Introduction
The energy demands on rapidly proliferating cancer
cells necessitate major metabolic adjustments that either precede
malignant transformation, pushing cells toward carcinogenesis, or
come about as adaptations to the stressful environmental conditions
to which tumor cells are subjected (1). One of the most common metabolic
alterations is the acquired ability of a cell to use glycolysis as
the primary ATP source even in the presence of oxygen, known as the
Warburg effect (2). The Warburg
effect has been demonstrated in multiple tumor types (3) and is generally accepted to be
necessary for the growth and survival of tumor cells and, hence,
glycolytic metabolites and glycolysis regulating enzymes could be
used as metabolic tumor markers (3). In fact, studies of colon tumor
metabolites and key glycolytic enzymes demonstrated the power of
these markers in discriminating tumor stage (4), location (5), malignancy and metastatic ability
(6).
The nucleoside triphosphate diphosphohydrolases
(NTPDases), also referred to as apyrases, make up a family of
enzymes responsible for intracellular and extracellular nucleotide
metabolism. This well-conserved family of nucleoside phosphate
cleaving enzymes has eight known members, which have been shown to
impact biological processes such as apoptosis, adhesion, and
differentiation (7). Knockout
studies have further indicated a role for NTPDases in tissue
development, cell proliferation, inflammatory response and
metabolism (8,9). NTPDase 5 is distinctive from the
other NTPDases as it is the only member described as a
protooncoprotein, better recognized as such by the name PCPH. The
proto-oncogene and oncogene were discovered when chemical
carcinogen treatment of Syrian hamster embryo fibroblasts activated
the ENTPD5/PCPH gene into its oncogene counterpart
(mt-PCPH) (10). Comparison
of the transforming and normal sequences revealed a single base
pair deletion in the open-reading frame (ORF) of the oncogene. As a
consequence of this mutation, the ORF was shifted resulting in a
truncated gene product relative to the normal protein (11). Although the ENTPD5/PCPH protein has
been reported to promote N-glycosylation, cell proliferation and
the Warburg effect (12), the
biological processes in which the PCPH and mt-PCPH proteins
participate and their molecular functions in mammalian cells remain
unclear.
The transforming activities of the overexpressed
normal PCPH and of the mt-PCPH oncoprotein were demonstrated in
various cell systems (11,13–15)
and, accordingly, their expression has been detected in multiple
cancers (16–18). Generally, expression resulted in
phenotypic changes shared by PCPH and mt-PCPH yet the effects were
consistently and substantially greater in cells expressing mt-PCPH
compared to PCPH. Two phenotypes normally associated with PCPH and
mt-PCPH expression are depleted cellular ATP levels (13, and
unpublished data) and increased resistance to stress (11,13–15).
It was demonstrated that reduced ATP levels play a part in the
concurrent chemo-resistance observed in mt-PCPH expressing cells.
ATP repletion experiments in BALB/3T3 cells expressing mt-PCPH
restored their sensitivity to cisplatin treatment (14). However, the mechanism by which
these proteins decrease cellular ATP levels remains unclear.
It was suggested that the intrinsic NTPDase activity
of PCPH (19) and mt-PCPH
(20) may be responsible for the
cleavage of ATP and hence for the concomitant stress-resistance
(15). Given that ATP is not a
favored substrate for hydrolysis by PCPH (19), this explanation seemed unlikely.
Still, direct evidence linking the NTPDase activity of PCPH or
mt-PCPH to ATP hydrolysis in the cell remains to be shown. Data in
this report demonstrate that mt-PCPH expression reduced ATP levels
and conferred oxaliplatin chemo-resistance in colorectal carcinoma
(CRC) cells. Results also demonstrate that the mt-PCPH oncoprotein
did not show any detectable NTPDase activity in assays performed
in vitro under a number of different experimental conditions
or when tested in situ in living cells, indicating that the
ATP depletion observed in mt-PCPH-expressing CRC cells most likely
did not result from the direct cleavage of endogenous ATP by
mt-PCPH. These findings strongly support the notion that the
mt-PCPH oncoprotein is catalytically inactive and imply that it may
regulate the cellular ATP content by NTPDase-independent
mechanisms. Understanding these alternative mechanisms will be
essential to devise strategies for the successful treatment of
predictably therapeutically resistant tumors expressing either
increased PCPH levels or, particularly, the mt-PCPH
oncoprotein.
Materials and methods
Reagents
RPMI-1640 medium, antibiotics, and general cell
culture reagents were from MediaTech, Inc. (Manassas, VA, USA).
Magnetic beads (protein G dynabeads), Novex®
Tris-Glycine Precast gels, Geneticin® (G418),
Optimem® and Lipofectamine™ 2000 transfection reagents
were from Invitrogen (Carlsbad, CA, USA). TrueBlot®
reagents were from eBioscience Inc. (San Diego, CA, USA), and the
myc-peptide was from AnaSpec Inc. (Fremont, CA, USA). Nucleotides
and other general chemicals were from Sigma-Aldrich (St. Louis, MO,
USA). The following antibodies were used: anti-myc-tag (Cell
Signaling, Danvers, MA, USA), anti-ENTPD5 (Sigma-Aldrich), and
HRP-conjugated secondary antibodies (Cell Signaling). The protease
inhibitor cocktail was from Roche Applied Science (Indianapolis,
IN, USA).
Cell culture and transfection
Human CRC HCT116 and prostate cancer PC-3 cells were
from the ATCC (Manassas, VA, USA), and human CRC HCT15 cells were
from our Cancer Center’s Tissue Culture Shared Resource. Cells were
maintained in RPMI-1640 medium containing 10% fetal bovine serum,
100 μg/ml streptomycin, and 100 IU penicillin, at 37°C, 5%
CO2 and 85% humidity. Hamster PCPH (AF084569) and
mt-PCPH (AF084568) ORFs were cloned into pcDNA3.1
(+)-myc-HisB mammalian expression vectors as described (20). Transient and stable transfections
were performed as described (18).
For chemical treatments, equal cell numbers were plated 24 h prior
to replacing the medium with chemical- or vehicle-containing
medium.
ATP replenishment
Liposome preparations (Encapsula NanoSciences,
Nashville, TN, USA) were composed of phosphatidylcholine,
cholesterol and dipalmitoylphosphatidylglycerol at 9:4:0.1 molar
ratio, with a 24.6 mg/ml total lipid concentration. The ATP content
of ATP-liposomes was determined for each batch before use. For ATP
replenishment experiments, actively dividing HCT116 cells
transfected with pcDNA, PCPH or mt-PCPH were counted and plated in
12-well dishes at 1.75×105 cells per well for ATP
analysis or in 96-well dishes at 1×104 cells per well
for cell viability assays. Following 24 h in culture, the medium
was replaced with fresh medium containing 0.5% of either
ATP-encapsulated or empty liposomes, continuing the incubation for
8 h. After this treatment, cells were washed thoroughly with PBS
and collected for ATP analysis. For chemosensitivity assays,
liposome-containing media were removed and cells were washed with
PBS and incubated with either vehicle- or oxaliplatin-containing
medium for 24 h before processing for cell viability
determinations.
Immunoblot and immunoprecipitation
Western immunoblot analyses were carried out
essentially as described (14),
being performed at least three times for each protein. Lysates for
immunoprecipitation (IP) were prepared in modified RIPA buffer (50
mM Tris pH 7.8, 140 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40)
containing protease inhibitors. IPs were performed using protein
G-conjugated dynabeads according to manufacturer’s recommendations
(Invitrogen). Tris-buffered saline (TBS) was used instead of PBS
and care was taken to prevent phosphate contamination. For
immunoblot analysis of IP samples, bead pellets were boiled in
Laemmli sample buffer (LSB) in the presence of β-mercaptoethanol.
Myc peptide elution was performed as described (21), pooling two incubations with 0.5
μg/μl of peptide followed by TCA precipitation and
resuspension in LSB containing β-mercaptoethanol.
Nucleotidase assay
NTPDase activity was detected by measuring the
release of inorganic phosphate in the presence of nucleotide
substrates as described (22).
Cell lysates were prepared in modified RIPA buffer containing
protease inhibitors. Briefly, whole-cell lysates or IP beads were
incubated at 37°C with 75 mM Tris-HCl pH 7.5, 10 mM
CaCl2, 10 mM MgCl2 and 0.1% Triton X-100,
with or without substrate. Reactions were stopped by adding
malachite green solution (1.2% malachite green, 4% sulfuric acid,
0.2% Tween-20). The optical density was measured at 630 nm and
reported in reference to phosphate standards.
Enzyme cytochemistry
Enzyme activity visualization in cultured cells was
performed using a lead phosphate detection method as described
(23). Transiently transfected
cells grown to 70% confluency in 6-well dishes were washed, fixed
with 4% paraformaldehyde in TBS solution, permeabilized with 0.1%
saponin, and incubated in the reaction solution [20 mM
Tris-maleate, 500 mM sucrose, 2 mM MgCl2, 2 mM
CaCl2 and 2 mM Pb(NO3)2] for 30
min with or without 2 mM GDP. Reactions were developed by
incubating with 1% ammonium sulfide. Samples were then blocked with
10% normal goat serum and then sequentially incubated with
dilutions of anti-myc-tag antibody and a fluorophore-conjugated
secondary antibody. Cells were imaged immediately using an Olympus
IX71 fluorescence microscope at our Cancer Center’s Microscopy and
Imaging Shared Resource. Phase-contrast and fluorescent images
collected from 5 random fields were densitometrically analyzed
using ImageJ software (24).
ATP measurements
Cells were counted, plated in 6-well dishes
(0.5–1×106 cells) and collected after 24 h. Cell pellets
were resuspended in boiling buffer and incubated at 100°C as
described (25). ATP measurements
were performed on supernatants using a luciferase/luciferin-based
kit (Invitrogen). Values are presented relative to an ATP standard
curve as the average ATP concentrations corrected by total protein
estimates of the supernatants performed using the BCA system.
Measurements were taken from at least two cell passages, in
triplicate.
Cell viability assays
Cells were counted, plated in 96-well plates
(1–30×103 cells), and after 24 h the medium was replaced
with fresh medium with or without oxaliplatin. Treated and
untreated cells were incubated again for 24 or 48 h, and cell
viability was measured using the Cell TiterGlo system (Promega,
Madison, WI, USA), according to manufacturer’s recommendations.
Luminescence was detected using a Berthold Microlumat Plus LV 96V
luminometer from our Cancer Center’s Genomics and Epigenomics
Shared Resource.
Statistical analysis
Data are presented as mean ± standard deviation (SD;
error bars) for a given experimental group or observation. One-way
ANOVA were performed to determine the statistical significance of
differences between experimental groups. p≤0.05 was regarded as
significant.
Results
ATP levels and chemo-resistance in CRC
cells expressing mt-PCPH
To understand the relationship between mt-PCPH
catalytic activity and the reduced ATP levels and enhanced
chemo-resistance conferred by mt-PCPH, ATP levels were determined
in extracts of HCT116 and HCT15 CRC cells, which express low
endogenous levels of PCPH or mt-PCPH, after stable transfection
with empty vector (pcDNA) or myc-tagged mt-PCPH (Fig. 1A). HCT116 cells expressing mt-PCPH
(HCT116/mt-PCPH) exhibited ATP levels around 28% lower than those
of pcDNA-transfected (HCT116/pcDNA) cells (Fig. 1B). Similarly, ATP levels were
nearly 40% lower in HCT15/mt-PCPH cells than in the HCT15/pcDNA
controls (Fig. 1B). In comparison
to the action of mt-PCPH, PCPH expression had little effect on the
ATP levels (∼5% decrease) in either HCT116 or HCT15 cells.
To establish the effect of mt-PCPH on their
chemo-response, HCT116/mt-PCPH and HCT15/mt-PCPH cells were treated
with several oxaliplatin concentrations for 24 and 48 h.
HCT116/mt-PCPH cells were more resistant to oxaliplatin (about 25%
after 24 h and 35% after 48 h) than control cells (Fig. 1C, left panel). Similarly,
HCT15/mt-PCPH cells were also more resistant to oxaliplatin (about
14% after 24 h and 31% after 48 h) than control cells (Fig. 1C, right panel).
To test the importance of reduced ATP levels in
mt-PCPH-induced chemo-resistance, the endogenous ATP levels of
HCT116/pcDNA, HCT116/PCPH and HCT116/mt-PCPH cells were replenished
by treatment with ATP-encapsulated liposomes for 8 h, and cells
were then analyzed for their response to oxaliplatin exposure.
HCT116/pcDNA cells treated with ATP-encapsulated liposomes showed a
nearly 2-fold increase in ATP levels relative to their ATP content
prior to the treatment (Fig. 1D).
The magnitude of the increase in ATP content of HCT116/mt-PCPH
cells after incubation with ATP-encapsulated liposomes was smaller
(∼1.5-fold) than in control HCT116/pcDNA cells (Fig. 1D), but reached ATP levels in the
same range as those found in HCT116/pcDNA and HCT116/PCPH cells
prior to liposome treatment (Fig.
1D, t=0). The increase in ATP levels rendered the
HCT116/mt-PCPH cells more sensitive to oxaliplatin than control
cells (Fig. 1E), reversing their
typical drug response and suggesting that ATP plays a role in
mt-PCPH-induced cellular chemo-resistance.
Generation of NTPDase-deficient PCPH and
mt-PCPH mutants
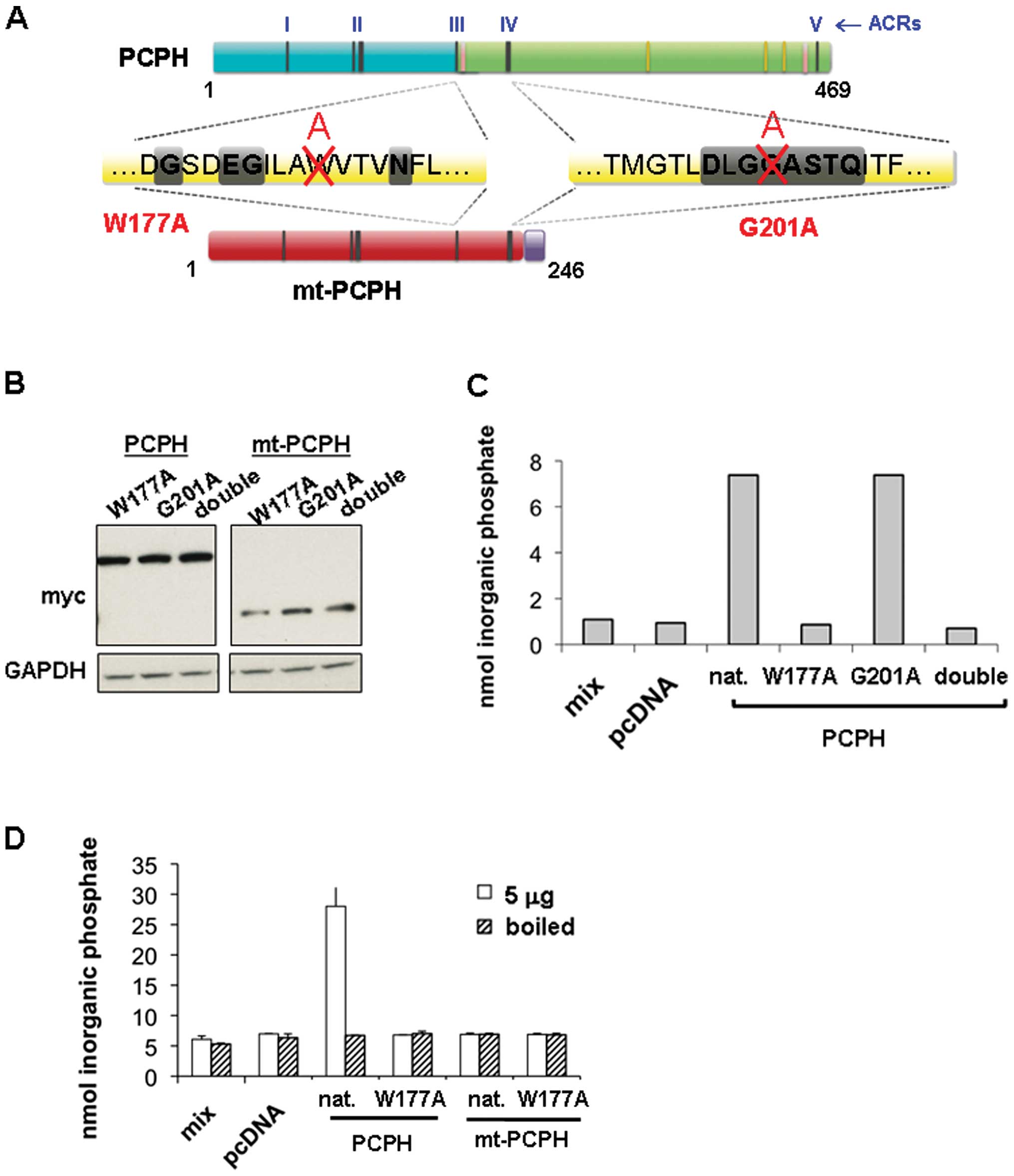
As the NTPDase activity of PCPH and mt-PCPH enzyme
activity had been previously reported (11,19,20,26–28),
to determine if the enzyme activity of mt-PCPH was directly
responsible for the cleavage of ATP and, therefore, for the
observed decrease in endogenous ATP levels, we generated inactive
apyrase mutants of both PCPH and mt-PCPH using a site-directed
mutagenesis approach. The tryptophan at position 177 (W177) and the
glycine at position 201 (G201) were selected for mutagenesis
(Fig. 2A) on the basis of
successful inactivating mutations of related NTPDases previously
reported in the literature (22,29,30).
The mutagenesis process converted both amino acid residues into
alanine, generating the W177A and G201A single mutants as well as
the double mutant of PCPH or mt-PCPH. All constructs were first
sequenced to confirm the successful generation of the expected
inactivating mutations, then transiently transfected into HCT116
cells, and whole extracts of the transfected cells were assayed for
NTPDase activity using GDP as the substrate (GDPase) as previously
described (31). GDP cleavage was
monitored by the detection of released inorganic phosphate using a
malachite green colorometric assay (32). Expression of the PCPH and mt-PCPH
NTPDase mutants in HCT116 cells was confirmed by immunoblot
analysis of cell lysates probed for the myc-tag (Fig. 2B). In comparison with extracts of
HCT116/PCPH cells, GDPase activity assays of extracts from cells
expressing mutated PCPH showed that the W177A mutation completely
abolished the GPDase activity, whereas the G201A mutation had no
effect on the activity (Fig. 2C).
Accordingly, the variant containing both mutations (double) did not
show GDPase activity. The GDPase activity of cell lysates from
HCT116/PCPH cells was lost when boiled prior to incubation with GDP
(Fig. 2D). However, when the
GDPase activity of the native PCPH and the PCPH/W177A apyrase
mutant was compared with that of native mt-PCPH and mt-PCPH/W177A,
it was revealed, as expected, that lysates prepared from cells
containing mt-PCPH/W177A did not show any activity. In addition,
most surprisingly, results showed that lysates of cells expressing
native mt-PCPH had no detectable GDPase activity either (Fig. 2D).
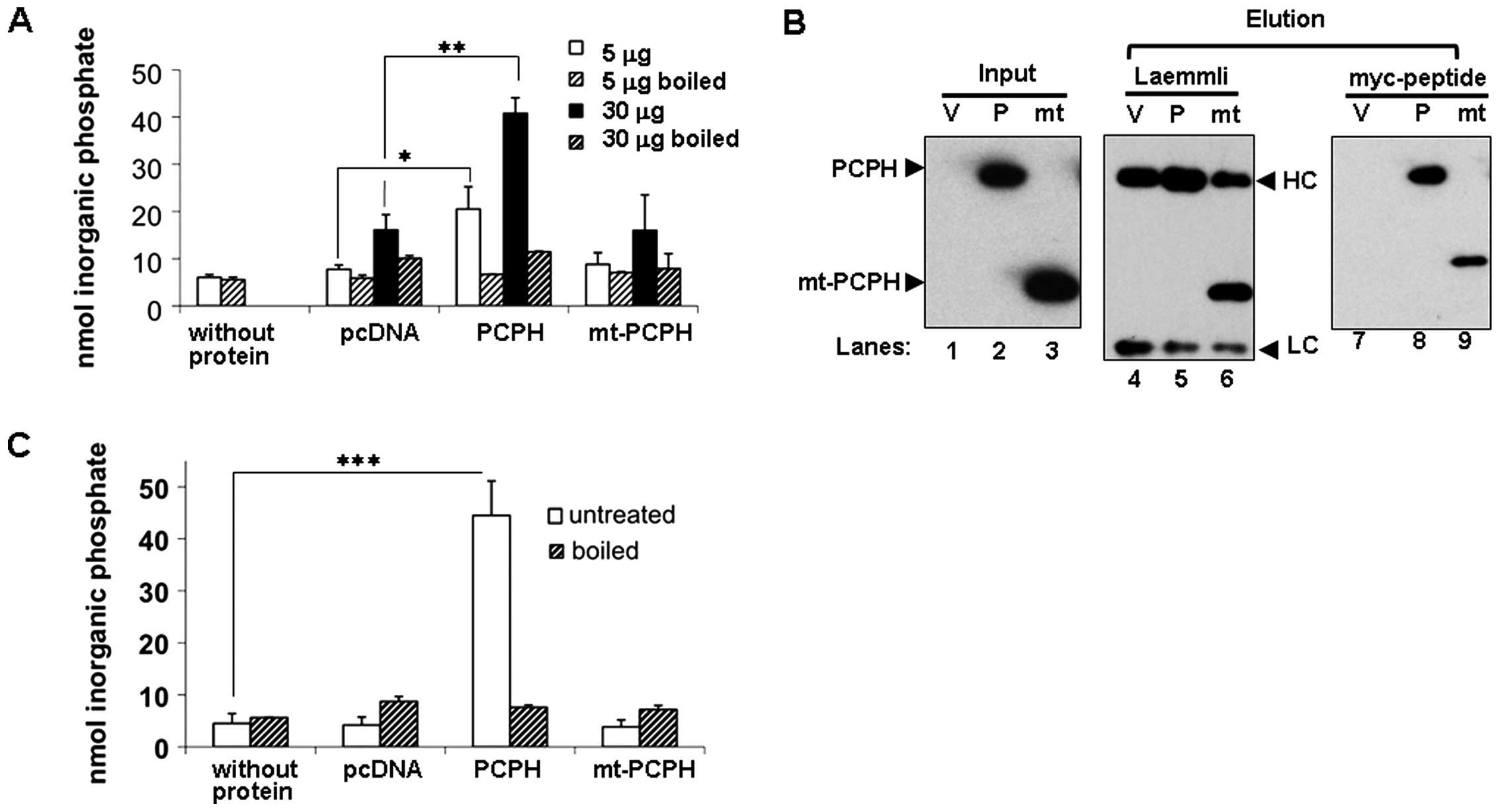
NTPDase activity of PCPH and mt-PCPH
expressed in mammalian cells
As the ATP depletion triggered by mt-PCPH expression
in CRC and other cell types was attributed to its NTPDase activity,
it became important to establish an enzyme activity baseline for
both PCPH and mt-PCPH in CRC cells by measuring their NTPDase
activity in vitro. Lysates from transiently transfected
HCT116 cells expressing myc-tagged PCPH or mt-PCPH at comparable
levels were incubated with GDP, as described above. Cell extracts
containing PCPH showed significant NTPDase activity relative to
controls (Fig. 3A), for two
different total protein concentrations (p≤0.05 for 5 μg and
p≤0.005 for 30 μg), which remained at background levels when
samples were boiled prior to incubation with the substrate.
However, boiled and non-boiled lysates from mt-PCPH-expressing
cells yielded phosphate signals indistinguishable from the
background levels detected in the corresponding controls (Fig. 3A). Similar results were observed
from NTPDase activity assays of whole cell lysates from PCPH
and mt-PCPH transfected PC-3 and HCT15 cells (data not
shown), indicating that the apparent lack of NTPDase activity by
mt-PCPH was not a cell type-specific observation. The fact that
negative results were obtained also when GDPase assays were
performed with extracts from HCT116 cells expressing an untagged
mt-PCPH construct (20)
confirmed that the presence of the myc-tag in the mt-PCPH protein
did not create a steric hindrance capable of preventing its NTPDase
activity.
To establish that the NTPDase activity observed in
cell lysates came specifically from the exogenously expressed
proteins, IPs were carried out using the myc-tag and the activity
of the proteins pulled down was tested on GDP. To confirm their
presence of PCPH and mt-PCPH in IPs, proteins were eluted from the
beads under reducing and denaturing conditions (Fig. 3B, lanes 4–6). As under these
conditions the antibody heavy chain co-migrated with PCPH,
precipitated proteins were also eluted using the myc peptide. Both
PCPH and mt-PCPH were detectable in immunoblots from myc
peptide-eluted IPs (Fig. 3B, lanes
7–9). Relative to controls, PCPH IPs showed a significant
(p≤0.0005) NTPDase activity (Fig.
3C), which was not detectable when IP beads were boiled prior
to the assays. On the contrary, boiled and non-boiled mt-PCPH IPs
showed no detectable NTPDase activity. To confirm that the
differences in NTPDase activity between PCPH and mt-PCPH were
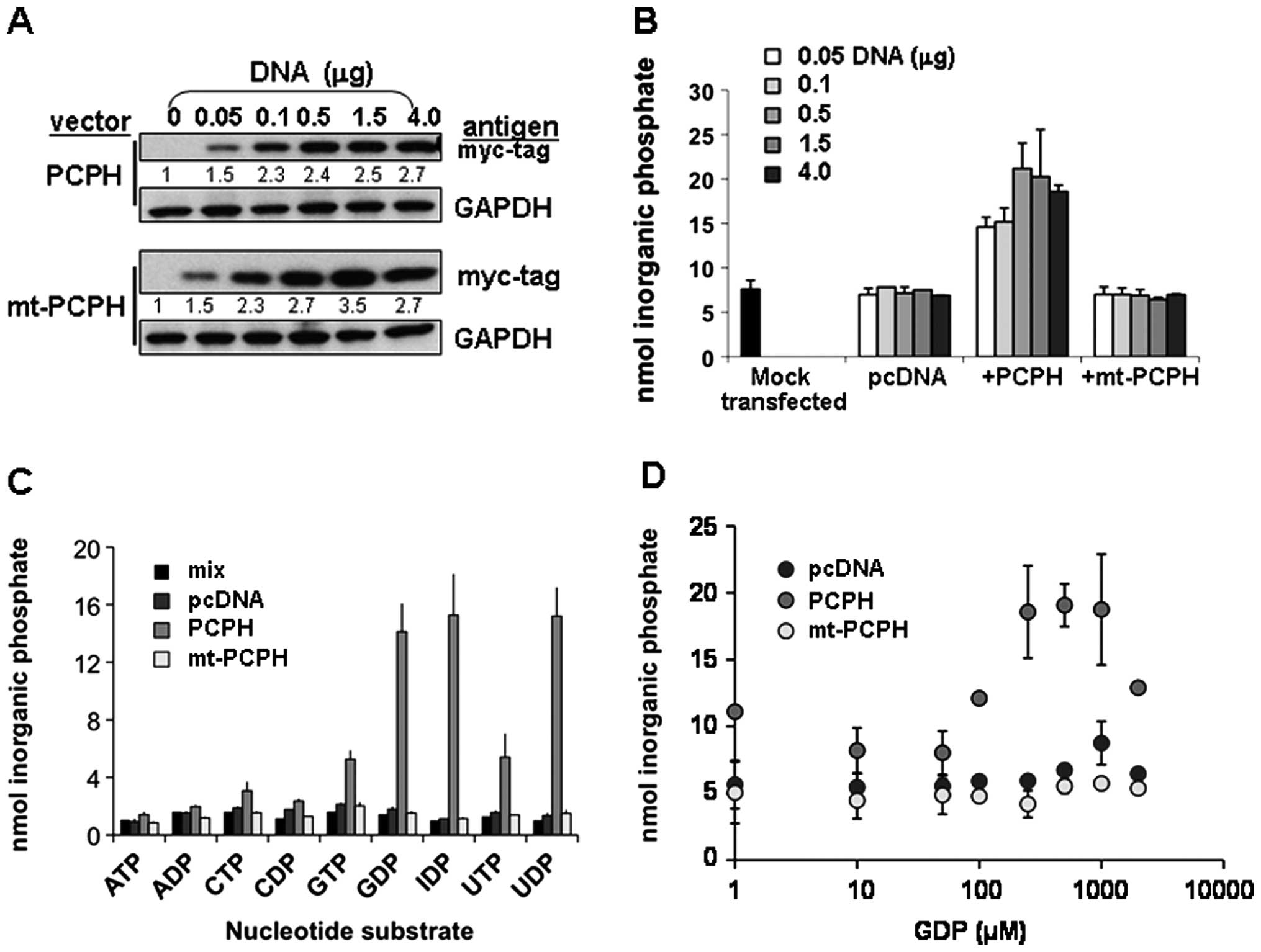
independent of their expression levels, activity assays were done
using lysates from cells transfected with different amounts of the
PCPH or mt-PCPH expression vectors and, therefore, having different
PCPH or mt-PCPH contents (Fig.
4A). Samples with up to 3.5-fold greater mt-PCPH levels
(Fig. 4A, lower panel) showed
NTPDase activity similar to those with low or no mt-PCPH (Fig. 4B), whereas the NTPDase activity of
PCPH correlated with protein levels in cells transfected with up to
1.5 μg DNA. These data suggested that PCPH does have NTPDase
activity in vitro, while mt-PCPH may either have severely
deregulated activity or lack NTPDase activity. Because differences
in primary sequence between PCPH and mt-PCPH could influence the
kinetic properties of mt-PCPH enzyme activity, NTPDase assays were
performed using different substrates and substrate concentrations.
The preferred PCPH substrates (Fig.
4C), were in agreement with previous reports (19,26,27),
but enzymatic activity remained undetectable in all assays
performed with mt-PCPH (Fig.
4D).
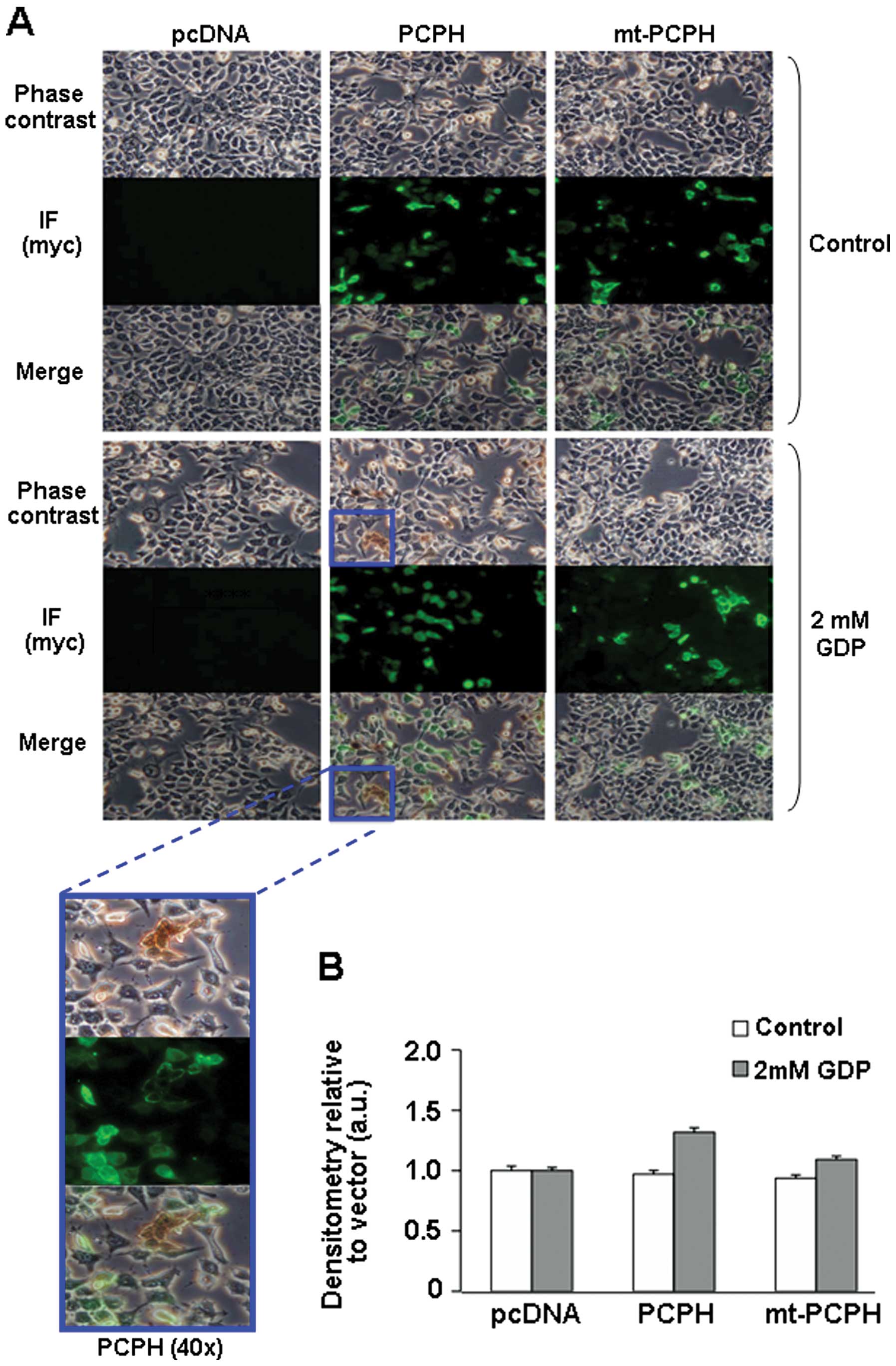
Because it seemed possible that the intrinsic
NTPDase activity of the mt-PCPH protein might be destroyed during
the process of preparation of the cell lysates, NTPDase activity
assays were also performed in situ, in actively
proliferating HCT116 cells, by combining the use of a phosphate
detection method that yields a brown precipitate visible under the
microscope, and indirect immunofluorescence to confirm whether the
precipitate forms on cells expressing either PCPH or mt-PCPH
(Fig. 5A, IF panels). No
precipitate formation was observed in control cultures to which no
GDP was added (Fig. 5A, top
panels). In the presence of GDP, subpopulations of cells containing
the brown precipitate could be observed in cultures of cells
expressing PCPH (Fig. 5A, middle
and magnified panels), but not in cultures of cells expressing
mt-PCPH or in the pcDNA-transfected control cells (Fig. 5A, side panels). Quantification of
in situ NTPDase activity demonstrated that PCPH, but not
mt-PCPH expressing cells, had significant activity compared to
vector-transfected controls (Fig.
5B). These data reiterated the notion that mt-PCPH most likely
lacks NTPDase activity, strongly suggesting that its intrinsic
NTPDase activity is not the functional determinant of the ATP
depletion and chemo-resistance phenotypes associated with
mt-PCPH-induced transformation.
Discussion
Acquisition of stress resistance has been reported
as an important component of the transforming activity of
overexpressed PCPH and of the mt-PCPH oncoprotein in different cell
systems (11,13–15).
It was suggested that such pro-survival role resulted from PCPH
and, particularly, mt-PCPH hydrolyzing ATP and consequently
depleting cellular energy levels (13,14).
However, the direct evidence linking the mt-PCPH NTPDase activity
to ATP depletion and induction of stress resistance was lacking.
This report represents the first description of mt-PCPH-induced
chemoresistance and ATP depletion in CRC cells, and demonstrates
the importance of the endogenous ATP levels in determining the
response of mt-PCPH expressing cells to chemotherapy.
The NTPDase activity, in vitro and in
situ, of PCPH and mt-PCPH expressed in human cells is also
described for the first time. Surprisingly, results showed that the
NTPDase activity of mt-PCPH was undetectable regardless of the cell
line, protein expression level, and substrate nature and
concentrations tested. These data do not agree with previous
studies demonstrating NTPDase activity for recombinant mt-PCPH in
non-human expression systems (20), and do not support the notion that
mt-PCPH catalyzed ATP hydrolysis. This discrepancy is likely due to
protein folding, dimerization ability and/or processing differences
in the expression systems used, as neither rabbit reticulocyte
in vitro transcription/translation (33) nor bacterial expression systems
perform post-translational modifications (PTMs) of the type that
NTPDase family members are known to undergo, such as
palmitoylation, acylation, phosphorylation and glycosylation
(7). In fact, glycosylation sites
are conserved across the NTPDase family (34) and, accordingly, PCPH glycosylation
was demonstrated (26). However,
NTPDase assays in systems where glycosylation does not occur, was
inhibited or removed showing that PCPH glycosylation was not
necessary for in vitro enzyme activity, although it
influenced multimer formation and protein solubility (19). The absence of glycosylation or
other PMTs in non-human systems (20) may have caused the observed
differences in NTPDase activity by altering native protein
processing. However, processing may also influence PCPH and mt-PCPH
NTPDase activity in mammalian cells as differential PCPH and
mt-PCPH expression was demonstrated for different species, tissues
and cell types (17,35).
Our findings agree with some structural features of
mt-PCPH. Fig. 2A shows the
N-terminus as conserved within the NTPDase family, including most
apyrase conserved regions (ACRs I–IV). PCPH and mt-PCPH also share
N-terminal amino acid sequence up to residue 213, after which
PCPH encodes 256 amino acids and mt-PCPH only 33
non-conserved residues (Fig. 2A).
Due to this truncation, mt-PCPH lacks the cofactor coordinating
residue within ACRV, four conserved cysteine residues presumably
involved in intra-protein disulfide bonds (36), and all residues comprising the
substrate-binding pocket (37),
strongly suggesting that the loss of these conserved features may
affect the tertiary structure and enzyme function of mt-PCPH. Other
C-terminally truncated NTPDase isoforms have been reported also to
be inactive (38,39).
The crystal structures for PCPH or mt-PCPH have not
been resolved, but NTPDase catalytic clefts and structural
properties are conserved across the family and are highly
homologous to other nucleotide hydrolyzing enzyme families
(37) and, therefore, can be used
to aid structural predictions. Using the reported crystal structure
of ENTPD2 as a reference for the likely similar structure of PCPH,
and not including the C-terminal tail of mt-PCPH, it seems that
mt-PCPH would lack nearly the entire second structural domain,
which comprises one of the lobes shaping the catalytic cleft and
described as containing key active site amino acids and structural
features involved in catalysis by other family members. Therefore,
although most of the ACRs essential for NTPDase activity are
retained in mt-PCPH, the fact that it is missing important
catalytic determinants strongly supports the overall soundness of
our conclusion that the mt-PCPH oncoprotein is a catalytically
inactive member of the NTPDase family of proteins. As indicated in
Fig. 2A, the differences in
primary sequence, domain organization and structural predictions
between mt-PCPH and PCPH could result in an mt-PCPH conformation
potentially exposing reactive residues that would otherwise be
protected by the second lobular domain present only in PCPH.
Consequently, we hypothesize that mt-PCPH might act through
protein-protein interactions rather than by way of NTPDase
activity.
The metabolic needs of cancer cells are different
from those of healthy cells. Cells adapt to the harsh conditions of
the tumor microenvironment and metabolic demands for cancer cell
survival and growth. Changes in ATP levels subsequent to mt-PCPH
expression may be one of these adaptations. In tumors, mt-PCPH
expression may be turned on in response to harsh tumor conditions,
as endogenous mt-PCPH expression was shown to be stress-induced
(11), and may increase with tumor
progression. Indeed, it was shown that PCPH and mt-PCPH expression
was increased in breast (17),
prostate (18), and testicular
(40) tumors compared to normal
tissues and PCPH has been described as a biomarker for colon tumors
(41). It seems that mt-PCPH
expression may be not only a powerful reporter for the metabolic
shift in tumor cells but also a possible predictor for tumor
chemotherapy responses. Understanding the NTPDase-independent
mechanisms by which mt-PCPH modifies cellular energy levels and
chemo-response may be critical to devise strategies for the
successful treatment of predictably therapeutically resistant
tumors expressing the mt-PCPH oncoprotein.
Abbreviations:
|
ACR
|
apyrase conserved region;
|
|
CRC
|
colorectal carcinoma;
|
|
IP
|
immunoprecipitation;
|
|
LSB
|
Laemmli sample buffer;
|
|
NTPDase
|
nucleoside triphosphate
diphosphohydrolase;
|
|
ORF
|
open reading frame;
|
|
PTM
|
post-translational modification;
|
|
TBS
|
Tris-buffered saline;
|
|
TCA
|
trichloroacetic acid
|
Acknowledgements
This study was supported by U.S.
Public Health Service grants RO1-CA64472 and RO1-CA134727 (to V.N.)
from the National Cancer Institute, NIH. This study utilized the
Tissue Culture, Microscopy and Imaging, and Genomics and
Epigenomics Shared Resources of the Lombardi Comprehensive Cancer
Center which are funded through USPHS grant P30-CA51008.
References
|
1.
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.PubMed/NCBI
|
|
3.
|
Sattler UG, Hirschhaeuser F and
Mueller-Klieser WF: Manipulation of glycolysis in malignant tumors:
fantasy or therapy? Curr Med Chem. 17:96–108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Hardt PD, Mazurek S, Toepler M, et al:
Faecal tumour M2 pyruvate kinase: a new, sensitive screening tool
for colorectal cancer. Br J Cancer. 91:980–984. 2004.PubMed/NCBI
|
|
5.
|
Chan EC, Koh PK, Mal M, et al: Metabolic
profiling of human colorectal cancer using high-resolution magic
angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy
and gas chromatography mass spectrometry (GC/MS). J Proteome Res.
8:352–361. 2009. View Article : Google Scholar
|
|
6.
|
Walenta S, Chau TV, Schroeder T, et al:
Metabolic classification of human rectal adenocarcinomas: a novel
guideline for clinical oncologists? J Cancer Res Clin Oncol.
129:321–326. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Robson SC, Sevigny J and Zimmermann H: The
E-NTPDase family of ectonucleotidases: Structure function
relationships and pathophysiological significance. Purinergic
Signal. 2:409–430. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Enjyoji K, Kotani K, Thukral C, et al:
Deletion of cd39/entpd1 results in hepatic insulin resistance.
Diabetes. 57:2311–2320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Read R, Hansen G, Kramer J, Finch R, Li L
and Vogel P: Ectonucleoside triphosphate diphosphohydrolase type 5
(Entpd5)-deficient mice develop progressive hepatopathy,
hepatocellular tumors, and spermatogenic arrest. Vet Pathol.
46:491–504. 2009. View Article : Google Scholar
|
|
10.
|
Notario V, Castro R, Flessate DM, Doniger
J and DiPaolo JA: Frequent activation of non-ras transforming
sequences in neoplastic Syrian hamster cells initiated with
chemical carcinogens. Oncogene. 5:1425–1430. 1990.PubMed/NCBI
|
|
11.
|
Velasco JA, Avila MA and Notario V: The
product of the cph oncogene is a truncated, nucleotide-binding
protein that enhances cellular survival to stress. Oncogene.
18:689–701. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Fang M, Shen Z, Huang S, et al: The ER
UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect,
and proliferation in the PTEN pathway. Cell. 143:711–724. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Recio JA, Páez JG, Sanders S, Kawakami T
and Notario V: Partial depletion of intracellular ATP mediates the
stress-survival function of the PCPH oncoprotein. Cancer Res.
62:2690–2694. 2002.PubMed/NCBI
|
|
14.
|
Tirado OM, Mateo-Lozano S, Sanders S,
Dettin LE and Notario V: The PCPH oncoprotein antagonizes the
proapoptotic role of the mammalian target of rapamycin in the
response of normal fibroblasts to ionizing radiation. Cancer Res.
63:6290–6298. 2003.PubMed/NCBI
|
|
15.
|
Villar J, Quadri HS, Song I, Tomita Y,
Tirado OM and Notario V: PCPH/ENTPD5 expression confers to prostate
cancer cells resistance against cisplatin-induced apoptosis through
protein kinase Cα-mediated Bcl-2 stabilization. Cancer Res.
69:102–110. 2009.PubMed/NCBI
|
|
16.
|
Rouzaut A, Recio JA and Notario V:
Expression of the protein product of the PCPH proto-oncogene in
human tumor cell lines. Radiat Res. 155:181–187. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Blánquez MJ, Arenas MI, Conde I, Tirado
OM, Paniagua R and Notario V: Deregulated expression of the PCPH
proto-oncogene in human breast cancers. Int J Oncol. 25:821–830.
2004.PubMed/NCBI
|
|
18.
|
Villar J, Arenas MI, MacCarthy CM,
Blánquez MJ, Tirado OM and Notario V: PCPH/ENTPD5 expression
enhances the invasiveness of human prostate cancer cells by a
protein kinase C delta-dependent mechanism. Cancer Res.
67:10859–10868. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Murphy-Piedmonte DM, Crawford PA and
Kirley TL: Bacterial expression, folding, purification and
characterization of soluble NTPDase5 (CD39L4) ecto-nucleotidase.
Biochim Biophys Acta. 1747:251–259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Recio JA, Páez JG, Maskeri B, Loveland M,
Velasco JA and Notario V: Both normal and transforming PCPH
proteins have guanosine diphosphatase activity but only the
oncoprotein cooperates with Ras in activating extracellular
signal-regulated kinase ERK1. Cancer Res. 60:1720–1728. 2000.
|
|
21.
|
Le Guezennec X, Vermeulen M, Brinkman AB,
et al: MBD2/NuRD and MBD3/NuRD, two distinct complexes with
different biochemical and functional properties. Mol Cell Biol.
26:843–851. 2006.PubMed/NCBI
|
|
22.
|
Smith TM and Kirley TL: Site-directed
mutagenesis of a human brain ecto-apyrase: evidence that the E-type
ATPases are related to the actin/heat shock 70/sugar kinase
superfamily. Biochemistry. 38:321–328. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Wachstein M and Meisel E: Histochemistry
of hepatic phosphatases of a physiologic pH; with special reference
to the demonstration of bile canaliculi. Am J Clin Pathol.
27:13–23. 1957.PubMed/NCBI
|
|
24.
|
Abramoff MD, Magelhaes PJ and Ram SJ:
Image processing with Image. J Biophotonics Int. 11:36–42.
2004.
|
|
25.
|
Li Y, Park JS, Deng JH and Bai Y:
Cytochrome c oxidase subunit IV is essential for assembly and
respiratory function of the enzyme complex. J Bioenerg Biomembr.
38:283–291. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Mulero JJ, Yeung G, Nelken ST and Ford JE:
CD39-L4 is a secreted human apyrase, specific for the hydrolysis of
nucleoside diphosphates. J Biol Chem. 274:20064–20067. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Trombetta ES and Helenius A: Glycoprotein
reglucosylation and nucleotide sugar utilization in the secretory
pathway: identification of a nucleoside diphosphatase in the
endoplasmic reticulum. EMBO J. 18:3282–3292. 1999. View Article : Google Scholar
|
|
28.
|
Mulero JJ, Yeung G, Nelken ST, Bright JM,
McGowan DW and Ford JE: Biochemical characterization of CD39L4.
Biochemistry. 39:12924–12928. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Yang F, Hicks-Berger CA, Smith TM and
Kirley TL: Site-directed mutagenesis of human nucleoside
triphosphate diphosphohydrolase 3: the importance of residues in
the apyrase conserved regions. Biochemistry. 40:3943–3950. 2001.
View Article : Google Scholar
|
|
30.
|
Smith TM, Lewis Carl SA and Kirley TL:
Mutagenesis of two conserved tryptophan residues of the E-type
ATPases: inactivation and conversion of an ecto-apyrase to an
ecto-NTPase. Biochemistry. 38:5849–5857. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Miura P, Thompson J, Chakkalakal JV,
Holcik M and Jasmin BJ: The utrophin A 5′-untranslated region
confers internal ribosome entry site-mediated translational control
during regeneration of skeletal muscle fibers. J Biol Chem.
280:32997–33005. 2005.
|
|
32.
|
Baykov AA, Evtushenko OA and Avaeva SM: A
malachite green procedure for orthophosphate determination and its
use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem.
171:266–270. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Zhang JT and Ling V: Involvement of
cytoplasmic factors regulating the membrane orientation of
P-glycoprotein sequences. Biochemistry. 34:9159–9165. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Massé K, Eason R, Bhamra S, Dale N and
Jones EA: Comparative genomic and expression analysis of the
conserved NTPDase gene family in Xenopus. Genomics.
87:366–381. 2006.PubMed/NCBI
|
|
35.
|
Recio JA, Zambrano N, de la Peña L, et al:
cDNA isolation, expression, and chromosomal localization of the
mouse pcph proto-oncogene. Mol Carcinog. 26:130–136. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Ivanenkov VV, Murphy-Piedmonte DM and
Kirley TL: Bacterial expression, characterization, and disulfide
bond determination of soluble human NTPDase6 (CD39L2) nucleotidase:
implications for structure and function. Biochemistry.
42:11726–11735. 2003. View Article : Google Scholar
|
|
37.
|
Zebisch M and Strater N: Structural
insight into signal conversion and inactivation by NTPDase2 in
purinergic signaling. Proc Natl Acad Sci USA. 105:6882–6887. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Crawford PA, Gaddie KJ, Smith TM and
Kirley TL: Characterization of an alternative splice variant of
human nucleoside triphosphate diphosphohydrolase 3 (NTPDase3): a
possible modulator of nucleotidase activity and purinergic
signaling. Arch Biochem Biophys. 457:7–15. 2007. View Article : Google Scholar
|
|
39.
|
Riewe D, Grosman L, Fernie AR, Wucke C and
Geigenberger P: The potato-specific apyrase is apoplastically
localized and has influence on gene expression, growth, and
development. Plant Physiol. 147:1092–1109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Regadera J, Blánquez MJ, González-Peramato
P, et al: PCPH expression is an early event in the development of
testicular germ cell tumors. Int J Oncol. 28:595–604.
2006.PubMed/NCBI
|
|
41.
|
Mikula M, Rubel T, Karczmarski J, Goryca
K, Dadlez M and Ostrowski J: Integrating proteomic and
transcriptomic high-throughput surveys for search of new biomarkers
of colon tumors. Funct Integr Genomics. 11:215–224. 2010.
View Article : Google Scholar : PubMed/NCBI
|