Introduction
More than half a million new cases of head and neck
squamous cell carcinoma (HNSCC) occurred in 2008 worldwide. Oral
squamous cell carcinoma (OSCC) is the most common HNSCC neoplasia,
with over a quarter of a million new cases reported in 2008 with a
mortality rate of ≤50% (1).
Despite advances in our knowledge of the disease and in
chemotherapy, radiotherapy and surgery, little improvement in the
relative survival has been seen in OSCC during the past several
decades (2). Therefore, a greater
understanding of the pathogenesis of OSCC is needed for the
development of more effective therapeutic approaches.
Cancer cells acquire abnormalities in multiple
oncogenes and tumor suppressor genes. Overexpression and
constitutive activation of several oncogenes support the
proliferation, invasion, and metastasis of cancer cells. However,
inactivation of a single critical oncogene can induce their
terminal differentiation or apoptosis and this has suggested that
this ‘oncogene addiction’ is an Achilles heel for cancers,
providing therapeutic opportunities in many human malignancies
(3). In HNSCC, including OSCC, the
overexpression of epidermal growth factor receptor (EGFR) has been
shown to be correlated with lymph node or distant metastasis, risk
of locoregional recurrence and poor prognosis (4). Cetuximab, which targets EGFR, is used
to treat patients with local advanced, recurrent or metastatic
HNSCC. Therapies combining radiotherapy or platinum-based
chemotherapy with cetuximab have improved locoregional control and
overall survival, and reduced mortality (5,6).
However, cetuximab is the only molecularly targeted drug available
for OSCC patients. In this study, we have attempted to identify
other target molecules by microarray analysis and to determine
whether targeting such molecules could provide novel, potential
therapeutic approaches for the treatment of OSCC patients.
Materials and methods
Cells and cell culture
In this study, we used 10 human oral cancer cell
lines, which were green fluorescent protein (GFP)-SAS (7), Ca9-22, HSC2, HSC3, HSC4, KB, SCC111,
SCC66, SCC9 and SCC25, and an immortalized non-neoplastic human
keratinocyte cell line, HaCaT, as described previously (8,9).
These cells were grown in Dulbecco's modified Eagle's medium (DMEM;
Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS;
Biosource, Camarillo, CA, USA), 100 U/ml penicillin and 100 μg/ml
streptomycin (Wako), referred to here as complete medium.
Primary cell cultures were established from
patients' OSCC tumors. After surgical excision, tumor tissue was
rinsed several times with complete medium, cut into small fragments
and dissociated with 0.1% collagenase (Wako) at 37°C for 2 h. Cell
suspensions were filtered through a 70-μm nylon mesh cell strainer
(BD, Franklin Lakes, NJ, USA). The cells were collected by
centrifugation, resuspended in keratinocyte serum-free medium
(K-SFM; Life Technologies, Carlsbad, CA, USA), and plated on a
plastic surface to grow. All cells were incubated in a humidified
atmosphere of 95% air and 5% CO2 at 37°C.
Samples from patients
OSCC and normal oral mucosa epithelial tissues were
obtained from patients at the Ehime University Hospital between
June 2009 and December 2012. Four primary OSCC cultured cells were
established, two from the tongue (a 70-year-old male with a T4N2cM0
stage tumor and a 40-year-old male with a T2N0M0 stage tumor) and
two from the gingiva (a 66-year-old male with a T4aN0M0 stage tumor
and an 80-year-old female with a T2N0M0 stage tumor). Clinical
stages were defined according to the tumor-node-metastasis (TNM)
classification of malignant tumors (Union Internationale Contre le
Cancer). The Institutional Review Board at Ehime University
Hospital approved this study (no. 0904009), and appropriate written
informed consent was obtained from each patient.
Microarray analysis
To identify oncogenic genes in OSCC, the Applied
Biosystems Chemiluminescent RT-IVT Labeling kit (Life Technologies)
was used to convert total RNA to digoxigenin (DIG)-labeled cRNA.
Total RNA was extracted by lysing the cells or tissues using ISOGEN
(NipponGene, Tokyo, Japan). Tissues were homogenized in 0.5 ml
ISOGEN using TissueLyser (Qiagen, Valencia, CA, USA).
Double-stranded cDNA was generated from 1 μg total RNA, transcribed
using DIG-labeled nucleotides (Roche Diagnostics, Basel,
Switzerland), fragmented, and hybridized to Human Genome Survey
Arrays (Life Technologies) according to the manufacturer's
instructions. After washing each array, the signal was developed
using a chemiluminescent detection kit (Life Technologies).
Processed arrays were scanned with a 1700 chemiluminescent
microarray analyzer (Life Technologies).
We clarified the molecular mechanisms of the growth
inhibitory effects using synthetic small interfering RNA (siRNA)
specific for Akt1 (siAkt1). Total RNA was extracted from GFP-SAS
cells treated with either 10 nM siAkt1 or non-targeting siRNA
(siNT), 100 ng total RNAs were labeled using a GeneAtlas™ 3′IVT
Express kit assay (Affymetrix, Santa Clara, CA, USA), and
hybridized onto Affymetrix Human Genome U219 Array Strips
(Affymetrix), which included >530,000 probes covering over
36,000 transcripts and variants, according to the manufacturer's
instructions. After washing and staining the array strips, they
were scanned using the GeneAtlas™ System (Affymetrix).
These results were analyzed using the GeneSpring GX
12.1 (Agilent Technologies, Santa Clara, CA, USA) and Ingenuity
Pathway Analysis software (IPA; Ingenuity® Systems,
www.ingenuity.com, Redwood City, CA, USA). Functional
analysis with IPA software identified the biological functions and
diseases that were most significant in our data set. Fischer's
exact test was used to calculate p-values for the probability that
each biological function or disease assigned to a data set was due
to chance alone. The raw microarray data have been deposited in
Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.-gov/geo, experiment nos.
GSE36090 and GSE56233) according to the minimum information about
microarray experiment (MIAME) guidelines.
Western blot analysis
Cells were grown in monolayers for 48 h and lysed
with lysis buffer of 0.5 M EDTA and 1% Triton X-100 (Sigma-Aldrich,
St. Louis, MO, USA) in phosphate-buffered saline (PBS) containing a
protease inhibitor cocktail and phosphatase inhibitor (Roche
Diagnostics). Tissues were homogenized in 500 μl lysis buffer using
a TissueLyser (Qiagen). The samples were centrifuged at 15,000 × g
for 15 min at 4°C, and the supernatants were electrophoresed on
SDS-polyacrylamide gels and transferred to polyvinylidene
difluoride membranes (Millipore, Bedford, MA, USA). The membranes
were blocked with 5% non-fat dried milk (Wako) in 25 mM Tris-HCl,
125 mM NaCl, and 0.1% Tween-20 (TBS-T; Sigma-Aldrich) for 1 h at
room temperature. They were then probed with monoclonal rabbit
anti-human Akt1 antibody (Cell Signaling Technology, Danvers, MA,
USA) or monoclonal mouse anti-β-tubulin antibody (BD) at a 1:1,000
dilution in 5% non-fat dried milk in TBS-T for 1 h at room
temperature, followed by horseradish peroxidase-conjugated
secondary antibodies against rabbit or mouse IgG (GE Healthcare,
Little Chalfont, UK) for 1 h at room temperature. The immune
complexes were visualized using the enhanced chemiluminescence
(ECL) Prime Western Blotting Detection Reagent (GE Healthcare). The
density of the visualized immune complexes was digitized using
RAS3000 (Fujifilm, Tokyo, Japan).
Immunohistochemistry
The surgically resected OSCC specimens were fixed in
phosphate-buffered 10% formalin and embedded in paraffin. A series
of 4-μm thick tissue sections were prepared from each sample.
Immunohistochemical staining was performed using the
avidin-biotin-peroxidase complex method. Briefly, the sections were
deparaffinized, pretreated with 10 mM citrate buffer (pH 6.0) at
95°C for 10 min, cooled at room temperature for 30 min, and
incubated with 0.3% H2O2 in distilled water
for 10 min to block endogenous peroxidase activity. After blocking
with 3% horse serum in PBS, the sections were incubated overnight
at 4°C with a 1:400 dilution of a monoclonal rabbit antibody
specific for human Akt1 (Cell Signaling Technology). After washing,
the sections were overlaid with biotinylated anti-rabbit antibody
(Vector Laboratories, Burlingame, CA, USA) at room temperature for
60 min, washed in PBS, and then labeled with
streptavidin-peroxidase complex (Vector Laboratories). The
peroxidase reaction was developed with 3′3-diaminobenzidine as a
chromogen. The sections were counterstained with hematoxylin,
dehydrated with ethanol, treated with xylene, and enclosed in
synthetic resin. Negative controls omitted the primary antibody.
Cases were considered to be Akt1-positive if the cell nuclei and
cytoplasm of >50% of the cancer cells were labeled.
Design and transfection of siRNA
We designed and synthesized five siAkt1s. The target
sequences were optimized for maximum target-gene silencing, minimum
sequence-specific cross reactivity (off-target effects) and
avoidance of single nucleotide polymorphisms (SNPs). Synthetic
siRNA targeting GFP (siGFP) or siNT were used as negative controls.
Transfection was performed using Lipofectamine RNAiMAX (Life
Technologies) mixed with 10 nM siRNAs for western blotting and the
cell growth assays. The sequences of synthetic siRNAs used were as
follows: siAkt1-6: 5′-GAG CGG GAG GAG UGG ACA ATT-3′ (sense) and
5′-UUG UCC ACU CCU CCC GCU CTT-3′ (antisense); siAkt1-22: 5′-CCA
UGA AGA UCC UCA AGA ATT-3′ (sense) and 5′-UUC UUG AGG AUC UUC AUG
GTT-3′ (antisense); siAkt1-33: 5′-CCA AGG AGA UCA UGC AGC ATT-3′
(sense) and 5′-UGC UGC AUG AUC UCC UUG GTT-3′ (antisense);
siAkt1-56: 5′-GGG UUU ACC CAG UGG GAC ATT-3′ (sense) and 5′-UGU CCC
ACU GGG UAA ACC CTT-3′ (antisense); siAkt1-58: 5′-GGA CAG AGG AGC
AAG GUU UTT-3′ (sense) and 5′-AAA CCU UGC UCC UCU GUC CTT-3′
(antisense); siGFP: 5′-CUA CAA CAG CCA CAA CGU CTT-3′ (sense) and
5′-GAC GUU GUG GCU GUU GUA GTT-3′ (antisense); siNT: 5′-UAC GUA CUA
UCG CGC GGA UTT-3′ (sense) and 5′-AUC CGC GCG ATA GUA CGU ATT-3′
(antisense).
Cell growth assay
Cells were seeded into 96-well plates in complete
medium with 10 nM synthetic siRNA and 0.2% Lipofectamine RNAiMAX in
a final volume of 100 μl. Cell growth was evaluated after 72 h
using WST-8 assays (Cell Counting Kit-8; Dojindo, Kumamoto,
Japan).
Three-dimensional collagen-gel assay
Eight volumes of rat tail type I collagen suspension
(BD) were mixed with one volume of 10-fold concentrated DMEM
(Sigma-Aldrich) and one volume of reconstruction buffer (2.2 g
NaHCO3 and 4.77 g HEPES in 100 ml of 0.05 N NaOH;
Sigma-Aldrich). The collagen gel and carcinoma cells
(5×104 cells/well) transfected with each siRNA were
together poured into a 24-well plate (0.5 ml/well). After
incubation for 30 min at 37°C to permit complete gelation, DMEM
complete medium was added; the medium was changed every other day.
After incubation for 4 days, the cells contained within the gel
were recovered by treatment with 0.1% collagenase and 0.5%
trypsin-5.3 mM EDTA (Invitrogen). The cells were counted with a Z1
Coulter® particle counter (Beckman Coulter, Fullerton,
CA, USA).
Xenograft model and tumor therapy
GFP-SAS cells (1×106/site) complexed with
Matrigel (BD) in 100-μl aliquots were injected subcutaneously at
two sites in the flanks of male athymic nude mice (CLEA Japan,
Tokyo, Japan). Two weeks later, tumor-bearing nude mice were
randomly divided into siGFP, siAkt1-22 or siAkt1-58 treatment
groups. Complexes of 40 μM siRNAs with atelocollagen (AteloGene;
Koken, Tokyo, Japan) were injected into mouse tail veins every 3
days (10–14). Tumor diameters were measured at
regular intervals with digital calipers, and tumor volumes
(mm3) were calculated using the following formula:
length × width × height × 0.523. Three mice were used in each
group. Four weeks after the first administration of siRNAs, the
mice were sacrificed humanely, the GFP-SAS xenografts, lungs,
livers and kidneys were dissected, and Akt1 protein expression
levels were determined by western blotting. The animal studies were
approved by the Ehime University Animal Care Committee (no.
MA-17-14).
Real-time quantitative reverse
transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues by
homogenization in 0.5 ml ISOGEN (NipponGene) using a TissueLyser
(Qiagen). The relative mRNA levels were quantified using the
comparative CT method (ΔΔCT method) by
qRT-PCR using the TaqMan® system. Hydroxymethylbilane
synthase (HMBS) was used as an internal control. PCR amplification
was performed in a 10-μl final reaction mixture containing 5 μl 2X
Quantitect RT-PCR Master Mix, 0.1 μl Quantitect RT mix (Qiagen),
0.5 μl TaqMan probe and primers (Life Technologies), and 100 ng
total RNA. Thermal cycling conditions comprised an initial step at
95°C for 10 min, followed by 45 cycles at 95°C for 10 sec, 60°C for
10 sec and 72°C for 5 sec. The TaqMan® probe and primers
for interferon response genes, interferon stimulated gene factor 3γ
(ISGF-3γ), 2′, 5′-oligoadenylate synthetase 2 (OAS2),
interferon-induced myxovirus resistance protein 1 (MX1) and HMBS
were purchased from Life Technologies. The 5′-fluorescent reporter
dye fluorescence was detected using a LightCycler (Roche
Diagnostics).
Statistical analysis
All in vitro experiments were performed in
triplicate and repeated 3 times. Student's t-test was used to
determine the significance of differences between the groups, with
values of P<0.05 considered statistically significant.
Results
Overexpression of Akt1 in human OSCC
cells and tissues
Using Human Genome Survey Arrays, we determined the
gene expression profiles in 10 primary OSCC tissues, 10 human OSCC
cell lines, 3 normal oral mucosal tissues and a human immortalized
non-neoplastic keratinocyte cell line. We evaluated gene expression
relative to the signal to noise (S/N) ratio, with an S/N ratio
<3 interpreted as lack of gene expression and an S/N ratio >3
interpreted as positive gene expression. The expression of 7,417
genes was undetectable in all normal oral mucosal tissues and in a
non-neoplastic epithelial cell line. The only gene we identified as
expressed in all OSCC tissues and cultured cells was Akt1 (Fig. 1A).
To confirm the microarray data, we examined the
expression of Akt1 protein in human OSCC cells and tissues by
western blot analysis. All human OSCC cells expressed Akt1 protein
at a much higher level than the non-neoplastic epithelial cell
line, HaCaT (Fig. 1B).
Subsequently, we compared the expression levels of Akt1 protein in
primary OSCCs with adjacent normal oral mucosal tissues from the
same patient and found that Akt1 protein was more highly expressed
in the tumor tissues than in the normal tissues (Fig. 1C). Furthermore, immunohistochemical
examination showed the expression of Akt1 protein in 59 of 63 (94%)
primary OSCC tissues. Akt1 staining was diffusely positive in OSCCs
but not in normal epithelium (Fig.
2). These results suggested that Akt1 was commonly
overexpressed in OSCC.
Effect of siAkt1 on the growth of human
OSCC cells in vitro
We designed five siRNAs, siAkt1-6, 22, 33, 56 and
58, and transfected them into GFP-SAS cells at a concentration of
10 nM, to avoid off-target effects and interferon responses.
Synthetic siAkt1-22 and 58 had potent RNA interference (RNAi)
effects (Fig. 3A). When the
effects of these two siRNAs on cell growth were tested in human
OSCC cells, we found that the knockdown of Akt1 expression by
siAkt1-22 and 58 significantly inhibited the growth of GFP-SAS
cells by between 49 and 94% compared with the control, siGFP
(Fig. 3B). We also examined their
effects on the invasive growth of OSCC cells using the collagen gel
culture system. Both siAkt1-22 and 58 reduced the invasive growth
of GFP-SAS cells by 52 and 63%, respectively (Fig. 3C).
Effect of siAkt1 on the in vivo growth of
human OSCC cells
We assessed the growth inhibitory effect of siAkt1
in vivo using a mouse model. We selected GFP-SAS cells for
the in vivo assay because, of the human OSCC cells we used,
only these cells showed stable tumorigenicity. We administered
siAkt1/atelocollagen complexes into mouse tail veins every 3 days,
on 9 occasions. We found that these complexes markedly reduced the
size of subcutaneously xenografted GFP-SAS tumors, compared with
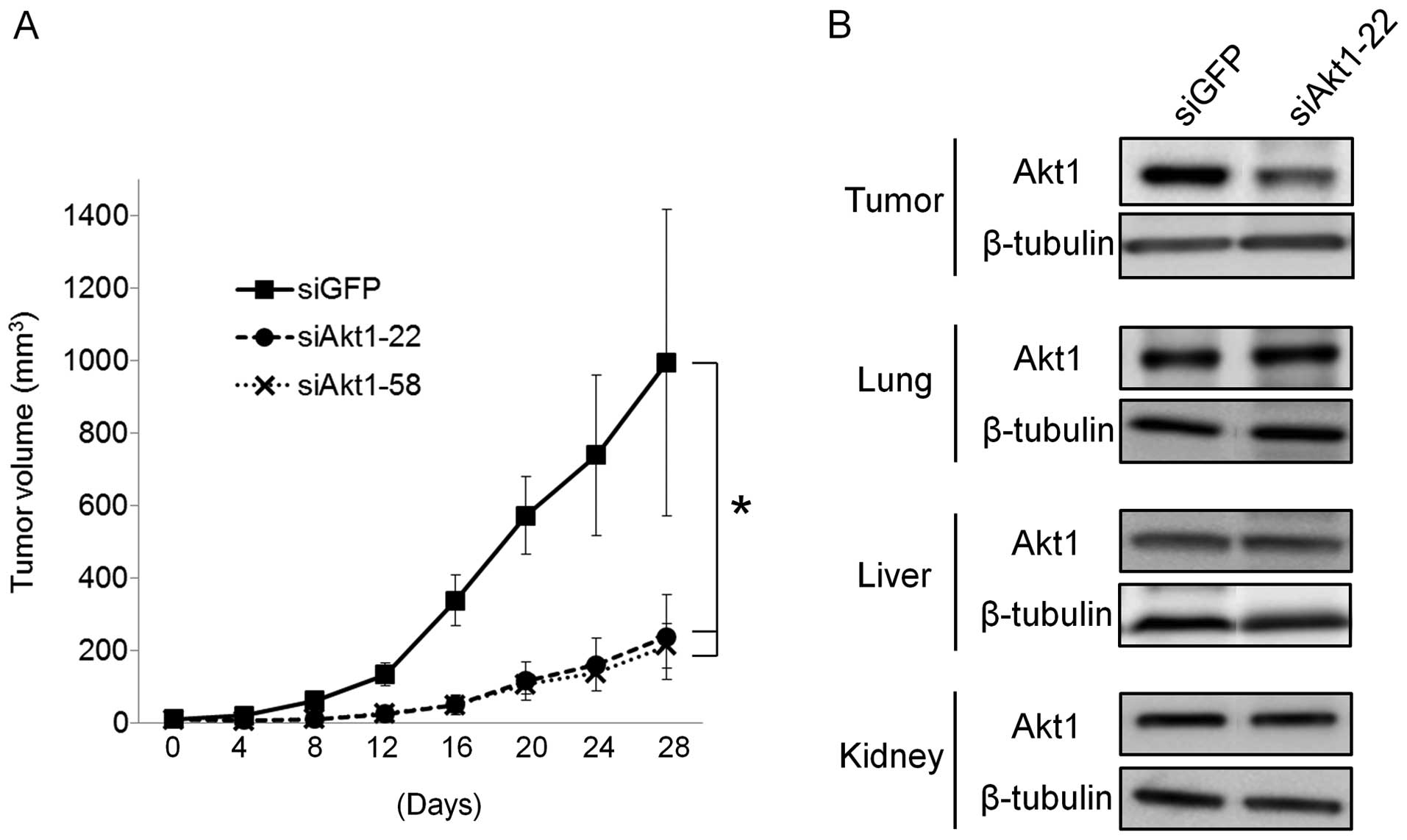
the siGFP controls (Fig. 4A).
During the administration of siAkt1, no reduction in food intake or
body weight was observed in the mice.
Only siAkt1-22 can target both human and mouse Akt1.
The expression of Akt1 in excised tumor tissues was markedly
suppressed by 58% in the groups injected with
siAkt1-22/atelocollagen complex systemically, but Akt1 expression
levels in the lung, liver and kidney did not change (Fig. 4B). We also checked whether the
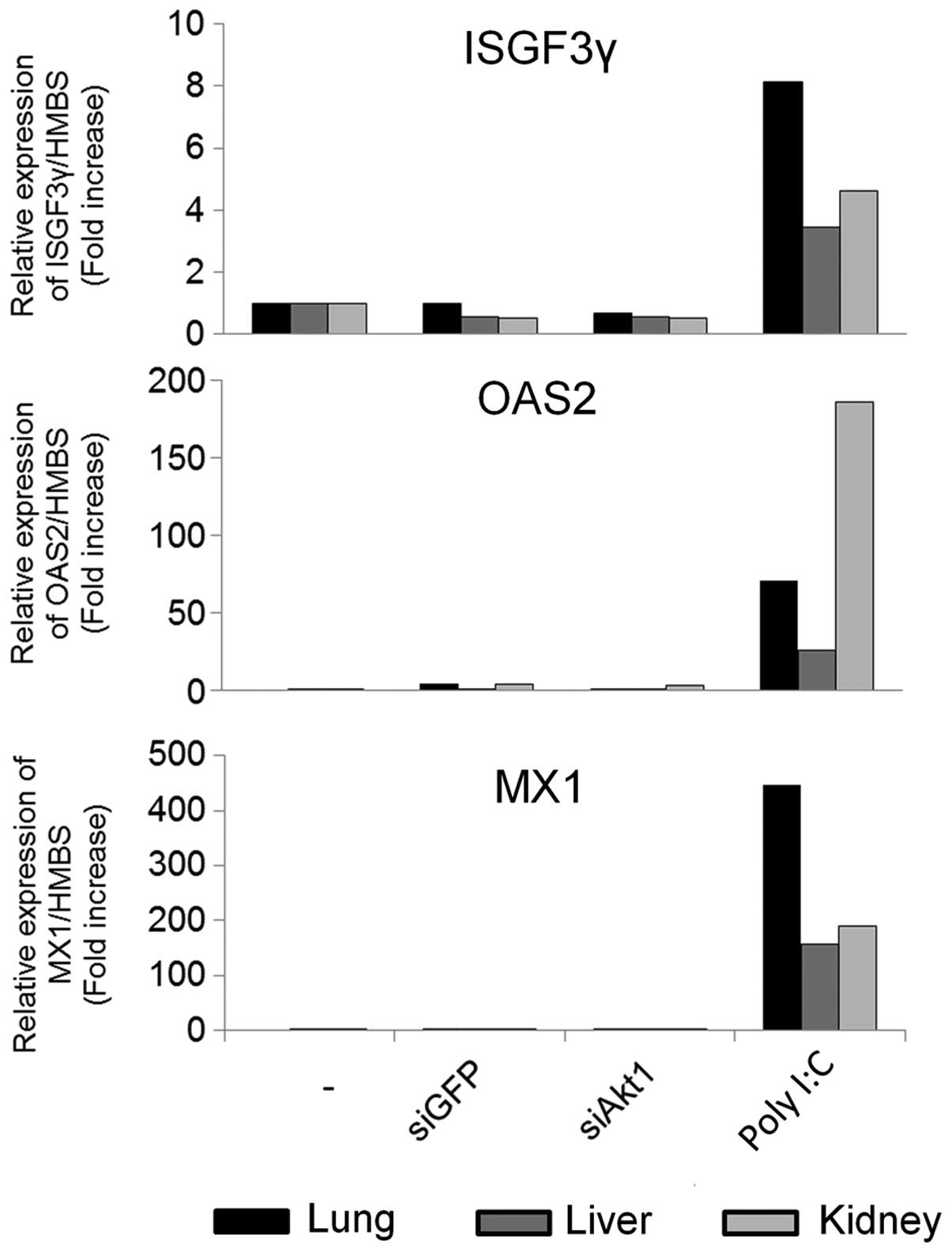
systemic delivery of atelocollagen-mediated siRNA induced
interferon responses in mice. After injecting the
siRNA/atelocollagen i.v., we evaluated mRNA expression of the
interferon response genes, ISGF3γ, OAS2, and MX1, in lung, liver
and kidney by qRT-PCR. We compared this to a positive control group
injected with poly(I:C), which induced a strong interferon
response, but the siRNAs did not (Fig.
5). These results indicated that atelocollagen specifically
delivered siRNAs into tumor tissues without inducing an interferon
response.
Effect of siAkt1 on the growth of human
OSCC primary cultured cells
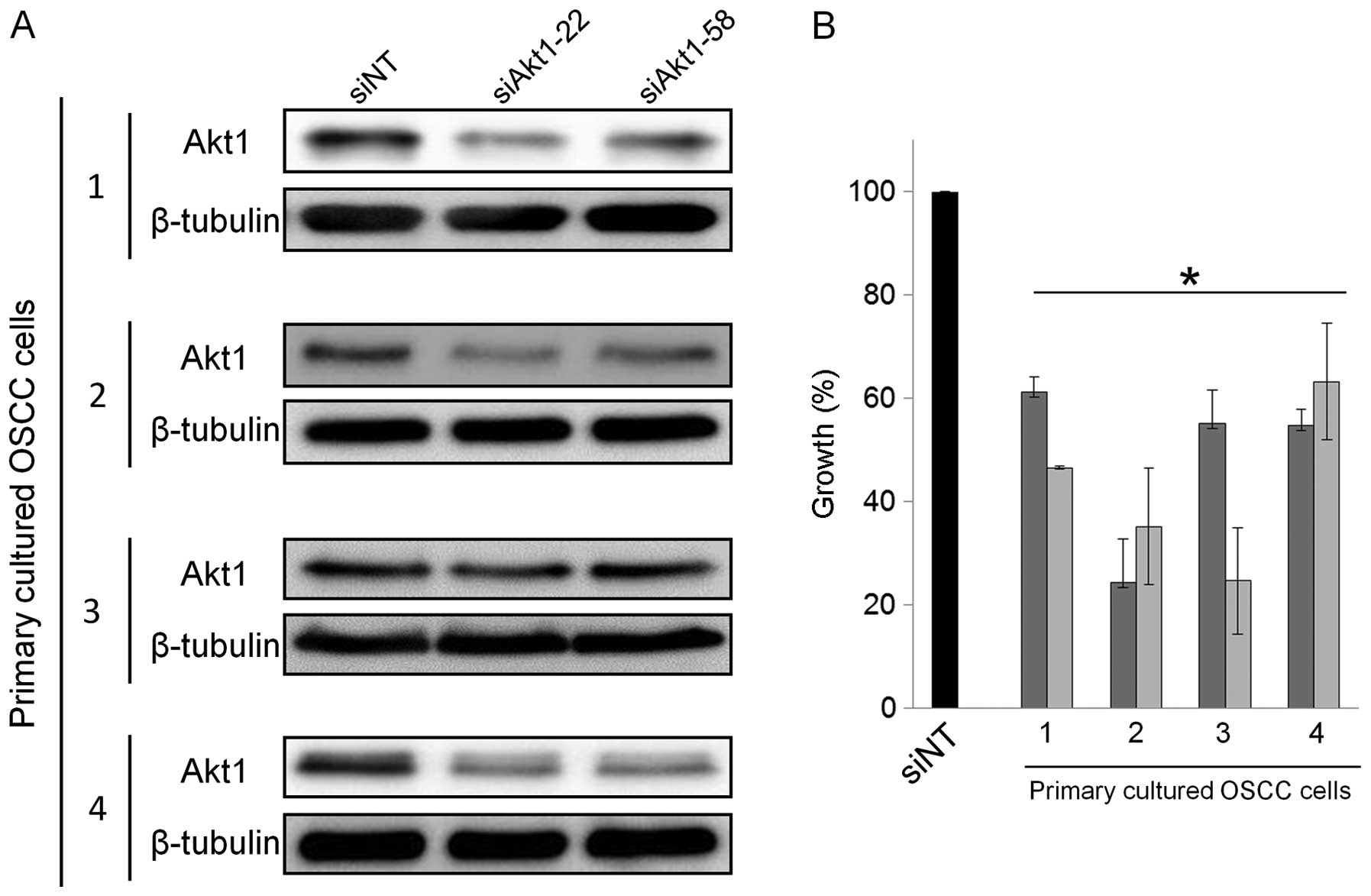
To confirm the usefulness of targeting Akt1 in OSCC,
we used primary cultured cells obtained from resected tumors from
four OSCC patients. Two of these primary cultured cells were
derived from tumors of the tongue and two from lower gingival
tumors. We then examined the in vitro effects of siAkt1 on
the growth of these primary cultured OSCC cells. As in GFP-SAS
cells, transfecting 10 nM siAkt1-22 or 58 into these primary cell
cultures reduced Akt1 expression and significantly suppressed cell
growth, by between 37 and 76%, compared with siNT (Fig. 6A and B).
Molecular mechanisms of the anti-tumor
effects of Akt1 knockdown in human OSCC cells
To understand the molecular mechanisms involved in
the growth inhibitory effect of siAkt1, we carried out an
oligonucleotide-based microarray analysis after transfecting siAkt1
into GFP-SAS cells. A total of 23 genes were differentially
upregulated by >3-fold in GFP-SAS cells transfected with either
siAkt1-22 or 58, whereas 82 were differentially downregulated by
>3-fold. Among these genes, cancer-related genes were identified
by functional analysis using IPA software (Table I). Akt1 knockdown induced the
expression of cyclin-dependent kinase inhibitor 2B (CDKN2B, p15), a
tumor suppressor gene, and reduced the expression of transforming
growth factor, β receptor 1 (TGFBR1), which supports malignant
phenotypes.
 | Table 1Cancer-related genes regulated by Akt1
knockdown in GFP-SAS cells. |
Table 1
Cancer-related genes regulated by Akt1
knockdown in GFP-SAS cells.
| Gene symbol | Gene name | Fold change |
|---|
| Up-regulated genes in
Akt1 knockdown cells |
| TXNIP | Thioredoxin
interacting protein | 7.400 |
| CDKN2B | Cyclin-dependent
kinase inhibitor 2B | 5.475 |
| PVRL4 | Poliovirus
receptor-related 4 | 4.389 |
| VAV3 | Vav 3 guanine
nucleotide exchange factor | 3.019 |
| Down-regulated genes
in Akt1 knockdown cells |
| CDC6 | Cell division cycle
6 | −3.198 |
| NFIB | Nuclear factor
I/B | −3.263 |
| IL6R | Interleukin 6
receptor | −3.282 |
| AHNAK2 | AHNAK nucleoprotein
2 | −3.350 |
| FST | Follistatin | −3.401 |
| BLM | Bloom syndrome, RecQ
helicase-like | −3.767 |
| CTGF | Connective tissue
growth factor | −4.137 |
| MYEOV | Myeloma
overexpressed | −4.422 |
| PLAGL1 | Pleiomorphic adenoma
gene-like 1 | −4.552 |
| IGFBP3 | Insulin-like growth
factor binding protein 3 | −4.689 |
| SOCS2 | Suppressor of
cytokine signaling 2 | −4.942 |
| LIFR | Leukemia inhibitory
factor receptor α | −6.629 |
| IL7R | Interleukin 7
receptor | −7.411 |
| AKT1 | v-akt murine thymoma
viral oncogene homolog 1 | −8.461 |
| TGFBR1 | Transforming growth
factor, β receptor 1 | −14.621 |
Discussion
In microarray analysis, we identified Akt1 as the
only candidate gene commonly overexpressed in OSCC tissues and cell
lines. Overexpression of Akt1 protein in OSCC was also confirmed by
western blot analysis and immunohistochemistry. Furthermore,
targeting Akt1 by RNAi significantly suppressed the growth of human
OSCC cells in vitro and in vivo. These data suggest
that Akt1 functions as a critical oncogene in OSCC and may be an
appropriate therapeutic target for patients with OSCC.
The proto-oncogene Akt, also known as protein kinase
B, is a well-characterized serine/threonine kinase that lies
downstream of phosphatidylinositol 3-kinase (PI3K). In many human
malignancies, activated Akt promotes cell proliferation, survival
and metastasis of cancer cells by regulating the activity of a
number of downstream molecules, such as mammalian target of
rapamycin (mTOR), glycogen synthase kinase 3 (GSK3) and
Bcl-2-associated death promoter (BAD) (15,16).
Akt activation has also been reported to be a significant
prognostic indicator in OSCC, with its inhibition being a possible
molecular approach to treatment (17). MK-2206 is a highly potent,
allosteric pan-Akt inhibitor. In vitro, MK-2206 has shown
synergistic inhibitory effects in combination with paclitaxel in
HNSCC cell lines. In a phase I clinical trial of MK-2206 in
combination with carboplatin/paclitaxel, in patients with advanced
solid tumors, there was a partial response in 1 patient with HNSCC
and disease was stabilized for >6 months in 6 patients (18).
Mammals have three homologous Akt serine/threonine
kinases, known as Akt1, Akt2 and Akt3. The phenotypes of knockout
mice have suggested a critical role for Akt1 in cell survival, a
role for Akt2 in glucose metabolism and a role for Akt3 in brain
development (19). In cancers,
overexpression of specific Akt isoforms, with or without gene
amplification, has been reported. For example, upregulation has
been seen in the expression of Akt1 in gastric and breast cancers,
of Akt2 in ovarian, pancreatic, liver, and colorectal cancers, and
of Akt3 in melanoma and breast cancers. Furthermore, a somatic
activating mutation in Akt1 has been identified in ovarian, breast,
and colorectal cancers. Although the correlation between the
development of specific cancers and the hyperactivation of specific
Akt isoforms remains unclear, a deficiency of Akt1 has been shown
to be sufficient to inhibit endometrial and prostatic neoplasia in
phosphatase and tensin homolog (PTEN) mice (19).
Whole-exome sequencing data from HNSCC primary
tumors has shown that 30.5% of mutations were in the PI3K pathway,
the most frequently mutated oncogenic pathway.
Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit α
(PIK3CA) was identified as the most commonly mutated gene (12.6%)
in the PI3K pathway, but mutations in the Akt1 gene were not
detected (20). In OSCC,
mutational analysis of the PI3K pathway has also shown that
missense mutations were only found in the PIK3CA gene (10.8%), but
not in the Akt1 or PTEN genes (21). Patient-derived tumorgrafts with
PI3KCA mutations growing in nonobese diabetic/severe combined
immunodeficient γ (NOD/SCIDγ) mice were sensitive to BEZ-235, an
mTOR/PI3K inhibitor, whereas PIK3CA-wild-type tumorgrafts were not.
Xenografts developed from an HNSCC cell line harboring a PIK3CA
mutation were more sensitive to a combination of BEZ-235 plus
cetuximab than to cetuximab alone (20). Most recently, Naruse and colleagues
reported that phosphorylated mTOR was overexpressed in
approximately half of OSCC cases and that its expression level was
correlated with the TN classification and survival rate.
Furthermore, everolimus, an mTOR inhibitor, suppressed the growth,
invasion and migration of human OSCC cells (22).
Our study has also shown that
atelocollagen-mediated, systemic administration of siAkt1
suppressed the expression of Akt1 in tumor tissues but not in
normal tissues and the growth of human OSCC cells in nude mice,
without severe side effects such as lung, liver, or renal damage.
This selective targeting may be due to the vascular abnormalities
of tumor tissues, called the enhanced permeability and retention
(EPR) effect (23). Furthermore,
atelocollagen is a highly biocompatible biomaterial having been
used in the field of healthcare as a component of local hemostatic
agents and for tissue regeneration and is unlikely to induce immune
reactions in vivo. These results suggest that nucleic
acid-based drugs, such as atelocollagen-complexed siRNA, may
provide novel therapeutic opportunities for human malignancies,
with minimal risks of adverse events.
In conclusion, overexpression of Akt1 plays a
critical role in OSCC progression and targeting the PI3K/Akt/mTOR
pathway appears an appropriate approach for novel therapies for
HNSCC, including OSCC.
Acknowledgements
This study was supported by Grant-in-Aid for Young
Scientists (A) 17689057 and Scientific Research (C) 21592557 in
Japan Society for the Promotion of Science.
Abbreviations:
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
OSCC
|
oral squamous cell carcinoma
|
|
siAkt1
|
small interfering RNA specific for
Akt1
|
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Gupta S, Kong W, Peng Y, Miao Q and
Mackillop WJ: Temporal trends in the incidence and survival of
cancers of the upper aerodigestive tract in Ontario and the United
States. Int J Cancer. 125:2159–2165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weinstein IB, Joe A and Felsher D:
Oncogene addiction. Cancer Res. 68:3077–3080. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang SF, Cheng SD, Chien HT, Liao CT,
Chen IH, Wang HM, Chuang WY, Wang CY and Hsieh LL: Relationship
between epidermal growth factor receptor gene copy number and
protein expression in oral cavity squamous cell carcinoma. Oral
Oncol. 48:67–72. 2012. View Article : Google Scholar
|
|
5
|
Bonner JA, Harari PM, Giralt J, Azarnia N,
Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al:
Radiotherapy plus cetuximab for squamous-cell carcinoma of the head
and neck. N Engl J Med. 354:567–578. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vermorken JB, Mesia R, Rivera F, Remenar
E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol
D, et al: Platinum-based chemotherapy plus cetuximab in head and
neck cancer. N Engl J Med. 359:1116–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shintani S, Mihara M, Nakahara Y, Aida T,
Tachikawa T and Hamakawa H: Lymph node metastasis of oral cancer
visualized in live tissue by green fluorescent protein expression.
Oral Oncol. 38:664–669. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klosek SK, Nakashiro K, Hara S, Goda H and
Hamakawa H: Stat3 as a molecular target in RNA interference-based
treatment of oral squamous cell carcinoma. Oncol Rep. 20:873–878.
2008.PubMed/NCBI
|
|
9
|
Shintani S, Hamakawa H, Nakashiro K,
Shirota T, Hatori M, Tanaka M, Kuroshita Y and Kurokawa Y: Friend
leukaemia insertion (Fli)-1 is a prediction marker candidate for
radiotherapy resistant oral squamous cell carcinoma. Int J Oral
Maxillofac Surg. 39:1115–1119. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mu P, Nagahara S, Makita N, Tarumi Y,
Kadomatsu K and Takei Y: Systemic delivery of siRNA specific to
tumor mediated by atelocollagen: Combined therapy using siRNA
targeting Bcl-xL and cisplatin against prostate cancer. Int J
Cancer. 125:2978–2990. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ochiya T, Takahama Y, Nagahara S, Sumita
Y, Hisada A, Itoh H, Nagai Y and Terada M: New delivery system for
plasmid DNA in vivo using atelocollagen as a carrier material: The
Minipellet. Nat Med. 5:707–710. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Minakuchi Y, Takeshita F, Kosaka N, Sasaki
H, Yamamoto Y, Kouno M, Honma K, Nagahara S, Hanai K, Sano A, et
al: Atelocollagen-mediated synthetic small interfering RNA delivery
for effective gene silencing in vitro and in vivo. Nucleic Acids
Res. 32:e1092004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Azuma K, Nakashiro K, Sasaki T, Goda H,
Onodera J, Tanji N, Yokoyama M and Hamakawa H: Anti-tumor effect of
small interfering RNA targeting the androgen receptor in human
androgen-independent prostate cancer cells. Biochem Biophys Res
Commun. 391:1075–1079. 2010. View Article : Google Scholar
|
|
14
|
Sasaki T, Nakashiro K, Tanaka H, Azuma K,
Goda H, Hara S, Onodera J, Fujimoto I, Tanji N, Yokoyama M, et al:
Knockdown of Akt isoforms by RNA silencing suppresses the growth of
human prostate cancer cells in vitro and in vivo. Biochem Biophys
Res Commun. 399:79–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo J, Manning BD and Cantley LC:
Targeting the PI3K-Akt pathway in human cancer: Rationale and
promise. Cancer Cell. 4:257–262. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lim J, Kim JH, Paeng JY, Kim MJ, Hong SD,
Lee JI and Hong SP: Prognostic value of activated Akt expression in
oral squamous cell carcinoma. J Clin Pathol. 58:1199–1205. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Beck JT, Ismail A and Tolomeo C: Targeting
the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of
rapamycin (mTOR) pathway: An emerging treatment strategy for
squamous cell lung carcinoma. Cancer Treat Rev. 40:980–989. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gonzalez E and McGraw TE: The Akt kinases:
Isoform specificity in metabolism and cancer. Cell Cycle.
8:2502–2508. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lui VWY, Hedberg ML, Li H, Vangara BS,
Pendleton K, Zeng Y, Lu Y, Zhang Q, Du Y, Gilbert BR, et al:
Frequent mutation of the PI3K pathway in head and neck cancer
defines predictive biomarkers. Cancer Discov. 3:761–769. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cohen Y, Goldenberg-Cohen N, Shalmon B,
Shani T, Oren S, Amariglio N, Dratviman-Storobinsky O,
Shnaiderman-Shapiro A, Yahalom R, Kaplan I, et al: Mutational
analysis of PTEN/PIK3CA/AKT pathway in oral squamous cell
carcinoma. Oral Oncol. 47:946–950. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Naruse T, Yanamoto S, Yamada S, Rokutanda
S, Kawakita A, Kawasaki G and Umeda M: Anti-tumor effect of the
mammalian target of rapamycin inhibitor everolimus in oral squamous
cell carcinoma. Pathol Oncol Res. 21:765–773. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takeshita F, Minakuchi Y, Nagahara S,
Honma K, Sasaki H, Hirai K, Teratani T, Namatame N, Yamamoto Y,
Hanai K, et al: Efficient delivery of small interfering RNA to
bone-metastatic tumors by using atelocollagen in vivo. Proc Natl
Acad Sci USA. 102:12177–12182. 2005. View Article : Google Scholar : PubMed/NCBI
|