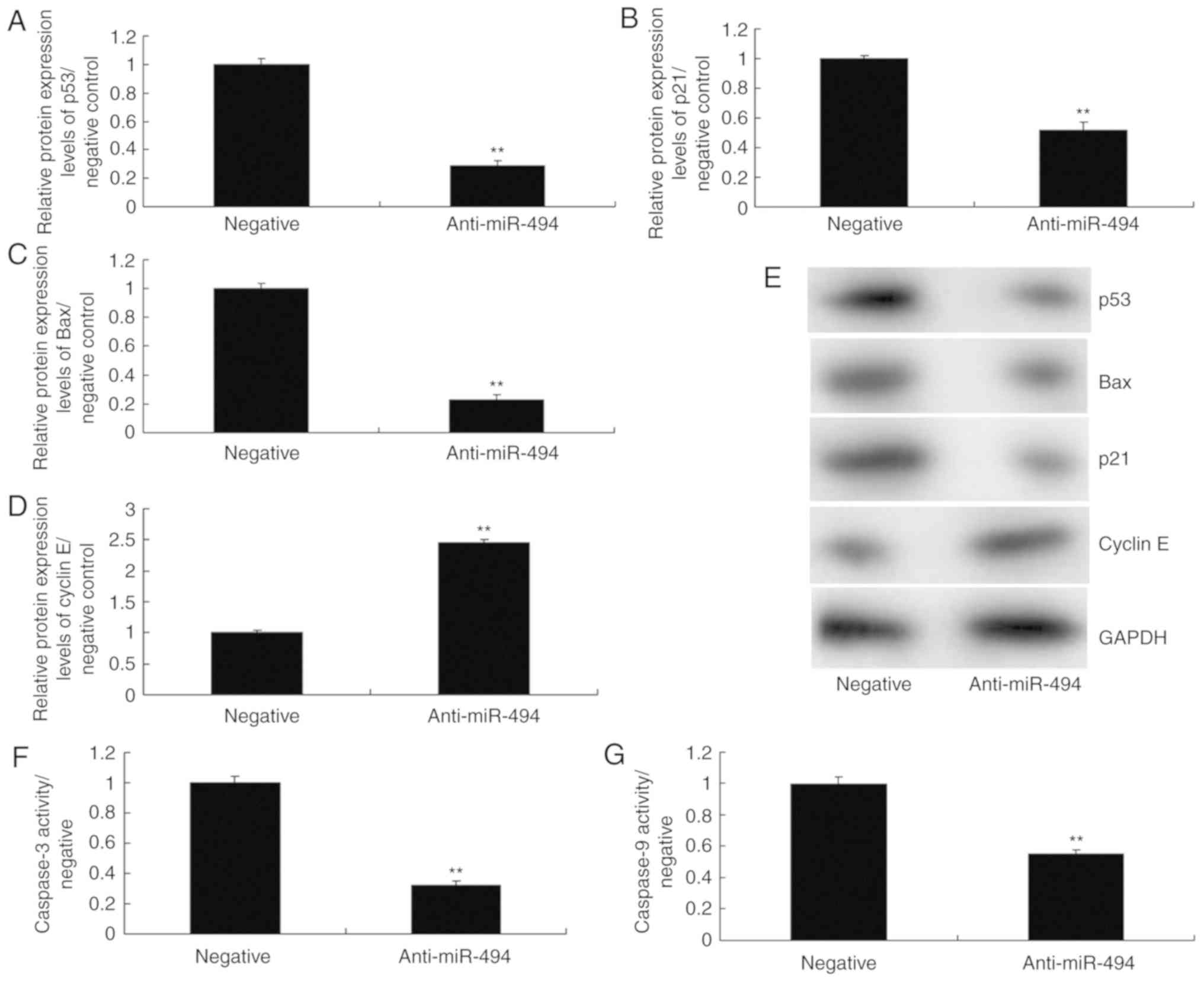
miR-494 regulates the p53 signaling
pathway in an in vitro model of SCI
To further explore the mechanism underlying the
effects of miR-494 on SCI, the p53 signaling pathway was analyzed
in the SCI model. As shown in Fig.
5, overexpression of miR-494 induced the protein expression of
p53, Bax, and p21, suppressed cyclin E protein expression, and
increased caspase-3/9 activity in the SCI model compared with in
the negative control group. Conversely, miR-494 knockdown
suppressed the protein expression of p53, Bax and p21, induced
cyclin E protein expression, and decreased caspase-3/9 activity in
the SCI model compared with in the negative control group (Fig. 6). Collectively, these results
indicated that miR-494 may mediate nerve cell apoptosis in SCI
through the p53 signaling pathway via SIRT1.
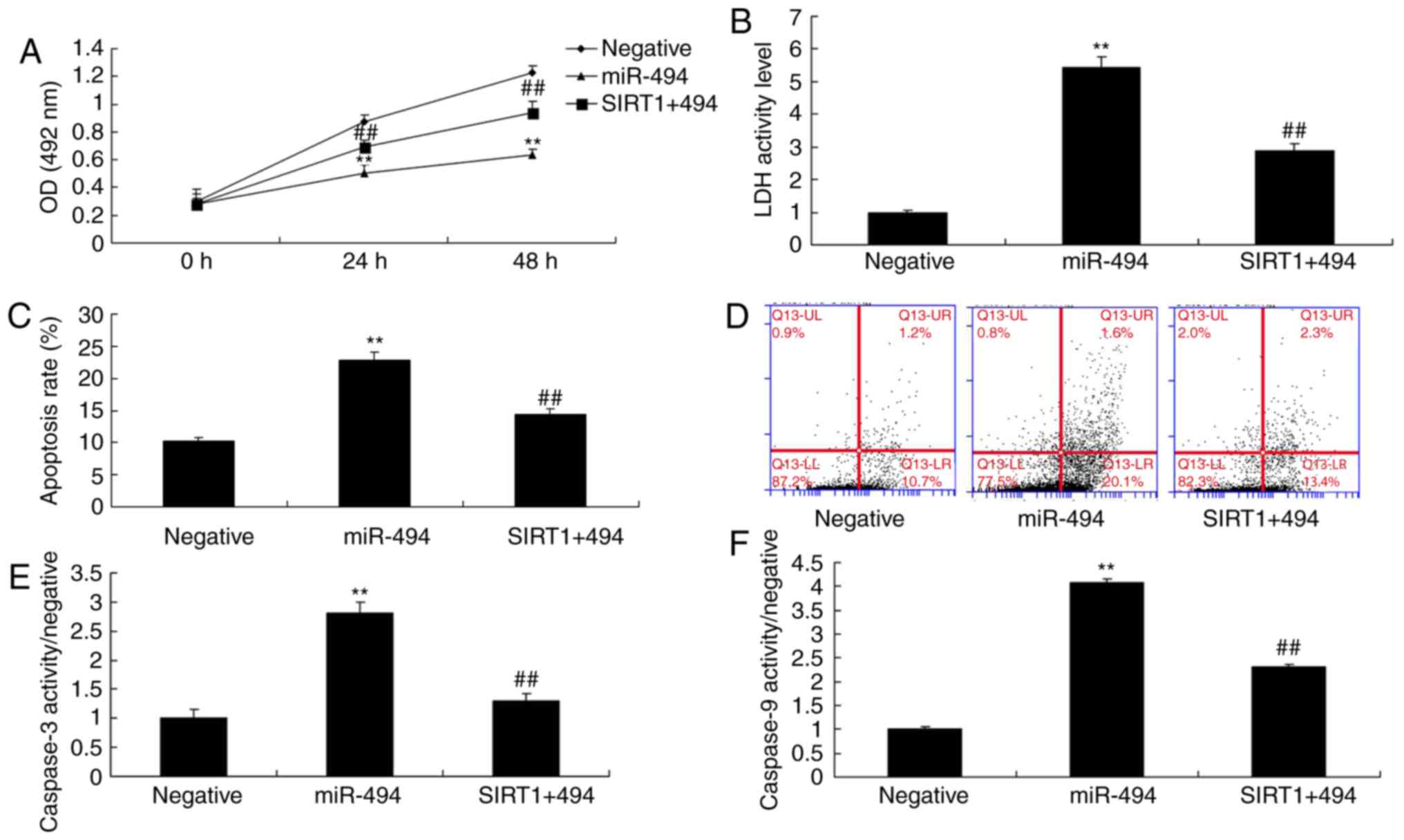
SIRT1 agonist attenuates the effects of
miR-494 on cell apoptosis in SCI
The present study explored the role of SIRT1 in the
effects of miR-494; the in vitro model of SCI was treated
with a SIRT1 agonist, CAY10602 (10 µM), following miR-494
transfection. As shown in Fig. 7,
the SIRT1 agonist increased the protein expression levels of SIRT1
and cyclin E, and suppressed p53, Bax and p21 protein expression in
the SIRT1 + miR-494 group compared with in the miR-494 group.
Treatment with the SIRT1 agonist also decreased cell apoptosis, LDH
activity and caspase-3/9 activity, and increased cell growth in the
SIRT1 + miR-494 group compared with in the miR-494 group (Fig. 8). Overall, these results indicated
that SIRT1 may be a direct target gene of miR-494, which regulates
nerve cell apoptosis in SCI.
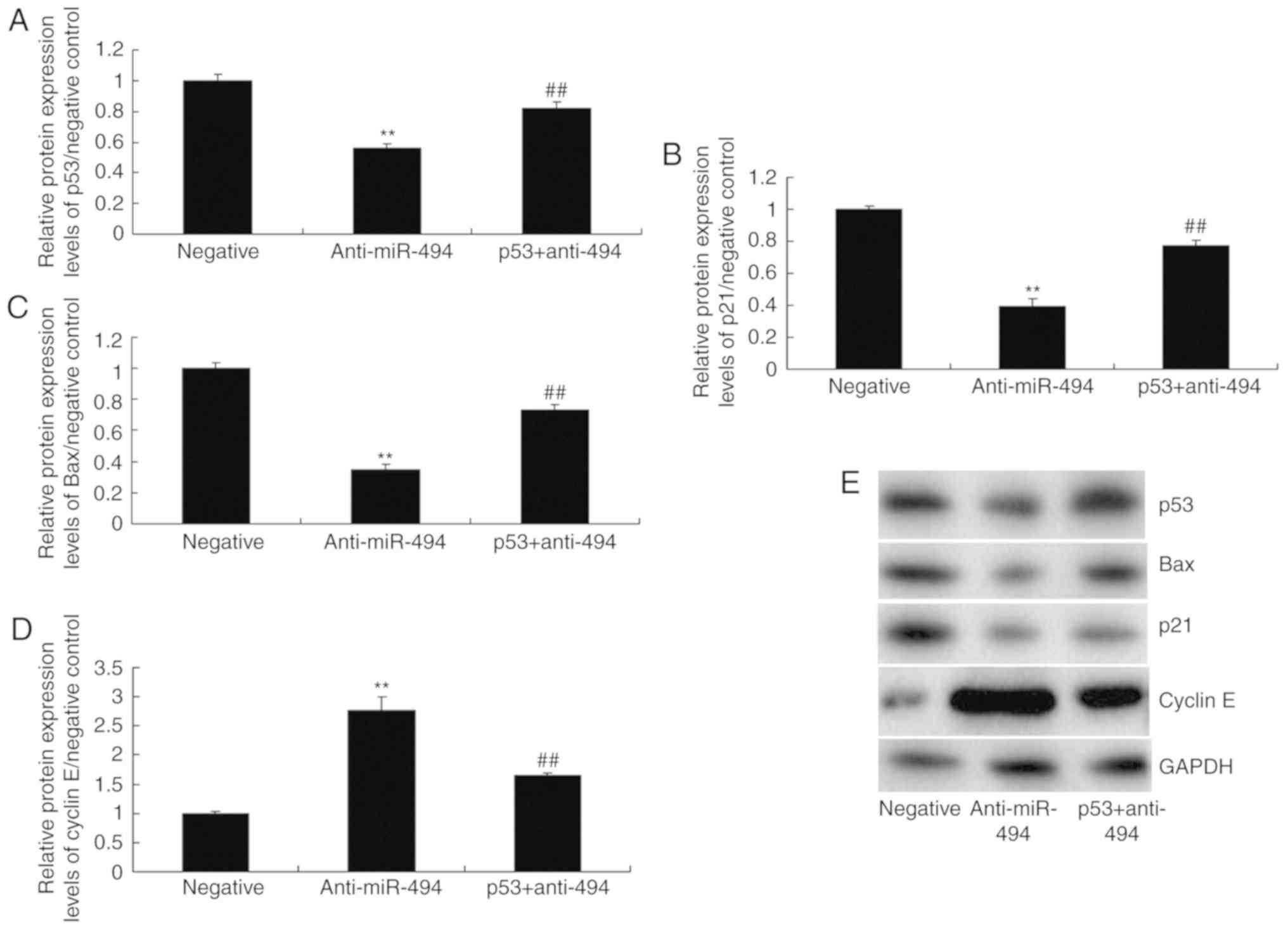
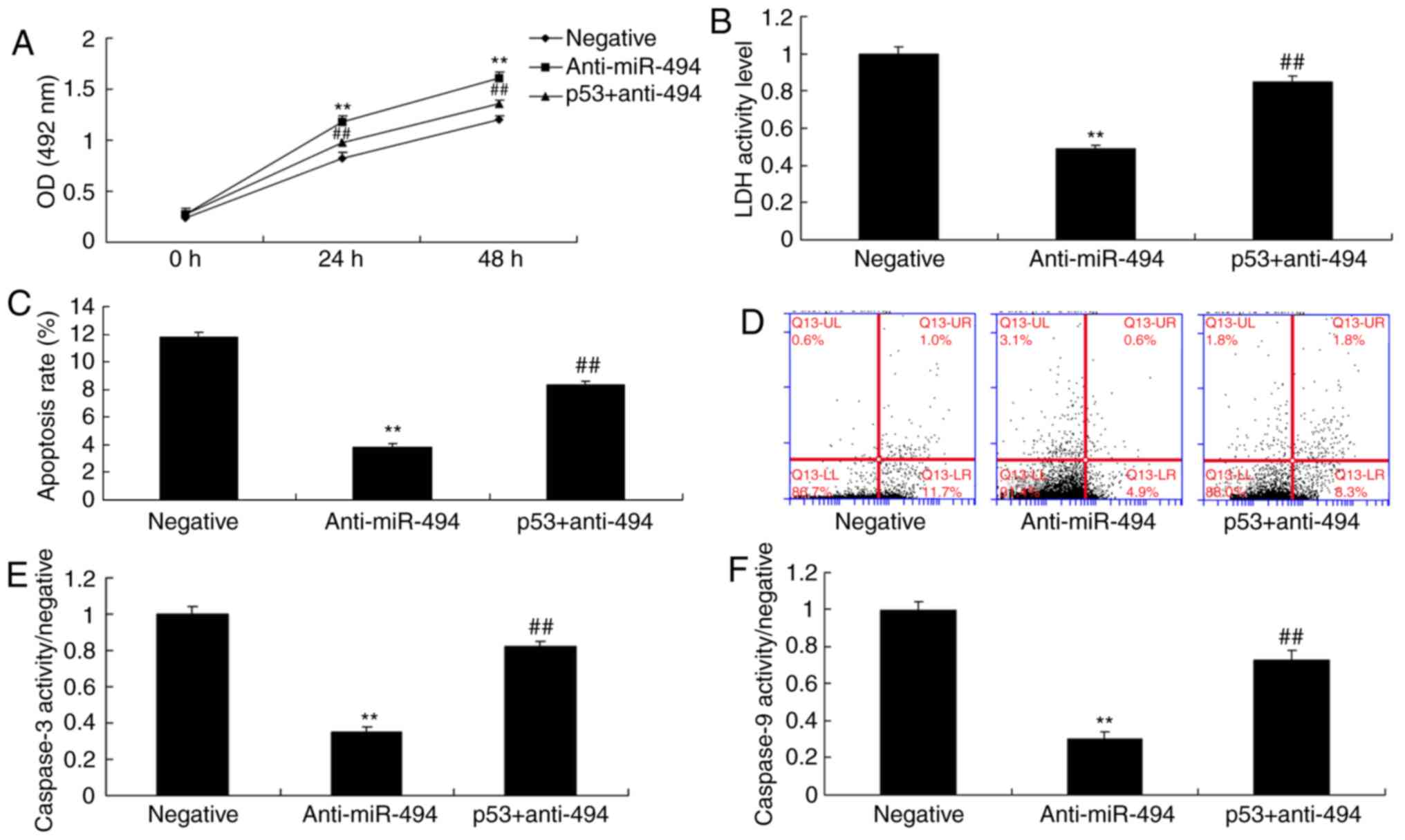
p53 agonist attenuates the effects of
anti-miR-494 on cell apoptosis in SCI
Finally, a p53 agonist, 20 nM Nutlin 3, was revealed
to increase the protein expression levels of p53, Bax and p21, and
suppress cyclin E protein expression in the p53 + anti-miR-494
group compared with in the anti-miR-494 group (Fig. 9). The p53 agonist also decreased
cell growth, and induced cell apoptosis, LDH activity and
caspase-3/9 activity in the p53 + anti-miR-494 group, compared with
in the anti-miR-494 group (Fig.
10). Taken together, these findings suggested that p53 may
promote nerve cell apoptosis in SCI via SIRT1/miR-494.
Discussion
SCI induces the cellular oxidative stress response,
necrosis and apoptosis. Furthermore, a large number of inflammatory
mediators are locally produced in response to SCI, thus resulting
in the immune inflammatory response (4). In such secondary injury, the
inflammatory response serves a crucial role in post-SCI recovery
(13). Control and inhibition of
the inflammatory response therefore affects post-SCI recovery.
miRNAs are small RNA molecules that regulate target gene expression
(14). In addition, miRNAs have a
vital role in gene expression during spinal development and SCI
(15); therefore, they may be
considered a novel target for therapeutic intervention in post-SCI
nerve regeneration and repair. Furthermore, miRNAs may be potential
biological markers of SCI. The present study demonstrated that
overexpression of miR-494 promoted cell apoptosis and LDH activity,
and inhibited cell growth in an SCI model compared with in the
negative control group. Kang et al (16) revealed that microRNA-494 promotes
apoptosis in degenerative human nucleus pulposus cells. Zhan et
al (17) suggested that
miR-494 inhibits breast cancer progression and induces apoptosis by
directly targeting p21 (RAC1)-activated kinase 1.
The SIRT1 anti-inflammatory mechanism is complex and
involves numerous pathways, including the mitogen-activated protein
kinase and NF-κB pathways (8).
Furthermore, it has been suggested that the cyclooxygenase pathway
may also be involved (8). At
present, the NF-κB pathway has been most extensively studied.
However, the inflammatory pathway is a multi-channel reticular
structure (18), as a result, it
is not determined by a certain pathway. The mechanism underlying
inflammation will become increasingly clear with research on each
pathway, and the reticular structure of the inflammatory pathway
will be elucidated (19);
therefore, it will be more easily controlled. SIRT1 can
downregulate the proapoptotic effects of p53 through its
deacetylation. At present, p53 is one of the most extensively and
profoundly studied tumor suppressor genes in human research
(20). Furthermore, p53
participates in cell differentiation, proliferation, aging and
mortality (20), and is involved
in gene transcription, DNA damage and repair, genomic stability and
cell cycle control (21). The
present study demonstrated that overexpression of miR-494
suppressed SIRT1 protein expression in an in vitro model
compared with in the control group. Liu et al (22) indicated that miR-494 inhibits the
proliferation, invasion and chemoresistance of pancreatic cancer
through SIRT1.
Neuronal apoptosis is suppressed following nervous
system injury (23). However,
several proteins and nuclear transcription factors are specifically
expressed in a delayed manner; p53 is one of the key molecules in
the cell apoptosis pathway. Upregulated p53 expression can directly
induce cell apoptosis (23). In
addition, it can induce cell apoptosis through regulating the
expression of other apoptosis-associated genes (19). The present study demonstrated that
overexpression of miR-494 induced p53, Bax and p21 protein
expression, suppressed cyclin E protein expression, and increased
caspase-3/9 activity in an SCI model. Comegna et al revealed
that direct targets of miR-494 are involved in senescence of human
diploid fibroblasts via p53 protein expression (24).
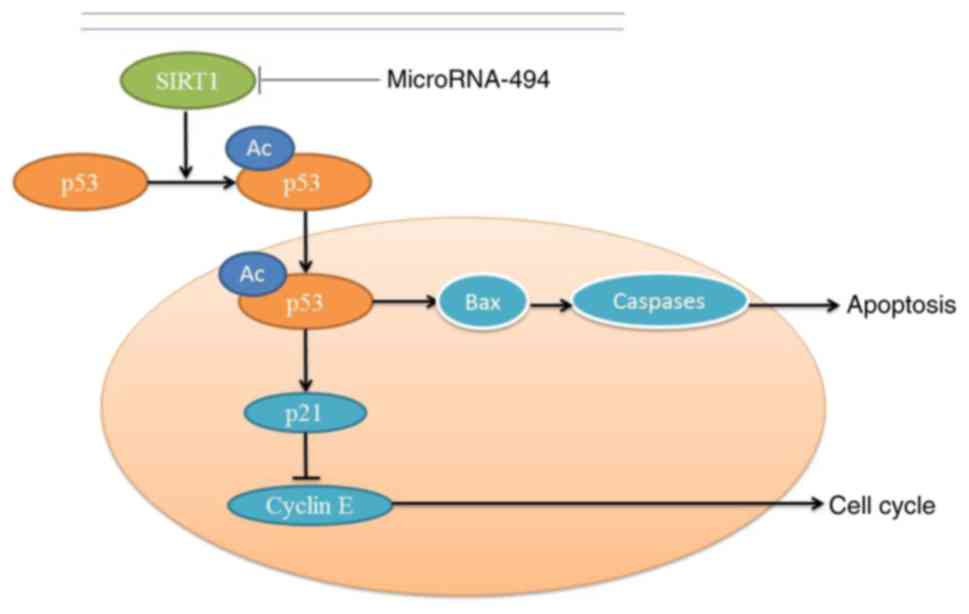
In conclusion, the present study indicated that
SIRT1 may inhibit apoptosis of SCI in vivo and in
vitro through the p53 signaling pathway, whereas miR-494 may
suppress SIRT1 and induce apoptosis (Fig. 11). Mechanistically, the present
study demonstrated that miR-494 promoted neurocyte apoptosis
through directly targeting the SIRT1/p53 signaling pathway. These
results suggested that the miR-494/SIRT1/p53 signaling pathway may
be a potential clinical biomarker and therapeutic target in
SCI.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
XY designed the experiment, analyzed the data and
wrote the manuscript. SZ, DZ, XZ, CX, TW, MZ, TL, WH and BW
performed the experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Scientific
Review Committee and the Institutional Review Board of Dalian
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ancha HR, Spungen AM, Bauman WA, Rosman
AS, Shaw S, Hunt KK, Post JB, Galea M and Korsten MA: Clinical
trial: The efficacy and safety of routine bowel cleansing agents
for elective colonoscopy in persons with spinal cord injury-a
randomized prospective single-blind study. Aliment Pharmacol Ther.
30:1110–1117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Apostolidis A, Thompson C, Yan X and
Mourad S: An exploratory, placebo-controlled, dose-response study
of the efficacy and safety of onabotulinumtoxinA in spinal cord
injury patients with urinary incontinence due to neurogenic
detrusor overactivity. World J Urol. 31:1469–1474. 2013. View Article : Google Scholar
|
|
3
|
Grossman RG, Fehlings MG, Frankowski RF,
Burau KD, Chow DS, Tator C, Teng A, Toups EG, Harrop JS, Aarabi B,
et al: A prospective, multicenter, phase I matched-comparison group
trial of safety, pharmacokinetics, and preliminary efficacy of
riluzole in patients with traumatic spinal cord injury. J
Neurotrauma. 31:239–255. 2014. View Article : Google Scholar :
|
|
4
|
Yang ML, Li JJ, So KF, Chen JY, Cheng WS,
Wu J, Wang ZM, Gao F and Young W: Efficacy and safety of lithium
carbonate treatment of chronic spinal cord injuries: A
double-blind, randomized, placebo-controlled clinical trial. Spinal
Cord. 50:141–146. 2012. View Article : Google Scholar
|
|
5
|
Celik EC, Erhan B, Gunduz B and Lakse E:
The effect of low-frequency TENS in the treatment of neuropathic
pain in patients with spinal cord injury. Spinal Cord. 51:334–337.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang Q, Dong S, Lei L, Liu J, Zhang J, Li
J, Duan J and Fan D: Protective effects of sparstolonin B, a
selective TLR2 and TLR4 antagonist, on mouse endotoxin shock.
Cytokine. 75:302–309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deng X, Zhang Y, Jiang F, Chen R, Peng P,
Wen B and Liang J: The chinese herb-derived sparstolonin B
suppresses HIV-1 transcription. Virol J. 12:1082015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Q, Li J, Liang Q, Wang D, Luo Y, Yu F,
Janicki JS and Fan D: Sparstolonin B suppresses rat vascular smooth
muscle cell proliferation, migration, inflammatory response and
lipid accumulation. Vascul Pharmacol. 67–69. 59–66. 2015.
|
|
9
|
Li XQ, Lv HW, Tan WF, Fang B, Wang H and
Ma H: Role of the TLR4 pathway in blood-spinal cord barrier
dysfunction during the bimodal stage after ischemia/reperfusion
injury in rats. J Neuroinflammation. 11:622014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wright KR, Mitchell B and Santanam N:
Redox regulation of microRNAs in endometriosis-associated pain.
Redox Biol. 12:956–966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
12
|
Li G, Chen T, Zhu Y, Xiao X, Bu J and
Huang Z: miR-103 alleviates autophagy and apoptosis by regulating
SOX2 in LPS-injured PC12 cells and SCI rats. Iran J Basic Med Sci.
21:292–300. 2018.PubMed/NCBI
|
|
13
|
Van Straaten MG, Cloud BA, Morrow MM,
Ludewig PM and Zhao KD: Effectiveness of home exercise on pain,
function, and strength of manual wheelchair users with spinal cord
injury: A high-dose shoulder program with telerehabilitation. Arch
Phys Med Rehabil. 95:1810–1817e1812. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Freria CM, Bernardes D, Almeida GL, Simoes
GF, Barbosa GO and Oliveira AL: Impairment of toll-like receptors 2
and 4 leads to compensatory mechanisms after sciatic nerve axotomy.
J Neuroinflammation. 13:1182016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lobenwein D, Tepekoylu C, Kozaryn R,
Pechriggl EJ, Bitsche M, Graber M, Fritsch H, Semsroth S, Stefanova
N, Paulus P, et al: Shock wave treatment protects from neuronal
degeneration via a Toll-Like receptor 3 dependent mechanism:
Implications of a first-ever causal treatment for ischemic spinal
cord injury. J Am Heart Assoc. 4:e0024402015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang L, Yang C, Song Y, Zhao K, Liu W, Hua
W, Wang K, Tu J, Li S, Yin H and Zhang Y: MicroRNA-494 promotes
apoptosis and extracellular matrix degradation in degenerative
human nucleus pulposus cells. On cotarget. 8:27868–27881. 2017.
|
|
17
|
Zhan MN, Yu XT, Tang J, Zhou CX, Wang CL,
Yin QQ, Gong XF, He M, He JR, Chen GQ and Zhao Q: MicroRNA-494
inhibits breast cancer progression by directly targeting PAK1. Cell
Death Dis. 8:e25292017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ak H, Gulsen I, Karaaslan T, Alaca İ,
Candan A, Koçak H, Atalay T, Çelikbilek A, Demir İ and Yılmaz T:
The effects of caffeic acid phenethyl ester on inflammatory
cytokines after acute spinal cord injury. Ulus Travma Acil Cerrahi
Derg. 21:96–101. 2015.PubMed/NCBI
|
|
19
|
Kim DI, Lee H, Lee BS, Kim J and Jeon JY:
Effects of a 6-week indoor hand-bike exercise program on health and
fitness levels in people with spinal cord injury: A randomized
controlled trial study. Arch Phys Med Rehabil. 96:2033–2040.e2031.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wadsworth BM, Haines TP, Cornwell PL,
Rodwell LT and Paratz JD: Abdominal binder improves lung volumes
and voice in people with tetraplegic spinal cord injury. Arch Phys
Med Rehabil. 93:2189–2197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Darouiche RO, Al Mohajer M, Siddiq DM and
Minard CG: Short versus long course of antibiotics for
catheter-associated urinary tract infections in patients with
spinal cord injury: A randomized controlled noninferiority trial.
Arch Phys Med Rehabil. 95:290–296. 2014. View Article : Google Scholar
|
|
22
|
Liu Y, Li X, Zhu S, Zhang JG, Yang M, Qin
Q, Deng SC, Wang B, Tian K, Liu L, et al: Ectopic expression of
miR-494 inhibited the proliferation, invasion and chemoresistance
of pancreatic cancer by regulating SIRT1 and c-Myc. Gene Ther.
22:729–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim Y, Jo SH, Kim WH and Kweon OK:
Antioxidant and anti-inflammatory effects of intravenously injected
adipose derived mesenchymal stem cells in dogs with acute spinal
cord injury. Stem Cell Res Ther. 6:2292015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Comegna M, Succoio M, Napolitano M, Vitale
M, D’Ambrosio C, Scaloni A, Passaro F, Zambrano N, Cimino F and
Faraonio R: Identification of miR-494 direct targets involved in
senescence of human diploid fibroblasts. FASEB J. 28:3720–3733.
2014. View Article : Google Scholar : PubMed/NCBI
|