Introduction
Malignant pleural mesothelioma (MPM) is an
aggressive cancer affecting the pleura. Despite recent advances in
chemotherapy, the median survival remains at approximately 12
months and the exploration of novel targets for therapeutic
intervention is required. Cyclooxygenase-2 (COX-2) is an inducible
enzyme which catalyses the conversion of arachidonic acid to
prostaglandins in response to proinflammatory or mitogenic signals.
It is overexpressed in many solid tumours and is a potential target
for therapeutic intervention (1–4).
Inhibition of COX-2 has been shown to have a significant
anti-neoplastic effect by reducing the production of prostaglandins
(4,5). Our previous work demonstrated that
COX-2 is overexpressed in 59% (51/86) of archival malignant pleural
mesothelioma tissue samples, a finding supported by similar studies
(6–10). The cytotoxic effect of COX-2
inhibitors has been demonstrated in mesothelioma cell lines
(8,11) and recently we reported that
specific COX-2 inhibitors, including DuP 697, induce
anti-proliferative effects in mesothelioma cell lines (12). Several COX-2 inhibitors which are
currently used in clinical practice, including celecoxib and
rofecoxib, are derived from DuP 697 (13).
The chemotherapy options for patients with MPM are
limited and improvements in survival are required. Pemetrexed, in
combination with cisplatin, has been approved for first line
chemotherapy in patients with MPM (14,15).
In mesothelioma cells, we have demonstrated that the cytotoxic
effect of pemetrexed chemotherapy can be enhanced by the addition
of DuP 697 (12). This compound is
therefore worthy of further clinical investigation, however, the
molecular mechanism of action of DuP 697 has not been widely
studied. In normal proliferating human umbilical vein endothelial
cells (HUVECs) expressing low levels of COX-2, DuP 697 was shown to
induce apoptosis and this was associated with the upregulation of
caspases 3, 8 and 9 (16). In the
K562 chronic myeloid leukaemia cell line, DuP 697 induced G1-S cell
cycle arrest and apoptosis with upregulation of caspase 8 (17). These hypothesis-driven studies
suggest that the mechanism of cytotoxic action of DuP 697 may be
via induction of apoptosis. We aimed to explore, using a novel
proteomic platform, the molecular mechanism of action of this
compound using cell lines derived from solid tumours.
Materials and methods
Cell line treatments
DuP 697 was previously demonstrated to have a
cytotoxic effect in the COX-2 positive mesothelioma cell line
MSTO-211H and in the lung cancer cell line A549, which was
originally selected as a COX-2 positive cell line (12). In order to induce a visible
cytotoxic effect in DuP 697 treated cells (50% reduction in cell
numbers compared to control cells treated with drug carrier only),
MSTO-211H and A549 cells were treated with 31.7 μM and 50
μM DuP 697 (#1430, Tocris Bioscience) respectively, for 72
h. Drug carrier (dimethyl sulfoxide; DMSO) only was added to
control cells. At the end of 72 h total protein lysates were
generated from DuP 697 treated and control cells, using both
antibody microarray buffer and immunoblotting buffer to yield at
least 1 mg of protein with a concentration of 1 mg/ml.
Antibody microarray analysis
The Panorama Xpress Profiler725 antibody microarray
kit (#XP725, Sigma Aldrich), which consists of 725 antibodies
spotted in duplicate on a nitro-cellulose-coated glass microscope
slide, was used for proteomic analysis as previously described
(18). Total protein lysates from
control (drug carrier only) samples were fluorescently labelled
with Cy3 (#PA23001, GE Healthcare) and lysates from DuP 697 treated
samples were labelled with Cy5 (#PA25001, GE Healthcare).
Dye-to-protein molar ratios of at least 2 were achieved prior to
protein binding. Equal amounts of protein from each sample were
incubated with the microarray slide for 45 min on an orbital
shaker. Normalisation and data analysis were performed as
previously described and in all experiments the ‘substances
matched’ value achieved was at least 90% (18). Differentially expressed proteins
(DEPs) were considered significant with a fold change ≥1.8, whilst
fold changes ≥1.5 were also recorded for each experiment for use as
supporting data (18).
Ingenuity Pathway Analysis
Gene identifiers which corresponded to the DEPs were
identified from the Ingenuity® Knowledge Base and the
dataset was analysed through the use of Ingenuity Pathway Analysis
(IPA; Ingenuity® Systems, www.ingenuity.com). The dataset containing gene
identifiers of the DEPs was uploaded into the application and each
identifier was mapped to its corresponding object in the
Ingenuity® Knowledge Base. Canonical Pathways Analysis
was used to identify pathways from the IPA library that were most
significant to the dataset.
Semi-quantitative immunoblotting
Proteins were extracted in Laemmli buffer [62.5 mM
Tris-HCl (pH 6.8), 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 1%
protease inhibitor mix and 0.00125% bromophenol blue] and 20
μg was electrophoresed on a 12% Precise gel (#25222, Pierce)
at a constant voltage of 140 V for 40 min. Proteins were
transferred using the iBlot dry transfer system (#IB3010-01,
Invitrogen) onto nitrocellulose membrane. The membrane was blocked
in 5% non-fat dry milk dissolved in Tris-buffered saline containing
0.05% Tween-20. A primary antibody against BCL2L1 (Bcl-xL; #B9429,
Sigma Aldrich) was applied at 1:5000 for 2 h. A primary antibody
against BID (#ab32060, Abcam) was applied at 1:300 for 16 h. As
loading control, a primary antibody against alpha tubulin (#ab7291,
Abcam) was applied at 1:2500 for 2 h. The relevant secondary
antibody (#SC-2030 or #SC-2031, Santa Cruz Biotechnology) was
applied at 1:1000 for 1 h and bands were detected using the
Supersignal West Pico Chemiluminscent Substrate Kit (#34078,
Pierce). Films were scanned using a GS800 calibrated densitometer
(Bio-Rad) with Quantity One software (Bio-Rad). Following data
normalisation against the loading control, differential expression
between samples was calculated.
Results
Antibody microarray analysis identified 32 unique
proteins which demonstrated ≥1.8-fold difference in expression in
at least one cell line, when comparing DuP 697 treated versus
control (drug carrier only) cells (Table I). Of these, 20 DEPs demonstrated
≥1.8-fold difference in 2/2 cell lines. The dataset of 32 DEPs was
submitted to IPA and the top relevant canonical pathway was
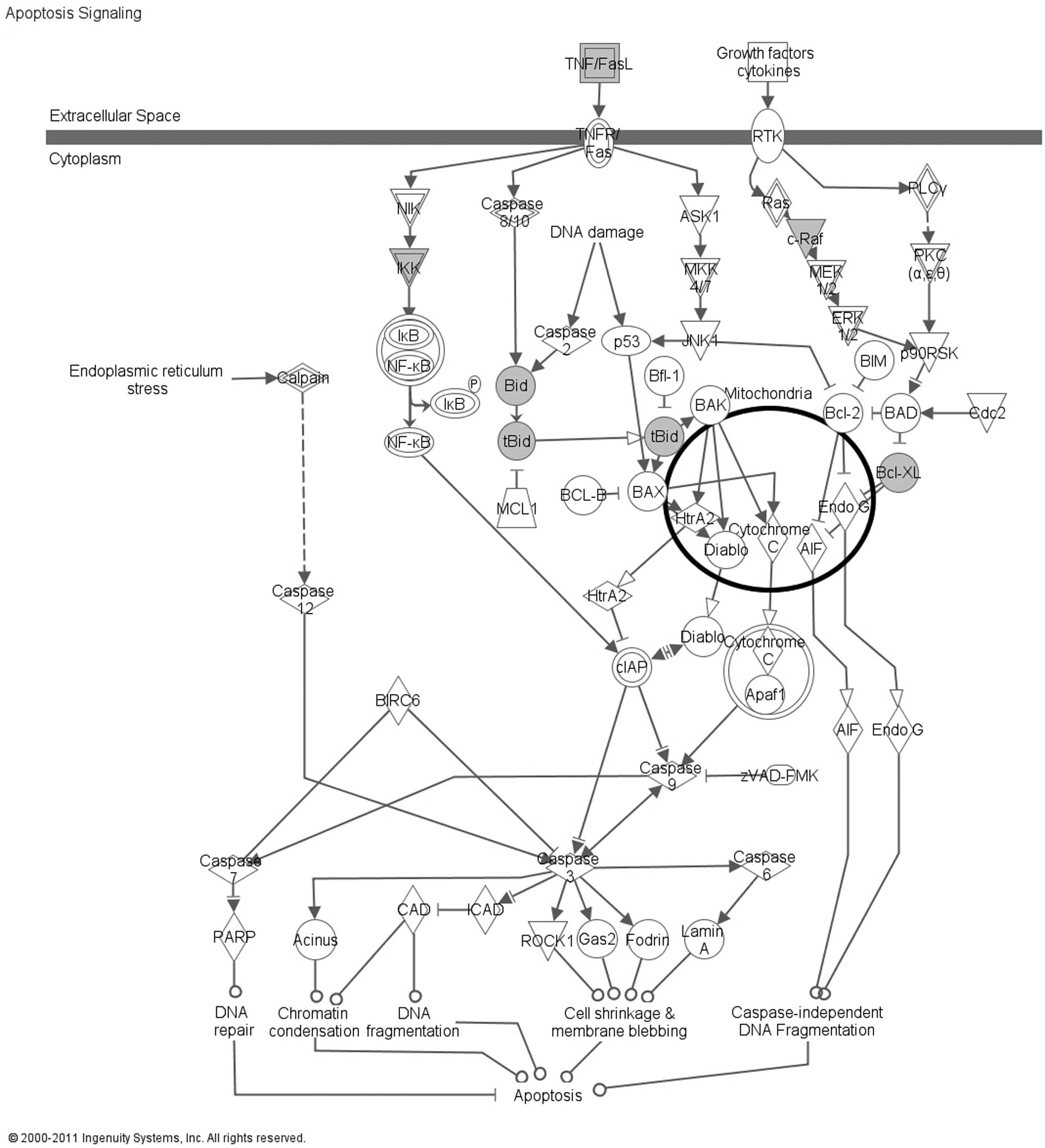
‘Apoptosis Signaling’, which involved 5 DEPs: BCL2L1 (Bcl-xL), BID,
CHUK (IKK), FASLG and RAF1 (Fig.
1). The BCL2L1 (Bcl-xL) and BID proteins were selected for
further analysis using immunoblotting. The anti-apoptotic BCL2L1
(Bcl-xL) protein was down-regulated by 2.48-fold in the MSTO-211H
cell line when treated with DuP 697 (Fig. 2). The anti-tBID antibody (B3183),
which was present on the antibody microarray, proved to be
unreliable in the immunoblotting application. However, full length
BID was found to be down-regulated in both the MSTO-211H and A549
cell lines by a fold change of 10.16 and 14.52 respectively,
following treatment with DuP 697 (Fig.
3).
 | Table IA total of 32 unique DEPs identified
using antibody microarray analysis following DuP 697 treatment of
MSTO-211H mesothelioma cells and A549 lung cancer cells.a |
Table I
A total of 32 unique DEPs identified
using antibody microarray analysis following DuP 697 treatment of
MSTO-211H mesothelioma cells and A549 lung cancer cells.a
| Ab # (Sigma
Aldrich) | Protein target | Gene identifier | A549 | MSTO-211H |
|---|
| P0084 | Pinin | PNN | 7.67 | 7.32 |
| Z0377 | Zxyin | ZYX | 4.39 | 4.74 |
| C1862 | Coilin | COIL | 4.6 | 3.46 |
| A5968 | AP-1 | JUN | 2.49 | 2.97 |
| B3183 | tBID | BID | 2.42 | 2.54 |
| C7736 | Centrin | CETN1 | 2.51 | 2.04 |
| S5446 | SUMO-1 | SUMO1 | 2.44 | 2.09 |
| C6219 | Connexin-43 | GJA1 | 2.44 | 2.01 |
| M0445 | MDMX | MDM4 | 2.13 | 2.39 |
| A0844 | AP-2a | TFAP2A | 2.37 | 2.28 |
| A7107 | AP2 | TFAP2A | 2.06 | 2.04 |
| I6139 | IKKa | CHUK | 2.37 | 1.99 |
| E8526 | E2F4 | E2F4 | 2.37 | 1.96 |
| S1190 | SLIPR/MAGI-3 | MAGI3 | 2.15 | 1.92 |
| B9429 | Bcl-xL | BCL2L1 | 2.13 | 2.24 |
| S9809 | Sp1 | SP1 | 2.13 | 1.95 |
| F3648 | Fibronectin | FN1 | 1.99 | 2.12 |
| F2051 | Fas ligand | FASLG | 1.81 | 2.14 |
| V7881 | Vitronectin | VTN | 1.99 | 1.82 |
| H9912 | Hsnf5/INI1 | SMARCB1 | 1.96 | 1.84 |
| A7833 | ATF-1 | ATF1 | 1.83 | 1.84 |
| R8274 | RIP receptor
interacting protein | RIPK1 | 2.11 | 1.74 |
| T5942 | 14-3-3
theta/tau | YWHAQ | 1.64 | 2.22 |
| T1075 | Tal | LRSAM1 | 2.05 | 1.72 |
| C3470 | Connexin-32 | GJB1 | 2.04 | 1.76 |
| R1151 | c-Raf pSer621 | RAF1 | 1.78 | 1.99 |
| R4904 | Reelin | RELN | 1.95 | 1.67 |
| S3934 | Smad4 (DPC4) | SMAD4 | 1.87 | 1.61 |
| R3529 | Rnase L | RNASEL | 1.59 | 1.82 |
| A5044 | Alpha actinin | ACTN1 | 1.82 | 1.66 |
| E8767 | c-erbB-3 | ERBB3 | 1.82 | - |
| C3956 | c-Myc | MYC | 1.81 | - |
| L1538 | LIN-7 | LIN7A | 1.8 | 1.57 |
Discussion
We have previously confirmed that COX-2 is
overexpressed in MPM samples which suggests that novel anticancer
therapies targeted at this pathway may be useful in mesothelioma
patients (10). In addition, we
have demonstrated that the COX-2 inhibitor DuP 697 enhanced the
cytotoxic effect of pemetrexed in mesothelioma cell lines,
including MSTO-211H (12). It is
important to understand the molecular mechanism of action of novel
agents before possible clinical testing and DuP 697 has not been
widely researched. In the present study we have explored the
molecular mechanism of action of DuP 697 using an antibody
microarray proteomic platform. We have identified 32 unique DEPs
which were associated with DuP 697 treatment for 72 h. Of these, 20
proteins demonstrated significant (≥1.8-fold) differential
expression in both the MSTO-211H mesothelioma and A549 lung cancer
cell lines. Using some of the data from these, and other,
experiments we have recently described Zyxin as the commonest
repeatedly identified DEP (RIDEP) when using this proteomic
platform (18) and therefore the
selection of proteins for further analysis must be carefully
considered. The analysis of the 32 DEPs using IPA indicated that 5
proteins, BCL2L1 (Bcl-xL), BID, CHUK (IKK), FASLG and RAF1, were
associated with the Apoptosis Signaling canonical pathway.
Following a positive signal for apoptosis, activated caspase 8
cleaves inactive, cytosolic, full length BID into active truncated
BID (tBID), which localises to the mitochondrial membrane (19–21).
The anti-apoptotic proteins BCL-2 and BCL2L1 (Bcl-xL) block the
escape of cytochrome C from the mitochondria, by preventing Bax
from forming channels in the mitochondrial membrane, until
activated tBID is localised to the membrane (19–21).
The onset of apoptosis may be associated with decreased levels of
full length BID, due to its cleavage into tBID, and decreased
levels of the anti-apoptotic protein BCL2L1 (Bcl-xL). Our
immunoblotting data would support these suggested protein changes
following administration of DuP 697 for 72 h.
The caspase pathway of apoptosis has previously been
implicated as the in vitro mechanism of action for DuP 697,
with upregulation of caspases 3, 8 and 9 being observed in
hypothesis-driven experiments in normal proliferating endothelial
cells or leukaemia cells (16,17).
At the 72-h time-point, which we examined here, we did not identify
differential expression of caspases 3, 4, 5, 6, 7, 8, 9, 10, 11, 12
or 13 or pro-caspase 8 in either MSTO-211H or A549 cells. However,
this may be due to the return of these proteins to basal levels
within 72 h since the upregulation of caspases 3, 8 and 9 was noted
within 8 h in HUVECs (16).
COX-2 is a key enzyme involved in the metabolism of
arachidonic acid resulting in the production of prostaglandins,
particularly PGE2, which plays an important role in tumour
progression. COX-2 inhibitors may act by inhibition of COX-2, but
the exact mechanism of how COX-2 inhibitors exert an
anti-neoplastic effect is currently unknown. Indeed, several
studies have suggested that COX-2 inhibitors may act independently
of COX-2 (22–25). In our antibody microarray
experiments, differential expression of COX-2 was not observed in
either cell line after treatment with DuP 697 for the duration
selected (72 h). In future work, the expression of COX-2 and the
individual proteins within the apoptosis signalling pathway, which
we have implicated here, could now be examined over a time-course
of treatment with DuP 697.
We have demonstrated that the antibody microarray
proteomic platform can be used to explore the molecular mechanism
of a COX-2 inhibitor. This will prove useful in gaining a more
thorough understanding of novel agents which may have clinical
applications. Specific COX-2 inhibitors, such as DuP 697, may have
a future therapeutic role in MPM. Our proteomic analysis suggests
that the anti-proliferative effect of DuP 697, which was previously
seen in mesothelioma cell lines, may be exerted via the induction
of apoptosis. DuP 697, or other COX-2 inhibitors such as celecoxib
or rofecoxib, may act as an effective apoptosis sensitiser when
combined with chemotherapy drugs such as Pemetrexed and further
studies are required to test this hypothesis.
Abbreviations:
|
COX-2
|
cyclooxygenase-2
|
|
DEP
|
differentially expressed protein
|
|
DMSO
|
dimethyl sulfoxide
|
|
HUVECs
|
human umbilical vein endothelial
cells
|
|
IPA
|
Ingenuity Pathway Analysis
|
|
MPM
|
malignant pleural mesothelioma
|
|
tBID
|
truncated BID
|
Acknowledgements
This study was kindly supported by
Yorkshire Cancer Research. The sponsor had no role in study design;
collection, interpretation or analysis of data; writing of the
manuscript; or decision to submit the article for publication.
References
|
1
|
Hull MA: Cyclooxygenase-2: how good is it
as a target for cancer chemoprevention? Eur J Cancer. 41:1854–1863.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gasparini G, Longo R, Sarmiento R and
Morabito A: Inhibitors of cyclo-oxygenase 2: a new class of
anticancer agents? Lancet Oncol. 4:605–615. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rao P and Knaus E: Evolution of
nonsteroidal antiinflammatory drugs (NSAIDS): cyclooxygenase (COX)
inhibition and beyond. J Pharm Pharm Sci. 11:81S–110S.
2008.PubMed/NCBI
|
|
4
|
Ghosh N, Chaki R, Mandal V and Mandal SC:
COX-2 as a target for cancer chemotherapy. Pharmacol Rep.
62:233–244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Menter DG, Schilsky RL and DuBois RN:
Cyclooxygenase-2 and cancer treatment: understanding the risk
should be worth the reward. Clin Cancer Res. 16:1384–1390. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baldi A, Santini D, Vasaturo F, et al:
Prognostic significance of cyclooxygenase-2 (COX-2) and expression
of cell cycle inhibitors p21 and p27 in human pleural malignant
mesothelioma. Thorax. 59:428–433. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Edwards JG, Faux SP, Plummer SM, et al:
Cyclooxygenase-2 expression is a novel prognostic factor in
malignant mesothelioma. Clin Cancer Res. 8:1857–1862.
2002.PubMed/NCBI
|
|
8
|
Marrogi A, Pass HI, Khan M, Metheny-Barlow
LJ, Harris CC and Gerwin BI: Human mesothelioma samples overexpress
both cyclooxygenase-2 (COX-2) and inducible nitric oxide synthase
(NOS2): in vitro antiproliferative effects of a COX-2 inhibitor.
Cancer Res. 60:3696–3700. 2000.
|
|
9
|
Cardillo I, Spugnini EP, Verdina A, Galati
R, Citro G and Baldi A: Cox and mesothelioma: an overview. Histol
Histopathol. 20:1267–1274. 2005.PubMed/NCBI
|
|
10
|
O’Kane SL, Cawkwell L, Campbell A and Lind
MJ: Cyclooxygenase-2 expression predicts survival in malignant
pleural mesothelioma. Eur J Cancer. 41:1645–1648. 2005.PubMed/NCBI
|
|
11
|
Catalano A, Graciotti L, Rinaldi L, et al:
Preclinical evaluation of the nonsteroidal anti-inflammatory agent
celecoxib on malignant mesothelioma chemoprevention. Int J Cancer.
109:322–328. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O’Kane SL, Eagle GL, Greenman J, Lind MJ
and Cawkwell L: COX-2 specific inhibitors enhance the cytotoxic
effects of pemetrexed in mesothelioma cell lines. Lung Cancer.
67:160–165. 2010.PubMed/NCBI
|
|
13
|
Blobaum AL and Marnett LJ: Structural and
functional basis of cyclooxygenase inhibition. J Med Chem.
50:1425–1441. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scagliotti GV, Parikh P, von Pawel J, et
al: Phase III study comparing cisplatin plus gemcitabine with
cisplatin plus pemetrexed in chemotherapy-naive patients with
advanced-stage non-small-cell lung cancer. J Clin Oncol.
26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vogelzang NJ, Rusthoven JJ, Symanowski J,
et al: Phase III study of pemetrexed in combination with cisplatin
versus cisplatin alone in patients with malignant pleural
mesothelioma. J Clin Oncol. 21:2636–2644. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Churchman A, Baydoun AR and Hoffman R:
Inhibition of angiogenic tubule formation and induction of
apoptosis in human endothelial cells by the selective
cyclooxygenase-2 inhibitor
5-bromo-2-(4-fluorophenyl)-3-(methylsulfonyl) thiophene (DuP-697).
Eur J Pharmacol. 573:176–183. 2007. View Article : Google Scholar
|
|
17
|
Peng HL, Zhang GS, Liu JH, Gong FJ and Li
RJ: Dup-697, a specific COX-2 inhibitor, suppresses growth and
induces apoptosis on K562 leukemia cells by cell-cycle arrest and
caspase-8 activation. Ann Hematol. 87:121–129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hodgkinson VC, Elfadl D, Drew PJ, Lind MJ
and Cawkwell L: Repeatedly identified differentially expressed
proteins (RIDEPs) from antibody microarray proteomic analysis. J
Proteomics. 74:698–703. 2011. View Article : Google Scholar
|
|
19
|
Danial NN: BCL-2 family proteins: critical
checkpoints of apoptotic cell death. Clin Cancer Res. 13:7254–7263.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song G, Chen G, Hu T and Lai PB: Bid
stands at the crossroad of stress-response pathways. Curr Cancer
Drug Targets. 10:584–592. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Strasser A, Cory S and Adams JM:
Deciphering the rules of programmed cell death to improve therapy
of cancer and other diseases. EMBO J. 30:3667–3683. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kern MA, Haugg AM, Koch AF, et al:
Cyclooxygenase-2 inhibition induces apoptosis signaling via death
receptors and mitochondria in hepatocellular carcinoma. Cancer Res.
66:7059–7066. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lou J, Fatima N, Xiao Z, et al: Proteomic
profiling identifies cyclooxygenase-2-independent global proteomic
changes by celecoxib in colorectal cancer cells. Cancer Epidemiol
Biomarkers Prev. 15:1598–1606. 2006. View Article : Google Scholar
|
|
24
|
Pang RP, Zhou JG, Zeng ZR, et al:
Celecoxib induces apoptosis in COX-2 deficient human gastric cancer
cells through Akt/GSK3beta/NAG-1 pathway. Cancer Lett. 251:268–277.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schonthal AH: Direct non-cyclooxygenase-2
targets of celecoxib and their potential relevance for cancer
therapy. Br J Cancer. 97:1465–1468. 2007. View Article : Google Scholar : PubMed/NCBI
|