Introduction
Thermal ablation therapy destroys cancer cells by
delivering electromagnetic or acoustic energy [e.g. radiofrequency
(1), laser (2), or focused ultrasound (3)] that is converted to heat in the
target tissue. The therapy raises the temperature of the ablation
site to between 56 and 100°C which results in irreversible damage
by coagulation necrosis. Preclinical study with conductive
interstitial thermal therapy (CITT) (4), (5)
has demonstrated decreased metastasis in rabbits with VX2 tumors
(6) and the reduction of hypoxia
in tumor tissue surviving partial CITT ablation of murine breast
carcinoma (7). Investigating the
molecular changes associated with CITT and with other thermal
ablation therapies may identify biomarkers that facilitate
development of methods for monitoring and predicting the potential
for therapy-associated enhancement of tumor re-growth, and
metastasis.
Histological analysis of thermal ablation lesions in
animal models has revealed three regions. The central region is
adjacent to the ablation (i.e. area of probe insertion, or focus of
ultrasound) and contains necrotic tumor tissue. Massive tumor
debris appears to result in release of immunogenic factors
(8). Adjacent to necrotic area is
a coagulation margin that defines a sharp transition zone between
necrotic and viable tissue. This highly hypoxic region was shown to
be a site of accelerated tumor re-growth after radiofrequency
ablation (9). A third region is
more distant from the center of heat delivery and contains viable
tumor tissue. In this region, we observed decreased hypoxia 72 h
after CITT ablation (7). Thus,
lesions of ablated tumor appear to possess areas with different
properties that may have the potential to initiate induction of
tumor re-growth (undesirable effect), stimulate antitumor immunity
and decrease hypoxia (desirable effects).
Our research is interested in the mechanisms by
which thermal tumor ablation affects communication between tumor
tissue, bone marrow and sites of metastasis. Recent progress in
understanding tumor biology has revealed crucial roles for the bone
marrow in carcinogenesis, tumor growth and metastasis. Cytokines
and growth factors secreted by tumor into circulation stimulate
release of a variety of cells from bone marrow. These molecules
contribute to tumor growth, vascularization, and regulation of
immune cells in the tumor microenvironment (10,11).
However, little is known about the effects of thermal ablation upon
the interactions between tumors, bone marrow, and site of
metastasis. Thus, characterization of the molecules that
potentially mediate these communications in the context of tumor
growth and treatment would be an important step towards
understanding the mechanisms underlying different outcomes of
thermal tumor ablation.
The study presented here reports the preliminary
results of quantitative analysis of gene expression and protein
levels for serum, marrow and surviving tumor collected 72 h after
partial ablation of 4T1 carcinoma in BALB/c mice. The CITT
treatment protocol was designed to result in only partial tumor
ablation to enable characterization of the remaining surviving
tumor tissue. The study hypothesized that tumor ablation would
broadly affect gene expression (Table
I) and protein (Table II)
levels for molecules involved in cell signaling, proliferation,
adhesion, angiogenesis and stress response. Candidate molecules
were chosen due to evidence in the literature for their role in
breast tumor development and spreading, bone marrow homeostasis and
tumor responses to thermal stress (12–16).
 | Table I.The genes analyzed in bone marrow and
4T1 tumors from BALB/c mice. |
Table I.
The genes analyzed in bone marrow and
4T1 tumors from BALB/c mice.
| Symbol | Molecule name | Description
(function in breast cancer) |
|---|
| Assay type:
Quantitative RT-PCR in bone marrow and tumor lysates |
| Bmp2 | Bone morphogenic
protein 2 | Growth factor, TGFβ
superfamily - tumor apoptosis, EMT |
| Ccl2 | Chemokine (C-C
motif) ligand 2 (MCP-1) | Signaling molecule
- TAM infiltration, cancer stem cell |
| Csf3 | Colony stimulating
factor 3 (granulocyte) (G-CSF) | Cytokine-apoptosis,
neutrophils release from BM |
| Csf3r | Colony stimulating
factor 3 receptor (granulocyte) | CD114,
hematopoietin receptor family - cell maturation in BM |
| Cxcl12 | Chemokine (C-X-C
motif) ligand 12 (SDF-1) | Signaling molecule
- hematopoietic cell homing and quiescence, communication between
tumor and BM |
| Cxcr4 | Chemokine (C-X-C
motif) receptor 4 (SDF-1 receptor) | CD184 (fusin),
α-chemokine receptor - metastasis to BM |
| Fgf2 | Fibroblast growth
factor 2 | Growth factor
(basicFGF) - myelopoiesis and angiogenesis in BM |
| Glg1 | Golgi apparatus
protein 1 (ESL-1) | Glycoprotein,
leukocyte ligand for E-selectin-metastasis |
| Hsf1 | Heat shock factor
1 | Nuclear protein
activating HSPs-growth, inhibition of apoptosis |
| Icam1 | Intercellular
adhesion molecule 1 | CD54, glycoprotein,
immunoglobulin family-metastasis |
| Id1 | Inhibitor of DNA
binding 1 | Nuclear protein -
metastasis, inhibition of differentiation |
| Il6 | Interleukin 6 | Pleiotropic
cytokine - metastasis to BM |
| Il17ra | Interleukin 17
receptor A | CD217, glycoprotein
- proinflamatory, associated with poor prognosis |
| Kdr | Kinase insert
domain protein receptor (VGFR2) | VGEF receptor -
angiogenesis, tumor progression |
| Klrk1 | Killer cell
lectin-like receptor subfamily number 1 | CD314 (NKG2D), NK
cell receptor-antitumor immunity |
| Lif | Leukemia inhibitory
factor | Pleiotropic
cytokine-tumor proliferation, chemoattractant for BM derived
cells |
| Lifr | Leukemia inhibitory
factor receptor | CD118, cytokine
receptor - suppressor of metastasis |
| Mmp2 | Matrix
metallopeptidase 2 | Type IV
collagenase, gelatinase A - metastasis |
| Mmp9 | Matrix
metallopeptidase 9 | Type IV
collagenase, gelatinase A - angiogenesis, metastasis |
| Ptgs2 |
Prostaglandin-endoperoxide synthase 2 | Cyclooxygenase 2
(COX2) enzyme - worse prognosis |
| Sele | E-selectin | CD62, adhesion
molecule, expressed only on activated endothelial cells -
metastasis |
| Serpin1 | Serine protease
inhibitor, nectin (PAI-1) | Inhibitor of tissue
plasminogen activator (tPA) - cancer progression |
|
Tnfrsf11a | Tumor necrosis
factor receptor superfamily, member 11a (RANK) | Expressed on
osteoclasts and dendritic cells - bone metastasis |
| Tnfsf11 | Tumor necrosis
factor (ligand) superfamily, member 11 (RANKL) | CD254, expressed on
osteoblasts, stromal and T cells - cancer cells
chemoattractant |
| Tnf | Tumor necrosis
factor | Cachectin,
TNFα-cytotoxin - breast cancer promoter |
| Ucp2 | Uncoupling protein
2 (mitochondrial proton carrier) | Mitochondrial ROS
regulator - tumor promoting factor |
| Vcam1 | Vascular cell
adhesion molecule 1 | CD106, mediator of
immune cell adhesion to endothelium - aberrantly expressed on
breast cancer cells, metastasis |
 | Table II.List of proteins analyzed in 4T1
tumors and serum from BALB/c mice. |
Table II.
List of proteins analyzed in 4T1
tumors and serum from BALB/c mice.
| Symbol | Molecule name | Description
(function in breast cancer) |
|---|
| Assay type: ELISA
in tumor lysates |
| CXCL12 | Chemokine (C-X-C
motif) ligand 12 (SDF-1) | Signaling molecule
- hematopoietic cell homing and quiescence, communication between
tumor and BM |
| Assay type: Western
immunoblotting in tumor lysates |
| HIF-1α | Hypoxia-inducible
factor 1A | Transcription
factor - adaptation to hypoxia, overexpression in invasive
cancers |
| HSP27 | Heat shock protein
27 | Stress response
protein - drug resistance, stem cell maintenance |
| HSP70 | Heat shock protein
70 | Stress response
protein - antigen binding, anti-apoptotic |
| MMP9 | Matrix
metallopeptidase 9 | Type IV
collagenase, gelatinase A - angiogenesis, metastasis |
| Ki67 | Antigen Ki-67 | Nuclear protein -
marker of proliferation |
| Assay type:
Antibodies multiplex of 16 murine chemokines in serum |
| IL-1α, IL-1β, IL-2,
IL-3, IL-4, IL-5, IL-6, IL-10, IL-12, Il-17, MCP-1, IFNγ, TNFα,
MIP-1α, GM-CSF, RANTES | Signaling molecules
with immunomodulatory and chemotaxic functions - altered in serum
of breast cancer patients |
Materials and methods
Growing 4T1 tumors in BALB/c mice and
CITT treatment
The 4T1 mammary carcinoma is a syngeneic mouse model
of breast cancer. The 4T1 cell line originated from spontaneously
grown, metastatic mammary tumor in BALB/c mice. This mammary tumor
has many characteristics of human invasive breast cancer (17). The thermal ablation procedures were
described in detail in our previous study (7). The use of BALB/c mice (Jackson
Laboratories, Bar Harbor, ME, USA) and the experimental protocol
was approved by the University of Arkansas for Medical Sciences
Institutional Animal Care and Use Committee. The 4T1 cancer cells
(2×105) were injected subcutaneously into the right rear
leg of each mouse. When tumor reached 10–12 mm in size, the mice
were anesthetized with 1–2% isofluorane and a CITT probe, without
pin deployment, was inserted in the center of a tumor. Temperatures
were monitored with two thermocouples, one next to the tip of the
CITT probe and another one, at the periphery of the tumor. Ablation
was performed for 10 min with temperature maintained within the
range 80–90°C (next to the CITT probe) while the peripheral tumor
temperature remained in the low to mid 40°C range. Each mouse had
one tumor. Five mice with tumors were ablated and four mice with
tumors were used as untreated control.
Sample preparation
The mice were scarified 72 h after ablation. At this
time point necrotic area is well established and the transition
zone separating necrotic area from viable perinecrotic zone is
clearly visible on tumor sections. The entire tumors were removed
immediately after necropsy. One half of the tumor was used to
prepare fresh sections in order to record the size of the ablated
areas with a viability staining, triphenyltetrazolium chloride
(TTC) as previously described (18). The other half of the tumor was used
to prepare tissue samples from viable portion of tumors which were
cut into pieces ranging from 50–100 mg, frozen in liquid
N2 and stored at −80°C until use. Femurs and tibiae were
carefully cleaned from adherent soft tissue and placed in cold,
sterile phosphate buffered saline, followed by transfer to Petri
dish filled with Dulbecco’s modified Eagle’s medium (DMEM). The
ends of bone were cut off with sterile scissors so that the bone
marrow could be flushed with DMEM using a 3cc syringe and 27G
needle. The cells suspensions were filtered through 70-mm BD Falcon
nylon mesh (Fisher Scientific Inc., Sewanee, GA, USA). Cells were
counted (Beckman Coutler Z series System, Hialeah, FL, USA),
centrifuged at 1410 rpm for 5 min at 4°C, and re-suspended in serum
free DMEM at a concentration 1×106 cells per ml.
Aliquots of 1.5 ml of cells were made, centrifuged and the medium
was removed so that cell pellets could be frozen in liquid
N2 and stored at −80°C until use.
Total RNA isolation and preparation of
cDNA
Total RNA was isolated from frozen tumor and bone
marrow cell pellets using the RNAqueous-4PCR kit (Ambion/Applied
Biosystems, Foster City, CA, USA) according the manufacturer’s
instructions. RNA concentration was determined with ND-1000
Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE,
USA) and RNA integrity was verified with Bioanalyzer (Applied
Biosystems). All RNA samples possessed an RNA integrity number
(RIN) >7. cDNAs were prepared from 2 μg of RNA using
iScript cDNA Synthesis kit (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). This kit contains blend of oligo dT and random hexamers
primers. The cDNAs were stored at −20°C in 5–10 μl aliquots.
A pooled sample of all cDNAs was also prepared to use in primer
testing and optimization of standard curves.
Quantititative RT-PCR
Primers used in the real-time polymerase chain
reaction (RT-PCR) were designed using the murine NCBI nucleotides
database and Primer Express v3.0 software (Table III). RT-PCR assays were performed
according to the manufacturer’s instructions using iTaqSYBR Green
Supermix with ROX (Bio-Rad Laboratories Inc.), 384-well plates, and
the ABI Prism 7900 Sequence Detection System (Applied Biosystems).
For each transcript of interest standard curves were evaluated in
order to optimize the primer concentration for maximum reaction
efficiency. Standard curves were prepared from five-fold serial
dilution of pooled cDNA. Gene expression was normalized to both 18S
ribosomal RNA and glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
The comparative CT (ΔΔCT) method for
calculating relative gene expression was used to evaluate
differences in levels of transcripts between control group (mice
with non-treated tumors) and treated group (mice with ablated
tumors) (19). The fold change of
transcripts was calculated using DataAssist v3.0 software (Applied
Biosystems).
 | Table III.The murine primers used. |
Table III.
The murine primers used.
| Gene | Accession no. | Forward primer | Reverse primer |
|---|
| Bmp2 | NM_007553 |
TGTCCCCAGTGACGAGTTTCT |
CCTGTATCTGTTCCCGGAAGAT |
| Ccl2 | NM_011333 |
CTGAAGCCAGCTCTCTCTTCCT |
CAGGCCCAGAAGCATGACA |
| Csf3 | NM_009971 |
CAGTACCCCCAAAAAATCAGTGA |
TGGGCCCCCCTGAGAT |
| Csf3r | NM_007782 |
TCCAGCGAGTCCCCAAAG |
CAGCATGGGAGGCTCCAAT |
| Cxcl12 | NM_021704 |
GCCTCCAAACGCATGCTT |
ATTGGTCCGTCAGGCTACAGA |
| Cxcr4 | NM_009911 |
TCGGCAATGGATTGGTGAT |
CCGTCATGCTCCTTAGCTTCTT |
| Fgf2 | NM_008006 |
TGGTATGTGGCACTGAAACGA |
TCCAGGTCCCGTTTTGGAT |
| Glg1 | NM_009149 |
CTCACTGCGCCCTCTAACG |
GGCACCTGATGCTGCTCTACT |
| Hsf1 | NM_008296 |
CATAAAAATACGCCAGGACAGTGT |
CCCCTTCATCAGCTGCACAT |
| Icam1 | NM_010493 |
TGGCGGGAAAGTTCCTGTT |
TCCAGCCGAGGACCATACA |
| Id1 | NM_010495 |
GAACGTCCTGCTCTACGACATG |
TGGGCACCAGCTCCTTGA |
| Il-17ra | NM_008359 |
CCCAGGCAAGAAGAATTCCA |
CACCAGTGAAACTTGCTTAGAGTGA |
| Il-6 | NM_031168 |
CCACGGCCTTCCCTACTTC |
TTGGGAGTGGTATCCTCTGTGA |
| Kdr | NM_010612 |
ACTGCAGTGATTGCCATGTTCT |
TCATTGGCCCGCTTAACG |
| Klrk1 | NM_033078 |
GGCAATTCGATTCACCCTTAAC |
ATACTGGCTGAAACGTCTCTTTGA |
| Lif | NM_008501 |
GCCACGGCAACCTCATG |
ATTGGCGCTGCCATTGA |
| Lifr | NM_013584 |
AGAACATCACTGACATATCCCAGAAG |
GTATAGGCTCGCAGGACCAGAT |
| Mmp2 | NM_008610 |
GGACCCCGGTTTCCCTAA |
CAGGTTATCAGGGATGGCATTC |
| Mmp9 | NM_013599 |
TGGTGGCAGCGCACG |
CTTCCGGCACGCTGGA |
| Ptgs2 | NM_011198 |
TGCCTCCCACTCCAGACTAGA |
CAGCTCAGTTGAACGCCTTTT |
| Sele | NM_011345 |
TCCTGCGAAGAAGGATTTGAA |
CCCCTCTTGGACCACACTGA |
| Serpin1 | NM_008871 |
CCGTGGAACAAGAATGAGATCAG |
CTCTAGGTCCCGCTGGACAA |
| Tgfb1 | NM_011577 |
GCAGTGGCTGAACCAAGGA |
AGCAGTGAGCGCTGAATCG |
| Tnf | NM_013693 |
CACAAGATGCTGGGACAGTGA |
TCCTTGATGGTGGTGCATGA |
|
Tnfrsf11a | NM_009399 |
TCGTCCACAGACAAATGCAAA |
GTGTGCTTCTAGCTTTCCAAGGA |
| Tnfsf11 | NM_011613 |
GGCCACAGCGCTTCTCA |
CCTCGCTGGGCCACATC |
| Ucp2 | NM_011671 |
GCCCCTTCACCTCTTTAGCA |
CCAAGCACTGGGAAGGTCTAAC |
| Vcam1 | NM_011693 |
CTCCCCTGAATACAAAACGATTG |
GCCCGTAGTGCTGCAAGTG |
Quantification of chemokines in
serum
Blood was collected by cardiac puncture of
anesthetized mice just before euthanasia. Isolated serum was
aliquoted for storage in −20°C. Cytokines in serum were quantified
with the Cytokine 16-plex panel (Quansys Biosciences, Inc., Logan,
UT, USA). This chemiluminescent Quansys Q-plex Array contained
IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, IL-12, IL-17,
MCP-1, IFNγ, TNFα, MIP-1α, GM-CSF and RANTES antibodies absorbed to
each well of a 96-well plate (Table
II). The lower level of detection was different for each
cytokine and ranged from 0.1 pg/ml (TNFα) to 5.0 pg/ml (MCP-1).
Serum samples were tested in triplicate. Data were collected using
Q-View Imager and software (Quansys Biosciences Inc.). Results are
presented in picograms of cytokine per ml of serum (pg/ml).
ELISA
Tumor lysates were prepared from frozen tumors by
homogenization in diluted Cell Lysis Buffer 10X (Cell Signaling
Technology, Inc., Beverly, MA, USA) containing 1 mM PMSF
(phenylmethanesulfonyl fluoride). The total protein concentration
of lysates was determined with Pierce BCA Protein Assay kit,
(Thermo Fisher Scientific Inc., Rockford, IL, USA), a Synergy HT
spectrophotometer and the Gene5 software (BioTech Instruments,
Winooski, VT, USA). The level of stromal derived factor 1 (SDF-1,
also known as chemokine CXCL12) in the lysates was determined using
Duo Set ELISA Development kit, according the manu facturer’s
instructions (cat # DY460, R&D Systems, Inc., Minneapolis, MN,
USA). Samples were assessed in triplicate. The results are
presented as picograms of SDF-1 protein per micrograms of total
protein content of the tumor lysate.
Western blotting
Tumor lysates for immunoblotting were prepared as
described above for ELISA. Novex Pre-Cast gradient gels (4–20%) and
NuPAGE Electrophoresis system (Invitrogen Life Technologies, Inc.,
Carlsbad, CA, USA) were used to resolve proteins from 8 μg
of total protein sample per well. Western transfer onto
polyvinylidene difluoride membranes (Amersham, Piscataway, NJ, USA)
was performed in an XCell II Blot Module (Invitrogen Life
Technologies, Inc.). After protein transfer, membranes were stored
at 4°C. For immunoblotting, membranes were incubated in 10%
fat-free powdered milk solution for 4 h at room temperature (RT).
Primary antibodies were applied for 1 h at RT, the membrane was
washed, and the secondary antibody was applied for 1 h at RT. The
following primary antibodies were used: monoclonal anti-HIF-1α (cat
# NB100-105, Novus Biologicals), polyclonal anti-HSP 27 (cat #
sc-1049, Santa Cruz Biotechnology), monoclonal anti-HSP 70 (cat #
sc-24, Santa Cruz Biotechnology), monoclonal anti-Ki67 (cat #
NBP1-40684, Novus Biologicals) rabbit polyclonal anti-MMP9 (cat #
ab38898, Abcam) and anti-actin (1–19)
(cat # sc1616, Santa Cruz Biotechnology). The secondary antibodies
were: anti-mouse (cat # 170-6516; Bio-Rad Laboratories, Inc.),
anti-goat (cat # sc-2020; Santa Cruz Biotechnology) and anti-rabbit
(cat # sc-2004, Santa Cruz Biotechnology).
Statistical analysis
The two-tailed Student’s t-test was used to analyze
RT-PCR and ELISA data. Mann-Whitney U test and Fisher’s exact test
were used to evaluate cytokines panel results. The software
DataAssist v3.0 (Applied Biosystems) and Sigma Plot v11 (Systat
Software Inc., San Jose, CA, USA) were applied.
Results
Effects on bone marrow gene
expression
The effects on the bone marrow of BALB/c mice 72 h
after tumor ablation by CITT therapy were measured by quantitative
real-time RT-PCR. Marrow RNA was assayed for 27 murine transcripts
(Table I). Four transcripts were
expressed at significantly higher levels for the mice treated with
tumor ablation (N=5) as compared to the control mice (N=4) with
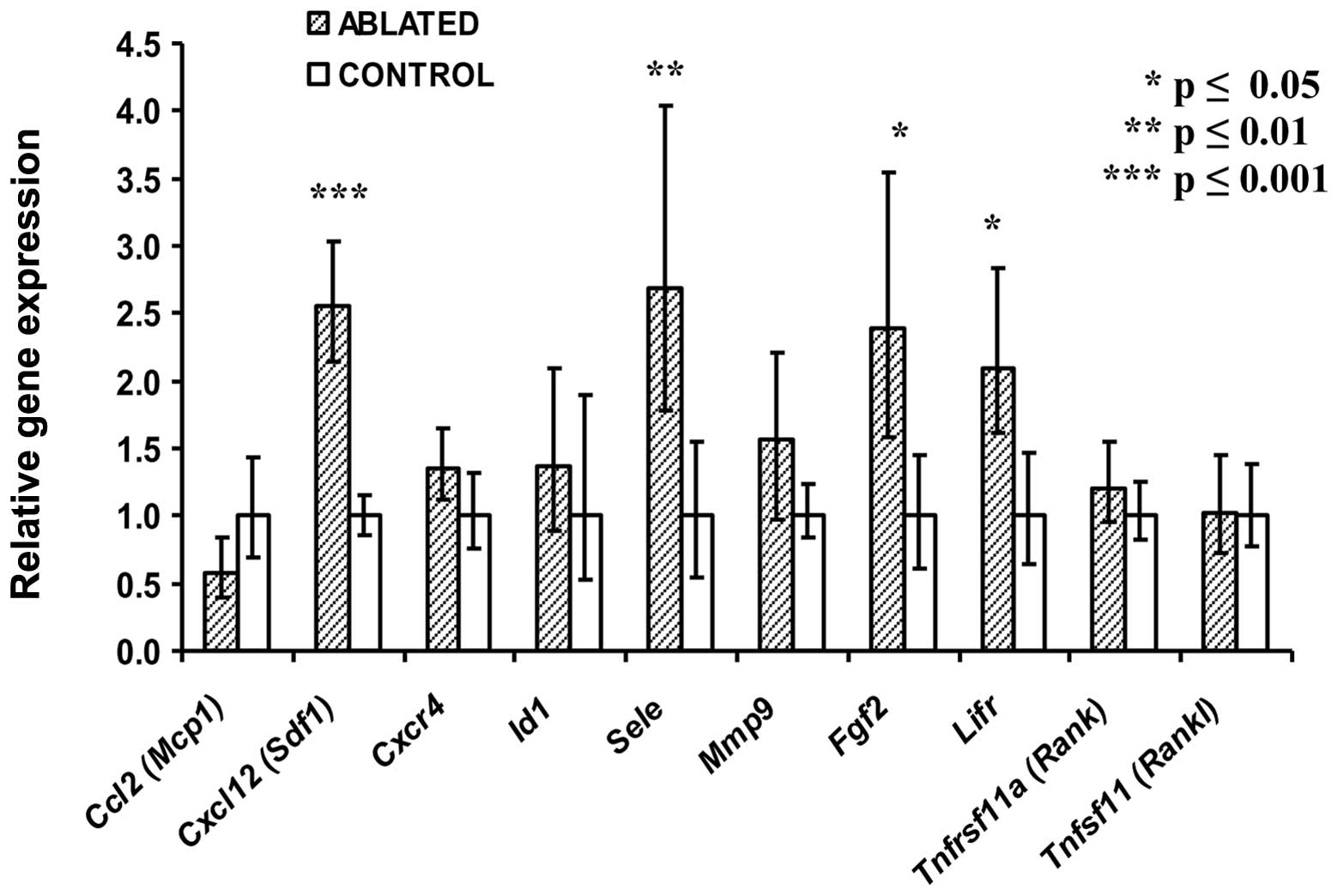
untreated tumors (Fig. 1).
Transcript levels for genes encoding stromal derived factor 1
(Cxcl12), E-selectin (Sele), leukemia inhibitory
factor 1 receptor (Lifr) and basic fibroblast growth factor
(Fgf2) ranged from an average of 2 to 2.5-fold higher
(P<0.05) in the treated as compared to the untreated mice.
Transcripts of other genes such as chemokine ligand-2 (Ccl2)
and chemokine receptor-4 (Cxcr4) were not affected in bone
marrow by tumor ablation (Fig. 1).
Four other examples of transcripts unaffected by the treatment are
also shown.
Effects on surviving tumor gene
expression
The effects on viable 4T1 tumor surrounding the site
of partial ablation were also measured by quantitative real-time
RT-PCR. Tumor RNA was assayed for 27 murine transcripts (Table I) for treated (N=5) and control
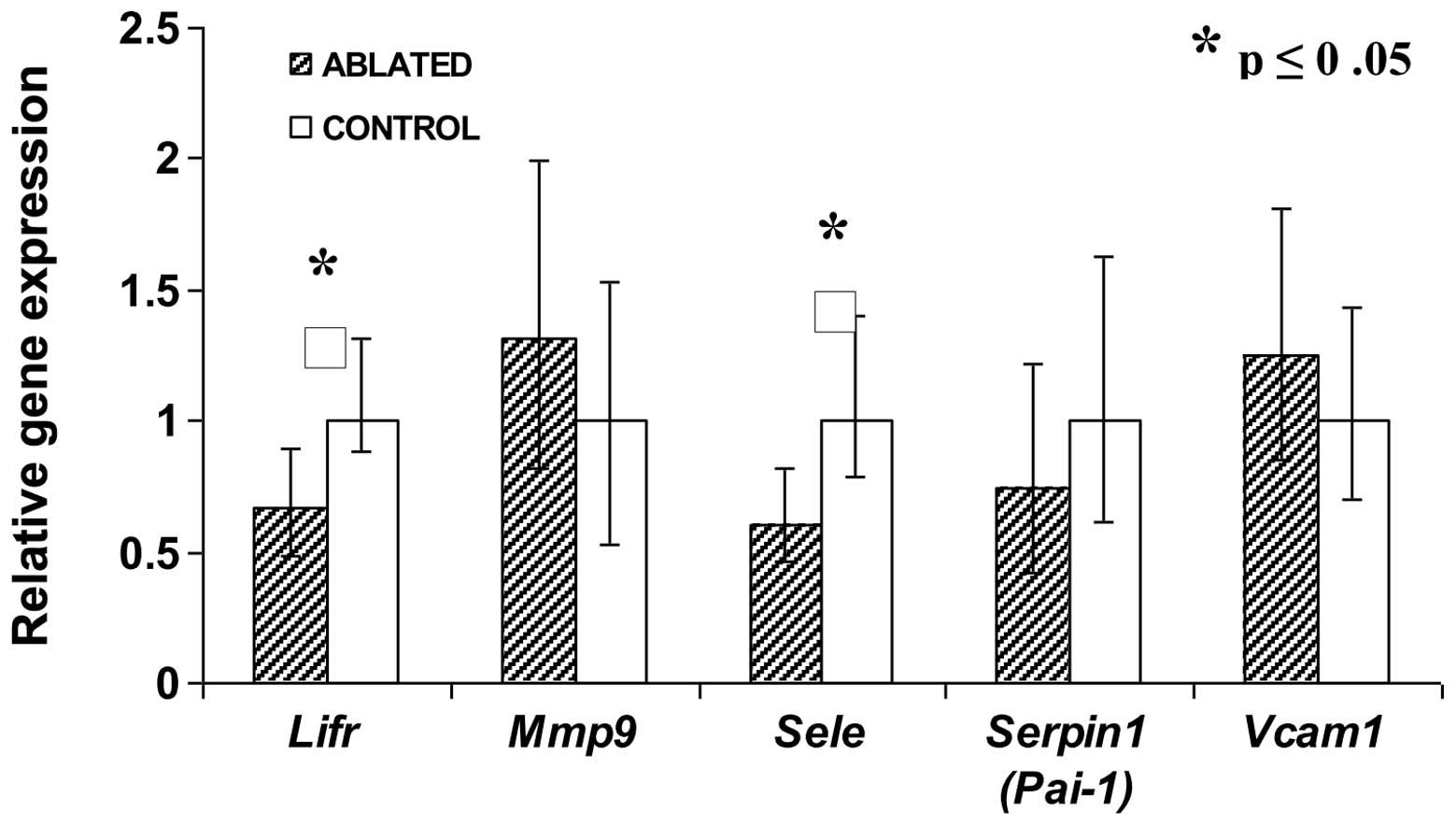
mice (N=4). Two transcripts were expressed at significantly lower
levels for the mice treated with tumor ablation (N=5) as compared
to the control mice (N=4) with untreated tumors (Fig. 2). Transcript levels of Lifr
and Sele genes for treated tumors was only ∼60% as high as
the control tumors (P<0.05). Transcripts of other genes such as
matrix metallopeptidase-9, serpin-1 and vascular cell adhesion
molecule-1 were not affected in tumor tissue that survived ablation
(Fig. 2).
Effects on surviving tumor protein
level
The effects of partial tumor ablation on protein
levels in viable tissue around the site of ablation 72 h after
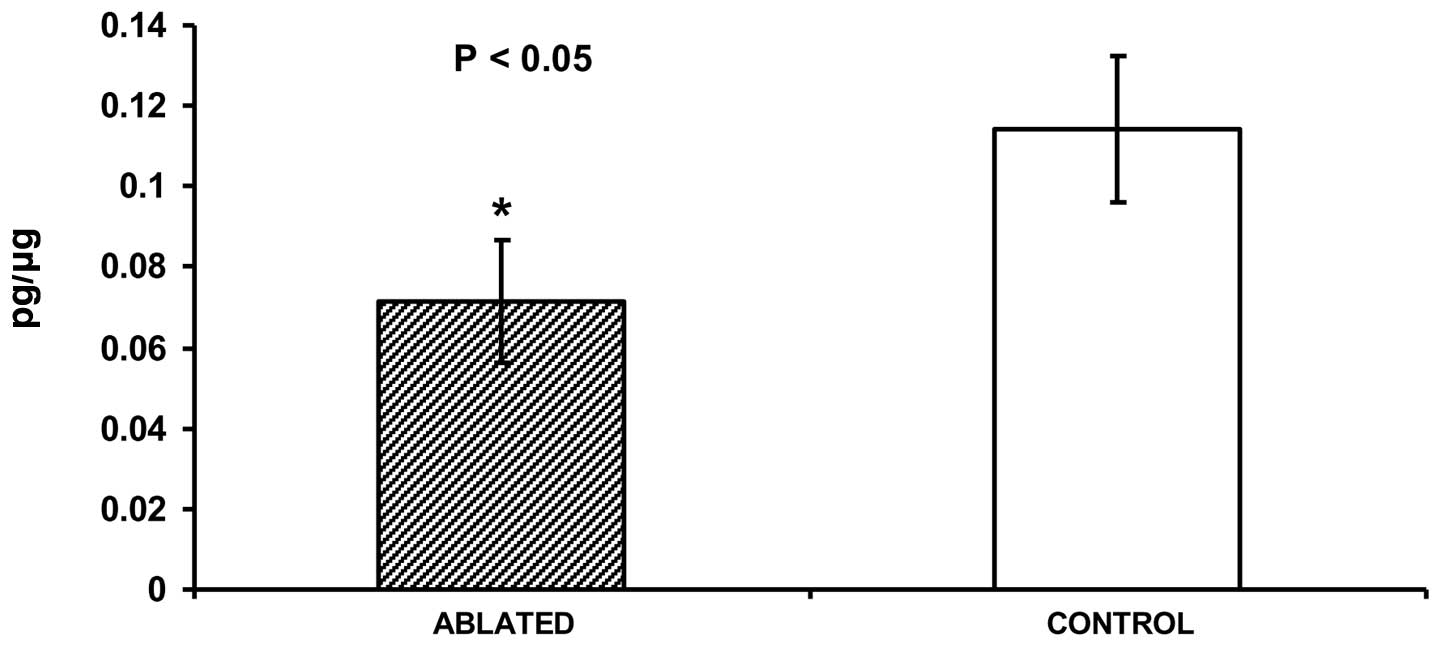
treatment were measured by immuno assays. As measured by ELISA,
levels of SDF-1 protein in tumor lysates were significantly lower
(0.071±0.016 pg/μg versus 0.113±0.017 pg/μg,
P<0.05) for ablated (N=4) as compared to control (N=3) tumor
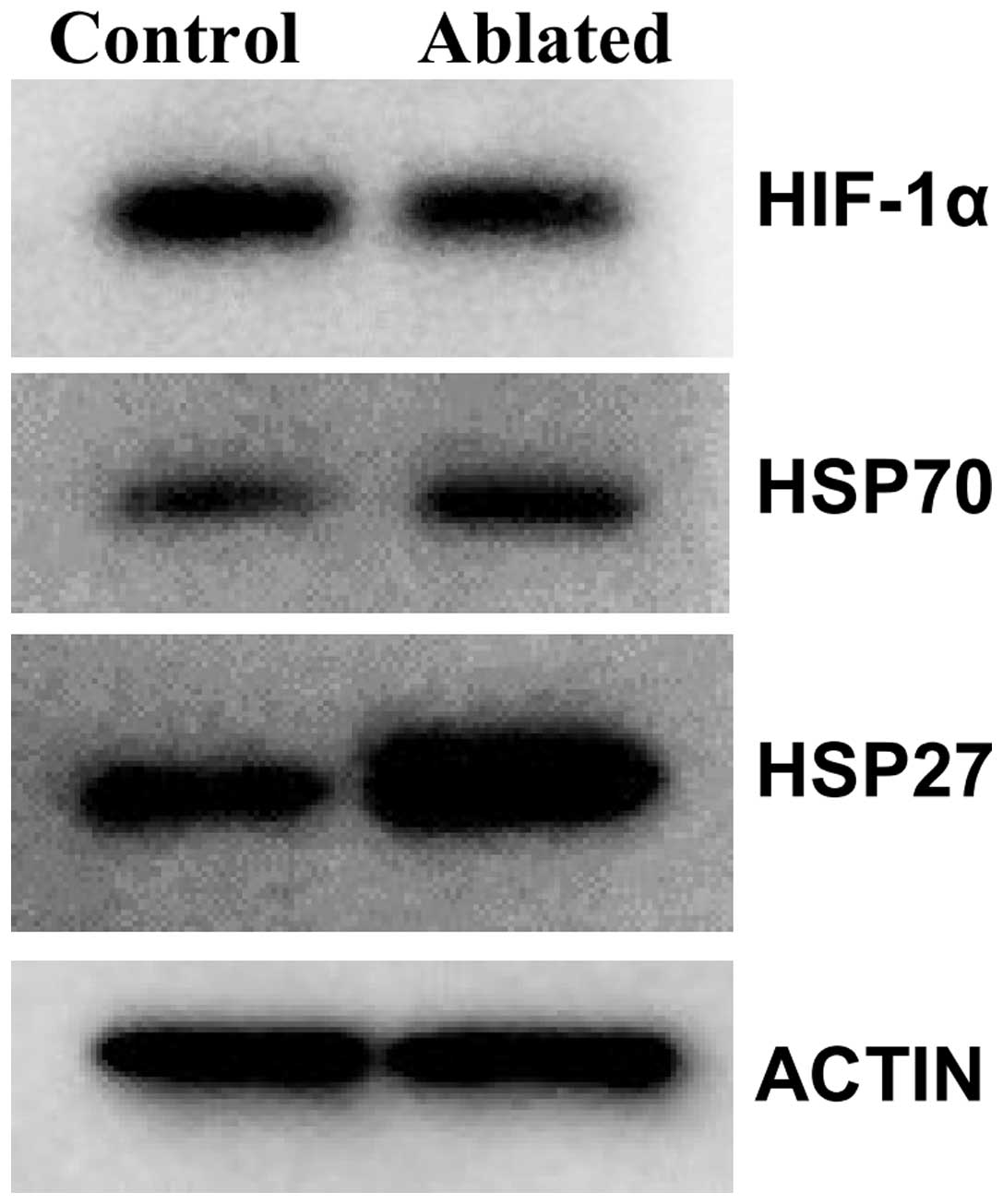
tissue (Fig. 3). Levels of six
proteins were measured in tumor lysates using western immuno
blotting (Table II). A
representative blot showing results for these proteins and actin, a
control protein, are presented (Fig.
4). Levels of heat shock proteins HSP70 and HSP27 were higher
for all individual samples for ablated (N=5) as compared to control
(N=4) tumor tissue; whereas levels of hypoxia inducible factor
(HIF-1α) were consistently lower for ablated tumors. Protein levels
for matrix metallopeptidase-9 and antigen Ki-67 were unaffected by
the treatment (data not shown).
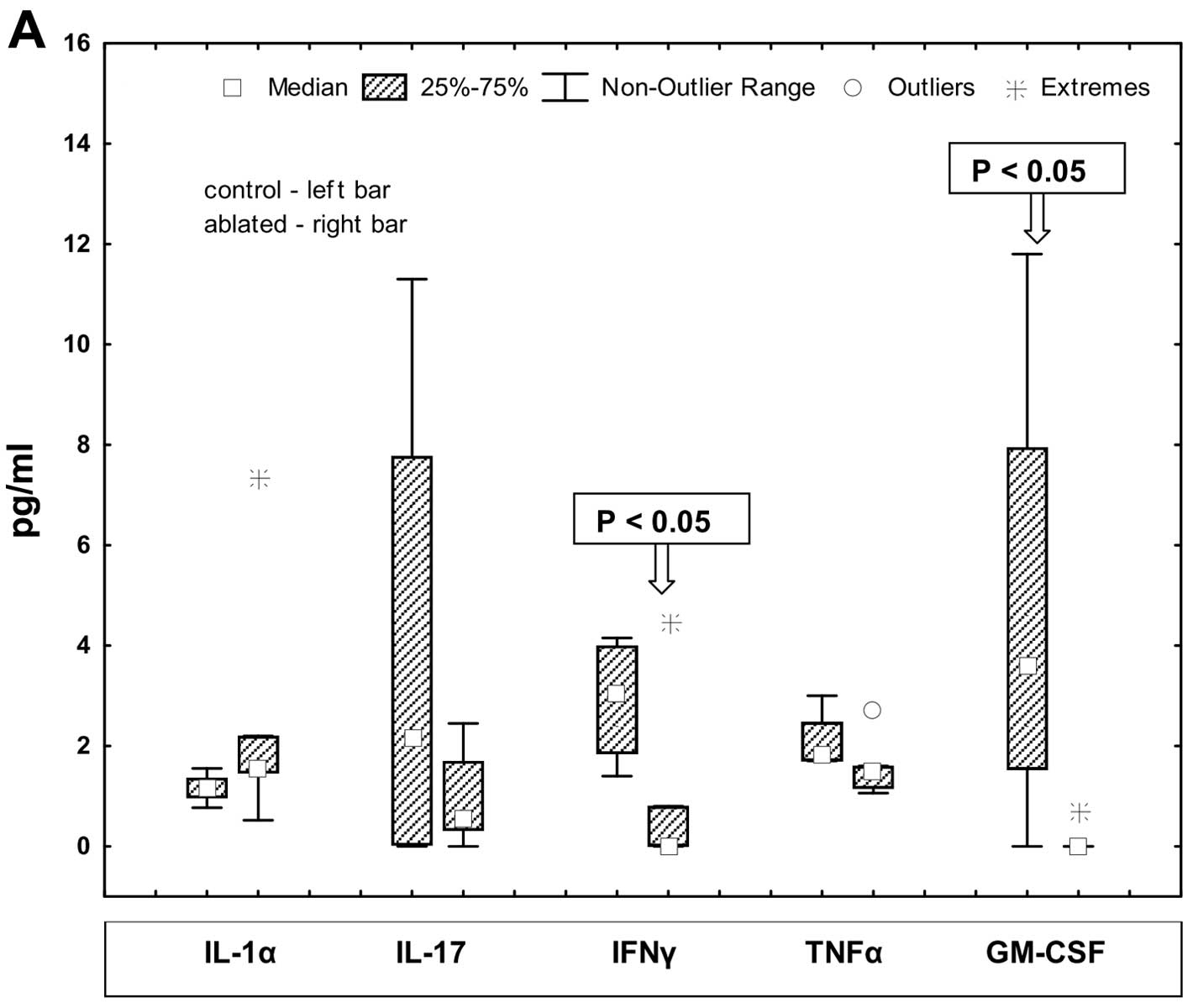
Effects on serum cytokine levels
The effects of partial tumor ablation on serum
cytokine levels 72 h after treatment were measured by multiplex
immunoassay. The results for 10 of the 16 cytokines are presented
(Fig. 5). The cytokines IL-1α,
IL-17, IFNγ, TNFα and GM-CSF were present in serum at low
concentrations. Other cytokines including IL-1β, IL-5, IL-6, MCP-1
and RANTES were present at higher concentrations. These ten
cytokines were present at detectable levels in serum from at least
five of the nine study mice and the inter-sample variability within
groups was large. Only two cytokines, interferon-γ (IFNγ) and
granulocyte-macrophage colony-stimulating factor (GM-CSF) showed
significant differences between groups. IFNγ and GM-CSF had a
significantly lower level in serum from the ablated mice as
compared to control mice (P<0.05). Notably, interleukin-1β
showed a trend towards lower values (P=0.154) in serum from ablated
mice.
Discussion
Thermal ablation has been used to treat tumors of
the liver (20), prostate
(21), and breast (22–24)
in humans and animal models. Results of these therapies can vary
greatly depending on many variables, including ablation modality,
operator techniques and tumor site. For example, radiofrequency
ablation can lead to accelerated perinecrotic outgrowth of
colorectal liver metastases (25);
whereas, we previously demonstrated that CITT therapy decreased
metastasis in rabbits with VX2 tumors (6). In the current study, a mouse mammary
carcinoma model was used to characterize the molecular changes that
occur in serum, bone marrow, and tumor surviving partial ablation.
At the transcriptional level, ablation increased Sele, Fgf2,
Lifr and Cxcl12 in marrow and decreased Lifr and
Sele in the surviving tumor. At the protein level, ablation
resulted in decreased levels of SDF1 and HIF-1α and increased
levels of HSP27 and HSP70 in the surviving tumor. In serum,
ablation decreased the concentration of the IFNγ and GM-CSF. Thus,
10 molecules have been identified that may be involved with
alteration of communication between the marrow and tumor as well as
alteration of homeostasis in surviving tumors.
The bone marrow responded to thermal ablation with
an increase in the Sele, Fgf2, Lifr and Cxcl12
transcripts. Sele encodes E-selectin, an adhesion molecule
expressed by activated endothelial cells while tumor cells express
E-selectin ligands. Interactions between these cell types through
E-selectin are thought to regulate cancer metastases (26). Thus, the possibility exists that
increased E-selectin could promote adhesion of cancer cells and
enhance seeding tumor cells into the marrow. Fibroblasts growth
factor 2 is a stromal cell mitogen and stimulates myelopoiesis in
marrow. However, in the presence of advanced, untreated human
breast and lung tumors, low levels of Fgf2 result in an arrest of
maturation of mesenchymal stromal cells in marrow (27). The response of Fgf2 to
ablation seen here may be an attempt of the marrow to re-establish
normal maturation for mesenchymal cells.
Leukemia inhibitory factor (LIF) is a cytokine that
also affects marrow mesenchymal cells (28,29).
Upregulation of the LIF receptor transcript (Lifr) in marrow
may indicate that tumor ablation influences not only maturation but
also differentiation of mesenchymal stromal cells. Stromal derived
factor 1 is a signaling molecule for communication between tumor
and marrow (30,31). The SDF1 receptor (CXCR4) is
expressed on breast and other epithelial cancer cells and has been
shown in multiple studies to be involved with metastasis (32–35).
Increase of expression of the Cxcl12 transcripts encoding
SDF1 in bone marrow may stimulate mobilization and recruitement of
immature hematopoietic cells, endothelial and smooth muscle
progenitor to neo-angiogenic niche (36,37).
However, it is unknown whether or not the elevation of
Cxcl12 transcript found in marrow post-ablation is evidence
of seeding premetastatic niches or the primary tumor site. In
summary, four transcripts have been identified which respond in
marrow to tumor ablation. Further investigation is needed to
confirm whether or not their encoded proteins play a role in
mesenchymal cells development or tumor dissemination after thermal
ablation.
Tumor tissue responded to thermal ablation with a
decrease in SDF1 and HIF-1α proteins, and an increase in HSP27 and
HSP70 proteins, and decreases in Lifr and Sele genes
transcripts. Our work previously showed that thermal ablation
decreases hypoxia in surviving tumor areas (7). The results presented here are
consistent with this effect in that SDF1 and hypoxia inducible
factor 1 (HIF-1α) are lower in treated than untreated tumors. These
changes could inhibit the metastatic potential of surviving tumor
cells or inhibit signaling between the marrow and tumor for the
recruitment of cells supportive of tumor growth such as
pro-angiogenic endothelial progenitor cells or marrow suppressor
cells (32,35,38,39).
The increase in heat shock proteins -70 and -27 after treatment may
be a stress response. Unfortunately, HSP70 may be acting to protect
the cancer cells as its expression has been associated with poor
prognosis (40). However, HSP70
may also be indicative of a boost in antitumor immune activity
post-ablation (41). Heightened
tumor HSP27 is also associated with poor prognosis and metastasis
(42) though evidence indicates
multiple potential roles in tumorigenesis including promotion of
tumor growth (43), regulation of
epithelial-mesenchymal transition (44), and conferral of chemo-resistance
(45). The increase of HSP27
implies that this heat shock protein should be closely monitored
during thermal ablation of tumors and may be a useful marker to
assist in determining appropriate treatment regimens in combination
with thermal ablation. The effects of ablation on the Sele
and Lifr genes were the opposite in tumor to that of marrow
where we proposed that these factors may be involved with
metastasis and mesenchymal cell development. It would be beneficial
if the decrease seen in tumor was the result of a dampened
potential for tumor cell proliferation and metastasis
post-treatment. In summary, two transcripts and four proteins have
been identified which respond in tumor tissue to ablation though
additional pre-clinical studies are needed to define their
functions and understand whether these molecules are involved with
pro- or anti-tumorigenic responses after thermal ablation.
Serum analysis revealed that thermal ablation
decreased concentrations of the granulocyte-macrophage stimulating
factor (GM-CSF) and interferon-γ (IFNγ). These molecules function
in inflammation and the immune response. Their levels are
influenced both by the presence of solid tumors (46,47)
and thermal ablation (48).
However, the significance of their low levels post-ablation is
unknown. These cytokines may be indicative of decreased need for
antitumor immune function, immunosuppression by the treatment, or
they may be associated with slowed tumor growth or smaller number
of tumor cells after partial ablation (49,50).
Future studies will be needed to determine the source, effects and
meaning of low systemic levels of these cytokines three days after
ablation therapy.
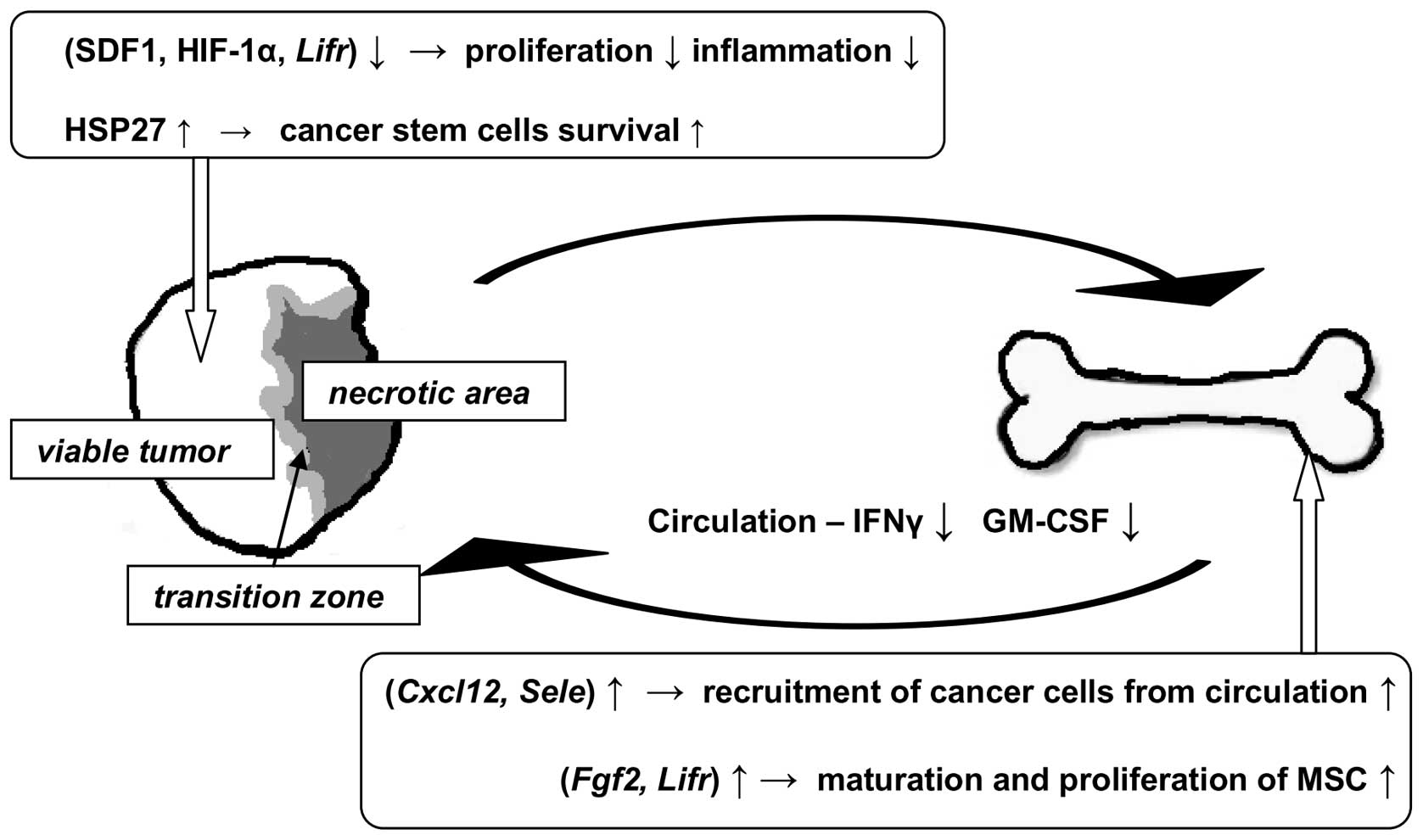
The major findings and proposed implications of the
study are summarized in Fig. 6.
Thermal ablation appears to decrease local and systemic decrease of
inflammation. Proliferation of viable tumor cells may decrease but
HSP27 may indicate the presence of a protected population of
surviving cancer cells that may be responsible for aggressive tumor
re-growth. Additionally, thermal ablation caused upregulation of
genes in bone marrow that are associated with maturation and
proliferation of mesenchymal stem and hematopoietic cells to
possibly replenish immune cells used during the initial response to
trauma. The increase of gene expression encoding SDF1, a ligand for
cancer cells, could increase the risk for circulating cancer cells
to establish themselves in marrow. We conclude that thermal
ablation clearly impacts transcript and protein levels of molecules
that may be involved in pro- and anti-tumor activity in the tumor
itself, in bone marrow and in serum. However, additional studies
are needed to determine the influence of these changes on cell
phenotype and their significance relative to cancer host or patient
outcomes.
Abbreviations:
|
CITT
|
conductive interstitial thermal
therapy
|
Acknowledgements
We thank Jessica S. Webber for
excellence in performing animal experiments and Azemat
Jamshidi-Parsian for help in preparation of western immunoblotting
assay. This work was supported, in part, by a grant from Fashion
Footwear Association of New York (FFANY/QVC) to G.S. and R.J.G. and
by NIH grants CA44114 (R.J.G).
References
|
1.
|
Shah DR, Green S, Elliot A, McGahan JP and
Khatri VP: Current oncologic applications of radiofrequency
ablation therapies. World J Gastrointest Oncol. 5:71–80. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Oto A, Sethi I, Karczmar G, et al: MR
imaging-guided focal laser ablation for prostate cancer: phase I
trial. Radiology. 267:932–940. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Merckel LG, Bartels LW, Kohler MO, et al:
MR-guided high-intensity focused ultrasound ablation of breast
cancer with a dedicated breast platform. Cardiovasc Intervent
Radiol. 36:292–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Shafirstein G, Hennings L, Kaufmann Y, et
al: Conductive interstitial thermal therapy (CITT) device
evaluation in VX2 rabbit model. Technol Cancer Res Treat.
6:235–246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Shafirstein G, Novák P, Moros EG, et al:
Conductive interstitial thermal therapy device for surgical margin
ablation: in vivo verification of a theoretical model. Int J
Hyperthermia. 23:477–492. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Shafirstein G, Kaufmann Y, Hennings L, et
al: Conductive interstitial thermal therapy (CITT) inhibits
recurrence and metastasis in rabbit VX2 carcinoma model. Int J
Hyperthermia. 25:446–454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Przybyla BD, Shafirstein G, Koonce NA,
Webber JS and Griffin RJ: Conductive thermal ablation of 4T1 murine
breast carcinoma reduces severe hypoxia in surviving tumour. Int J
Hyperthermia. 28:156–162. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Dromi SA, Walsh MP, Herby S, et al:
Radiofrequency ablation induces antigen-presenting cell
infiltration and amplification of weak tumor-induced immunity.
Radiology. 251:58–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
van der Bilt JD, Soeters ME, Duyverman AM,
et al: Perinecrotic hypoxia contributes to
ischemia/reperfusion-accelerated outgrowth of colorectal
micrometastases. Am J Pathol. 170:1379–1388. 2007.PubMed/NCBI
|
|
10.
|
Peters BA, Diaz LA, Polyak K, et al:
Contribution of bone marrow-derived endothelial cells to human
tumor vasculature. Nat Med. 11:261–262. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Bunt SK, Sinha P, Clements VK, Leips J and
Ostrand-Rosenberg S: Inflammation induces myeloid-derived
suppressor cells that facilitate tumor progression. J Immunol.
176:284–290. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Di Rosa F: T-lymphocyte interaction with
stromal, bone and hematopoietic cells in the bone marrow. Immunol
Cell Biol. 87:20–29. 2009.PubMed/NCBI
|
|
13.
|
Evans SS, Fisher DT, Skitzki JJ and Chen
Q: Targeted regulation of a lymphocyte-endothelial-interleukin-6
axis by thermal stress. Int J Hyperthermia. 24:67–78. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Peer AJ, Grimm MJ, Zynda ER and Repasky
EA: Diverse immune mechanisms may contribute to the survival
benefit seen in cancer patients receiving hyperthermia. Immunol
Res. 46:137–154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Baronzio G, Gramaglia A and Fiorentini G:
Hyperthermia and immunity. A brief overview. In Vivo. 20:689–695.
2006.PubMed/NCBI
|
|
16.
|
Chantrain CF, Feron O, Marbaix E and
DeClerck YA: Bone mrrow microenvironment and tumor progression.
Cancer Microenviron. 1:23–35. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Heppner GH, Miller FR and Shekhar PM:
Nontransgenic models of breast cancer. Breast Cancer Res.
2:331–334. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Lippold HJ: Quantitative succinic
dehydrogenases histochemistry. A comparison of different
tetrazolium salts. Histochemistry. 76:381–405. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Schmittgen TD, Lee EJ and Jiang J:
High-throughput real-time PCR. Methods Mol Biol. 429:89–98. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wiggermann P, Zeman F, Niessen C, et al:
Percutaneous irreversible electroporation (IRE) of hepatic
malignant tumours: contrast-enhanced ultrasound (CEUS) findings.
Clin Hemorheol Microcirc. 52:417–427. 2012.
|
|
21.
|
Uchida T, Nakano M, Hongo S, et al:
High-intensity focused ultrasound therapy for prostate cancer. Int
J Urol. 19:187–201. 2011. View Article : Google Scholar
|
|
22.
|
Zhao Z and Wu F: Minimally-invasive
thermal ablation of early-stage breast cancer: a systemic review.
Eur J Surg Oncol. 36:1149–1155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Yoon J, Cho J, Kim N, et al:
High-frequency microwave ablation method for enhanced cancer
treatment with minimized collateral damage. Int J Cancer.
129:1970–1978. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Hung WK, Mak KL, Ying M and Chan M:
Radiofrequency ablation of breast cancer: a comparative study of
two needle designs. Breast Cancer. 18:124–128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Nijkamp MW, van der Bilt JD, de Bruijn MT,
et al: Accelerated perinecrotic outgrowth of colorectal liver
metastases following radiofrequency ablation is a hypoxia-driven
phenomenon. Ann Surg. 249:814–823. 2009. View Article : Google Scholar
|
|
26.
|
St Hill CA: Interactions between
endothelial selectins and cancer cells regulate metastasis. Front
Biosci. 16:3233–3251. 2011.PubMed/NCBI
|
|
27.
|
Hofer EL, Labovsky V, La Russa V, et al:
Mesenchymal stromal cells, colony-forming unit fibroblasts, from
bone marrow of untreated advanced breast and lung cancer patients
suppress fibroblast colony formation from healthy marrow. Stem
Cells Dev. 19:359–370. 2010. View Article : Google Scholar
|
|
28.
|
Oskowitz AZ, Lu J, Penfornis P, et al:
Human multipotent stromal cells from bone marrow and microRNA:
regulation of differentiation and leukemia inhibitory factor
expression. Proc Natl Acad Sci USA. 105:18372–18377. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Whitney MJ, Lee A, Ylostalo J, Zeitouni S,
Tucker A and Gregory CA: Leukemia inhibitory factor secretion is a
predictor and indicator of early progenitor status in adult bone
marrow stromal cells. Tissue Eng Part A. 15:33–44. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
McAllister SS and Weinberg RA: Tumor-host
interactions: a far-reaching relationship. J Clin Oncol.
28:4022–4028. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Castano Z, Tracy K and McAllister SS: The
tumor macro-environment and systemic regulation of breast cancer
progression. Int J Dev Biol. 55:889–897. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Hirbe AC, Morgan EA and Weilbaecher KN:
The CXCR4/SDF-1 chemokine axis: a potential therapeutic target for
bone metastases? Curr Pharm Des. 16:1284–1290. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Meads MB, Hazlehurst LA and Dalton WS: The
bone marrow microenvironment as a tumor sanctuary and contributor
to drug resistance. Clin Cancer Res. 14:2519–2526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Cronin PA, Wang JH and Redmond HP: Hypoxia
increases the metastatic ability of breast cancer cells via
upregulation of CXCR4. BMC Cancer. 10:2252010. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Kucia M, Jankowski K, Reca R, et al:
CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol
Histol. 35:233–245. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Petit I, Jin D and Rafii S: The
SDF-1-CXCR4 signaling pathway: a molecular hub modulating
neo-angiogenesis. Trends Immunol. 28:299–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Du R, Lu KV, Petritsch C, et al: HIF1alpha
induces the recruitment of bone marrow-derived vascular modulatory
cells to regulate tumor angiogenesis and invasion. Cancer Cell.
13:206–220. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Sceneay J, Chow MT, Chen A, et al: Primary
tumor hypoxia recruits CD11b+/Ly6Cmed/Ly6G+
immune suppressor cells and compromises NK cell cytotoxicity in the
premetastatic niche. Cancer Res. 72:3906–3911. 2012.PubMed/NCBI
|
|
40.
|
Murphy ME: The HSP70 family and cancer.
Carcinogenesis. 34:1181–1188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Teng LS, Jin KT, Han N and Cao J:
Radiofrequency ablation, heat shock protein 70 and potential
anti-tumor immunity in hepatic and pancreatic cancers: a
minireview. Hepatobiliary Pancreat Dis Int. 9:361–365.
2010.PubMed/NCBI
|
|
42.
|
Ciocca DR and Calderwood SK: Heat shock
proteins in cancer: diagnostic, prognostic, predictive, and
treatment implications. Cell Stress Chaperones. 10:86–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Gibert B, Hadchity E, Czekalla A, et al:
Inhibition of heat shock protein 27 (HspB1) tumorigenic functions
by peptide aptamers. Oncogene. 30:3672–3681. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Wei L, Liu TT, Wang HH, et al: Hsp27
participates in the maintenance of breast cancer stem cells through
regulation of epithelial-mesenchymal transition and nuclear
factor-kappaB. Breast Cancer Res. 13:R1012011. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Hsu HS, Lin JH, Huang WC, et al:
Chemoresistance of lung cancer stemlike cells depends on activation
of Hsp27. Cancer. 117:1516–1528. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Lin EY, Gouon-Evans V, Nguyen AV and
Pollard JW: The macrophage growth factor CSF-1 in mammary gland
development and tumor progression. J Mammary Gland Biol Neoplasia.
7:147–162. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Tsavaris N, Kosmas C, Vadiaka M,
Kanelopoulos P and Boulamatsis D: Immune changes in patients with
advanced breast cancer undergoing chemotherapy with taxanes. Br J
Cancer. 87:21–27. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Haen SP, Pereira PL, Salih HR, Rammensee
HG and Gouttefangeas C: More than just tumor destruction:
immuno-modulation by thermal ablation of cancer. Clin Dev Immunol.
2011:1602502011.PubMed/NCBI
|
|
49.
|
Li J, Bouton-Verville H, Holmes LM, et al:
Inhibition or promotion of tumor growth by granulocyte-macrophage
colony stimulating factor derived from engineered tumor cells is
dose-dependent. Anticancer Res. 24:2717–2721. 2004.PubMed/NCBI
|
|
50.
|
Eubank TD, Roberts RD, Khan M, et al:
Granulocyte macrophage colony-stimulating factor inhibits breast
cancer growth and metastasis by invoking an anti-angiogenic program
in tumor-educated macrophages. Cancer Res. 69:2133–2140. 2009.
View Article : Google Scholar
|