Introduction
Colorectal cancer (CRC) is the third most frequently
diagnosed cancer in males and the second most frequently diagnosed
cancer in females (1). Although
current radio-chemical therapies and surgery have shown great
progress, the morbidity and mortality rate of CRC has still
increased over the years (2).
Therefore, there is an urgent need to understand the molecular
mechanisms underlying CRC tumorigenesis and to identify new
therapeutic targets for CRC.
With advances in sequencing technologies, non-coding
RNAs, which account for 70% of the human genome, have shown great
potential in biological research and clinical diagnostics.
Non-coding RNAs (3) are divided
into two major classes: small non-coding RNAs (sncRNAs) and long
non-coding RNAs (lncRNAs), based on the transcript size. Although
many studies proved that sncRNAs, especially microRNAs (miRNAs)
(4,5), played important roles in the
pathogenesis of many diseases, little is known about lncRNAs.
LncRNAs (>200 nucleotides), which were once regarded as
‘transcriptional noise’ in the genomic RNA, have been proved to
play important roles in regulating gene expression at the
epigenetic, transcriptional and post-transcriptional levels
(6,7). In the last few decades, a large
number of lncRNAs [e.g., H19 (8,9),
MALAT1 (10,11) and HOTAIR (12)] have been identified and studied in
a variety of diseases. It is expected that lncRNAs could be used in
clinical applications as prognostic or predictors of cancer. In
CRC, emerging evidence (13–15)
revealed that aberrant expression of particular lncRNAs could
represent novel cancer biomarkers. PVT-1 (16), generates anti-apoptotic activity in
CRC, was a prognostic indicator for CRC patients. LncRNA 91H
(17) was considered as a
prognosis indicator that contributed to tumor metastasis and
predicted patient survival in CRC. However, the relationship
between lncRNA expression level and progression of CRC is still
elusive.
To screen tumor initiation and
progression-associated lncRNAs in CRC, we profiled the expression
of lncRNAs in six pairs of CRC tissues and peritumoral tissues
using microarray analysis. We found that the levels of UCA1
in CRC tissues were 7.104 times higher than peritumoral tissues.
UCA1, which was identified as a novel bladder transitional
cell carcinoma (TCC) transcript (18), played a key role in cellular
proliferation, metastasis and oncogenesis, and was identified as a
novel therapeutic target (19,20).
However, the role of UCA1 in CRC is not well studied,
therefore, we focused our attention on UCA1 and investigated
the clinical values and biological roles of UCA1 in CRC.
Materials and methods
Study subjects and sample collection
The Institutional Review Board of Sun Yat-Sen
University approved the study protocol. In the present study, 54
CRC samples and paired peritumoral samples, deposited between 2010
March to 2010 July in Tissue bank of the Sixth Affiliated Hospital,
Sun Yat-Sen University, were used. Informed consent was obtained
from participants for the use of their tissues in the present
study. All the diagnoses of CRC were histopathologically confirmed.
The data of clinicopathological characteristics (include age,
gender, tumor size, tumor location, differentiation, histological
stage, tumor invasion and lymph node metastasis) were collected
from medical records and pathology reports. The stage of CRC was
evaluated based on the American Joint Committee on Cancer staging
Manual. Follow-up was performed according to the National
Comprehensive Cancer Network (NCCN) guidelines.
Cell lines and cell culture
All the human CRC cell lines (HCT116, SW480, RKO,
HCT8, LoVo, T84, HT29, DLD1, HCT15, Colo205 and Caco2) were
purchased in March 2013 from the Culture Collection of Chinese
Academy of Science, Shanghai, China. They were routinely cultured
in Dulbecco's modified Eagle's medium (DMEM) or RPMI-1640 medium,
supplemented with 1% penicillin/streptomycin and 10% fetal bovine
serum (FBS; Life Technologies, Carlsbad, CA, USA). The cells were
grown at 37°C with 5% CO2 in a humidified incubator.
Microarray and computational
analysis
Six CRC tissues and paired peritumoral tissues were
used to investigate the expression of both protein coding mRNAs
(~26109) and lncRNAs (~30586) using the Human 8×60K LncRNA
Microarray V3.0 (Arraystar, Rockville, MD, USA). The raw signal
intensities were normalized and hierarchical clustering of
differentially expressed lncRNAs was performed using GeneSpring GX
v11.5.1 software (Agilent Technologies, Santa Clara, CA, USA).
Kangcheng Biology Engineering Co., Ltd., (Shanghai, China)
performed the microarray analysis.
siRNA transfection
HCT116 and DLD1 cells were transfected with siRNA
oligonucleotides using the Lipofectamine RNAiMAX transfection
reagent (Invitrogen, Carlsbad, CA, USA) for 48 h in 6-well plates.
Both the UCA1 siRNA and the scrambled control siRNA were
synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Each
well contained 3×105 cells, 5 μl siRNA, 5 μl
Lipofectamine RNAiMAX and 500 μl Opti-MEM (Invitrogen). Both the
UCA1 siRNA and the scrambled control siRNA were synthesized
by Guangzhou Ribobio. We selected one of the siRNA sequences from
three candidates based on the highest knockdown efficiency, as
confirmed by qPCR. The sequences of these siRNAs are listed in
Table I.
 | Table IPrimers used in the present
study. |
Table I
Primers used in the present
study.
| Gene | Sequence
(5′-3′) | Experimental
use |
|---|
| UCA1 siRNA |
GCAUCCAGGACAACACAAAdTdT
dTdTCGUAGGUCCUGUUGUGUUU | siRNA |
| UCA1 mRNA |
CTCTCCATTGGGTTCACCATTC
GCGGCAGGTCTTAAGAGATGAG | qRCR/RT-PCR |
| GAPDH mRNA |
GACAGTCAGCCGCATCTTCTT
AATCCGTTGACTCCGACCTTC | qRCR/RT-PCR |
| UCA1 promoter |
CGGGGTACCTCCTAAGGGGTTTCCTTT
CCCAAGCTTGGCTGTTAATTCACTTGGG | PCR |
| Ets-2 binding
site |
AGATCTAAATGACCCAGGAGCTGATA
AGAGGACAGCCTGAGATGTGCCTGTGG | Site-directed
mutagenesis |
| CEBP binding
site |
CTCAAGTTAGGGAACTGTCAGGCCTCTGA
CGTTACAGGGTGATGTGACCTCAGCCCAC | Site-directed
mutagenesis |
| C-myb binding
site |
CGCCTGAAACCCAGGACCAGGAAAAGA
TGCCTGGGGCTCATCTGAGATGCCCAC | Site-directed
mutagenesis |
| GATA1 binding
site |
AGCGAGAGGGTGGGCTGAGGTCACATC
TTTTCCTGGTCCTGGGTTTCAGAACTG | Site-directed
mutagenesis |
| CREB/α binding
site |
GAGCTCCACATCACCCTGTAACGTTTCC
GCCCACCCTCTTATCTTTTCCTGGTCCT | Site-directed
mutagenesis |
Reverse transcription and quantitative
real-time PCR (qPCR)
RNAs from cells and tissues were extracted by using
the TRIzol reagent (Invitrogen), according to the manufacturer's
instructions. Reverse transcription was carried out with ReverTra
Ace qPCR RT Master Mix with gDNA remover (Toyobo Co., Ltd., Osaka,
Japan). qPCR was conducted using SYBR-Green real-time PCR Master
Mixes (Applied Biosystems, Foster City, CA, USA), using the
following conditions: 95°C for 10 min; and 40 cycles of 95°C for 15
sec; 60°C for 1 min. The PCR products were subjected to 1% agarose
gel and the relative UCA1 expression levels were quantified
by using Quantity One software (Bio-Rad Laboratories, Hercules, CA,
USA). In each qPCR assay, amplification of the housekeeping gene
encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was
performed as the internal control. qPCR was performed in
triplicate, including no-template controls. The amplification of
the appropriate product was confirmed by melting curve analysis
following amplification. The relative expression of UCA1 was
calculated using the comparative cycle threshold (CT)
(2−ΔΔCT) method with glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) as the endogenous control to normalize the
data. The PCR primers used in the present study are listed in
Table I.
Cell proliferation assays
Cell proliferation was performed on the xCELLigence
real-time cellular analysis DP device (ACEA Biosciences, San Diego,
CA, USA). Cells were seeded in cell culture E-plates at a density
of 6000 cells/well and incubated at 37°C and 5% CO2. The
plate was automatically monitored and recorded every 15 min for a
total of 120 h. Three replicate cells were used and three
independent experiments were conducted. The results were expressed
as the parameter ‘Cell Index’.
Cell migration and invasion assays
Cell invasion assays were performed using a
24-Multiwell insert plate with an 8.0-micron pore size membrane (BD
Biosciences, Bedford, MA, USA) chamber containing a Matrigel-coated
membrane. Cells were prepared by suspending them in serum-free DMEM
and adding them to apical chambers whose reverse sides had been
covered by fibronectin (BD Biosciences). Cells at a density of
40,000 cells/100 μl of DMEM media were placed into the upper
chamber of the Transwell plate and 700 μl of DMEM medium containing
20% FBS was to the basal chambers for better cell access. After
incubating the plate for 48 h at 37°C, 5% CO2
atmosphere, invasive cells were stained with DAPI and counted using
a fluorescence microscope. The cell migration assay was the same
but without Matrigel. Experiments were repeated independently three
times.
Cell cycle and apoptosis assays
Cells were stained by using cell cycle staining
solution (Lianke, Hangzhou, China) and analyzed by flow cytometry.
Approximately 2×105 cells were pelleted by
centrifugation and washed with PBS. Cold 75% ethanol (1 ml) was
added to the cells at −20°C overnight. The ethanol was discarded
and PBS was added to rehydrate the cells for 15 min. The cells were
incubated with 1 ml DNA staining solution at room temperature, then
sorted by FACSCalibur (BD Biosciences, Flanklin Lakes, NJ, USA).
The results were analyzed using ModFit 3.0 software (Verity
Software House, Topsham, ME, USA). Cell apoptosis was detected
using an Annexin V/PI apoptosis kit (Lianke). Cells
(2×105) were collected by centrifugation and resuspended
in 500 μl of 1X binding buffer. Annexin V-FITC (5 μl) and 10 μl
propidium iodide were added and the cells were incubated for 5 min
in the dark, at room temperature before being examined using flow
cytometry.
Plasmid constructs and site-directed
mutagenesis
The UCA1 promoter (−500 to +200 bp) was
obtained by PCR (PrimeSTAR Max DNA polymerase; Takara, Shiga,
Japan). The PCR products were subjected to 1% agarose gel and
isolated from the gel using a Gel/PCR Extraction kit (Biomiga, San
Diego, CA, USA). The pGL3-UCA1-promoter vector was
constructed from the pGL3 basic vector and the purified UCA1
promoter using Ligation high (Toyobo). Site-directed mutagenesis
was performed using a KOD-plus-mutagenesis kit (Toyobo), according
to the manufacturer's instructions. The pGL3-UCA1-promoter
vector was used as the template. The mutation sites were designed
at the 3′ region of the primers. All the primers are listed in
Table I.
Luciferase assays
Lipofectamine 3000 (Invitrogen) was used to
co-transfect CRC cells with the pGL3-UCA1-promoter
(mutation), in combination with the pRL Renilla luciferase
reporter vectors (Promega, Madison, WI, USA) as an internal control
reporter. After incubating cells for 48 h at 37°C, luciferase
activities were detected using a Dual-luciferase reporter assay
(Promega), according to the manufacturer's instructions. Firefly
luciferase luminescence was measured using Varioskan Flash (Thermo
Fisher Scientific, Waltham, MA, USA).
Statistical analysis
Statistical analyses were performed with the SPSS
statistical package (version 16.0; SPSS, Inc., Chicago, IL, USA).
Data are presented as means ± standard deviation (SD). Differences
between groups were compared using paired t-tests, unpaired
Student's t-tests or the Mann-Whitney U test. Overall survival
curves were plotted according to the Kaplan-Meier method, with the
log-rank test applied for comparison. A Cox proportional hazards
model univariate and multivariate analysis were performed to
evaluate the association between UCA1 expression and
clinicopathological parameters on the overall survival. In all
analyses, a probability (P) of ≤0.05 was considered statistically
significant.
Results
UCA1 is highly expressed in CRC tissues
and cell lines
To determine the effects of lncRNAs on CRC, we
profiled the expression of lncRNAs in six CRC tissues and paired
peritumoral tissues using microarray analysis. Hierarchical
clustering showed systematic variations in the expression of
lncRNAs (fold-change ≥4) between CRC and paired peritumoral tissues
(Fig. 1A). We noted that
UCA1 was remarkably upregulated (fold-change, 7.104,
P=0.0066) in CRC according to microarray data. To validate the
microarray analysis findings, the expression of UCA1 was
detected by qPCR from 54 pairs of CRC tissues compared with paired
peritumoral tissues. The results showed that UCA1 expression
was significantly higher in the tumor tissues than in the paired
peritumoral tissues (P<0.001) (Fig.
1B and C). PCR assays were further developed to quantify the
UCA1 expression in 11 CRC cell lines (HCT116, SW480, RKO,
HCT8, LoVo, T84, HT29, DLD1, HCT15, Colo205 and Caco2 cells).
Almost all the cell lines showed positive, high expression, except
RKO (Fig. 1D). These results
revealed that the expression of UCA1 is upregulated in CRC
tissues.
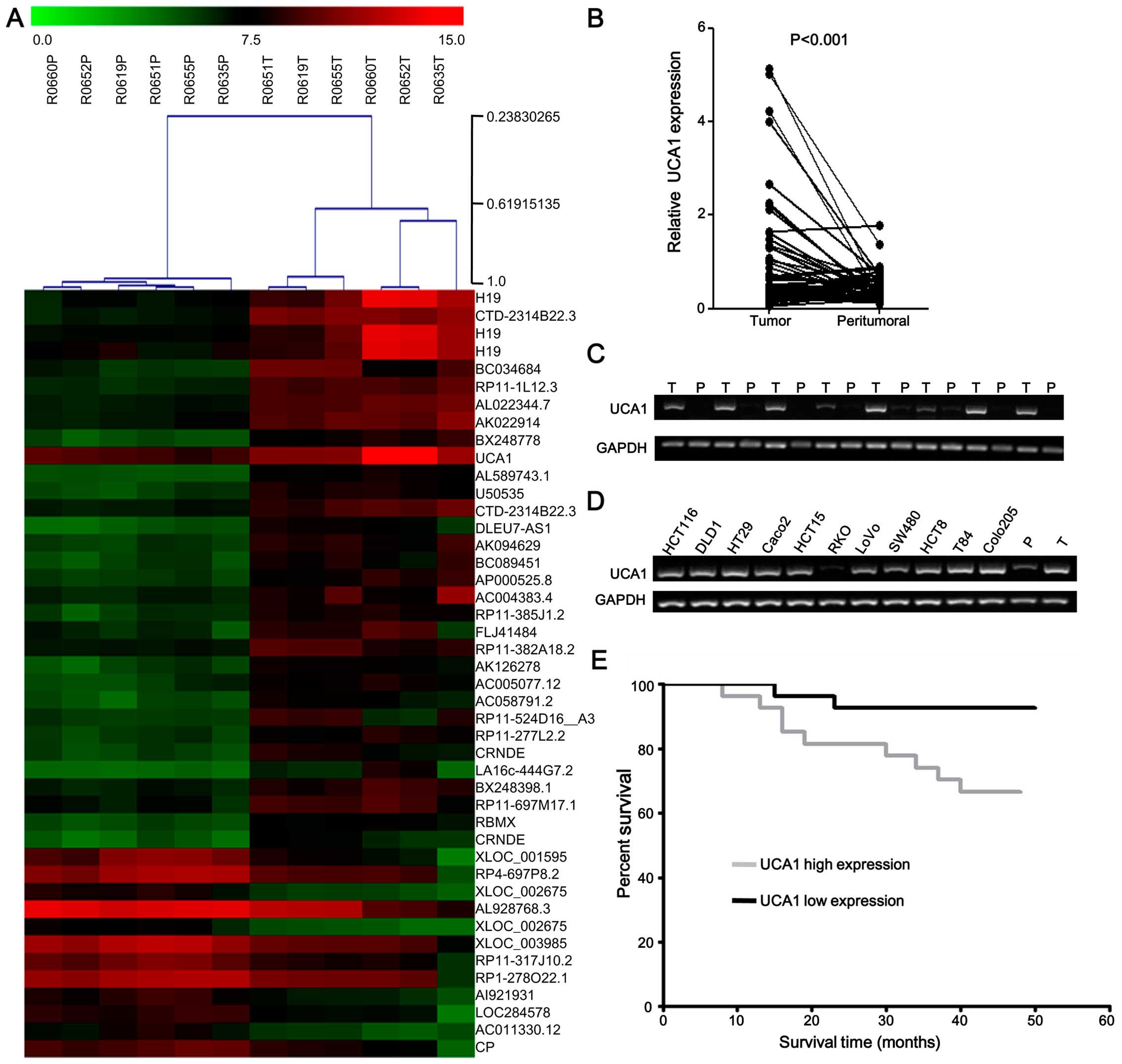 | Figure 1Expression of UCA1 in CRC
tissues and cell lines. (A) Hierarchical clustering showed
systematic variations in the expression of lncRNAs (fold-change ≥4)
between CRC and paired peritumoral tissues. The red and green
indicate high and low expression, respectively. (B) qPCR analyses
of UCA1 expression levels in tumor tissues and peritumoral
tissues from 54 pairs of CRC samples. (C) Representative results
for expression of UCA1 in tumor tissues (T) and peritumoral
tissues (P). UCA1 levels were normalized to that of GAPDH.
(D) PCR analyses of UCA1 expression levels in 11 CRC cell
lines (HCT116, SW480, RKO, HCT8, LoVo, T84, HT29, DLD1, HCT15,
Colo205 and Caco2 cells). The CRC tissue and peritumoral tissue as
the positive control and the negative control, respectively. (E)
Kaplan-Meier overall survival curves for 54 patients with CRC
classified according to UCA1 expression level. The overall
survival of the high UCA1 expression group (n=27) was
significantly higher than that of the low expression group (n=27;
log rank test; P=0.021). |
UCA1 is a predictor of poor outcome in
patients with CRC
To assess the correlation of UCA1 expression
with clinicopathological characteristics, the expression levels of
UCA1 in CRC tissues were categorized as low or high in
relation to the median value. The clinicopathological
characteristics are summarized in Table II. UCA1 expression in CRC
was significantly correlated with lymphatic metastasis (P=0.040),
distant metastasis (P=0.043) and tumor stage (P=0.010). However,
there was no significant association between UCA1 expression
and age, gender, differentiation, lymphatic invasion or venous
invasion (all P>0.05). With regard to overall survival, patients
with high UCA1 expression had a significantly poorer
prognosis than those with low UCA1 expression (Log-rank
P=0.021) (Fig. 1E). Unvariate
analysis of overall survival revealed that the relative level of
UCA1 expression (P=0.038), lymphatic invasion (P=0.001),
lymph node metastasis (P=0.015), distant metastasis (P=0.001) and
tumor stage (P=0.002) were prognostic indicators (Table III). The other
clinicopathological characteristics, such as age, gender,
histological grade, venous invasion, were not statistically
significant prognosis factors (P>0.05; Table III). Multivariate analysis showed
that UCA1 expression was an independent prognostic indicator
of poor survival in CRC (P=0.023) in addition to the presence of
lymphatic invasion (P=0.016) (Table
III).
 | Table IIUCA1 expression and
clinicopathological factors in 54 CRC cases. |
Table II
UCA1 expression and
clinicopathological factors in 54 CRC cases.
| High expression
(N=27) | Low expression
(N=27) | |
|---|
|
|
| |
|---|
| Factors | N (%) | N (%) | P-value |
|---|
| Age (years) |
| ≤60 | 9 (33.3) | 10 (37.0) | 0.776 |
| >60 | 18 (66.7) | 17 (63.0) | |
| Gender |
| Male | 17 (63.0) | 22 (81.5) | 0.129 |
| Female | 10 (37.0) | 5 (18.5) | |
| Histological
grade |
| Well and
moderately | 15 (55.6) | 17 (63.0) | 0.580 |
| Poorly and
other | 12 (44.4) | 10 (37.0) | |
| Lymphatic
invasion |
| Absent | 22 (81.5) | 24 (88.9) | 0.440 |
| Present | 5 (18.5) | 3 (11.1) | |
| Venous
invasion |
| Absent | 22 (81.5) | 22 (81.5) | 1.00 |
| Present | 5 (18.5) | 5 (18.5) | |
| Lymph node
metastasis |
| N0 | 15 (55.6) | 22 (81.5) | 0.040 |
| N1–2 | 12 (44.4) | 5 (18.5) | |
| Distant
metastasis |
| Absent | 21 (22.2) | 26 (96.3) | 0.043 |
| Present | 6 (77.8) | 1 (3.7) | |
| AJCC stage |
| I, II | 13 (48.1) | 22 (81.5) | 0.010 |
| III, IV | 14 (51.9) | 5 (18.5) | |
 | Table IIIUnivariate and multivariate analysis
for overall survival (Cox proportional hazards regression
model). |
Table III
Univariate and multivariate analysis
for overall survival (Cox proportional hazards regression
model).
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Factors | RR | P-value | RR | P-value |
|---|
| Age (years) | 0.650 | 0.477 | | |
| Gender | 0.549 | 0.443 | | |
| Histological
grade | 0.497 | 0.302 | | |
| Lymphatic invasion
(absent/present) | 13.71 | 0.001a | 11.91 | 0.016a |
| Venous invasion
(absent/present) | 1.678 | 0.445 | | |
| Lymph node
metastasis (N0/N1–2) | 4.574 | 0.015a | 1.119 | 0.926 |
| Distant metastasis
(absent/present) | 25.65 | 0.001a | 3.984 | 0.157 |
| AJCC stage | 11.17 | 0.002a | 3.62 | 0.443 |
| UCA1 expression
(low/high) | 5.068 | 0.038a | 3.137 | 0.023a |
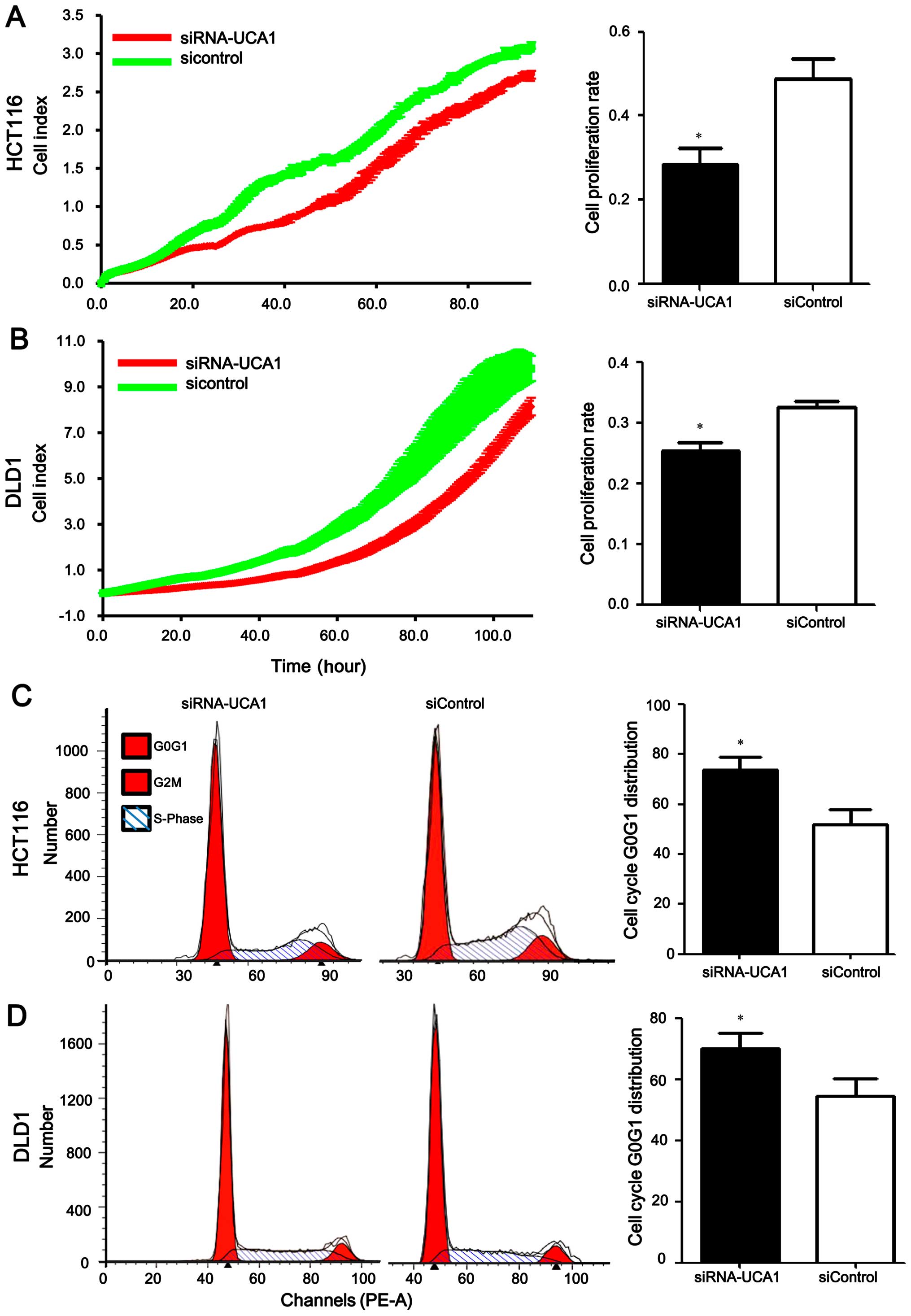
Knockdown of UCA1 suppresses cell
proliferation and prevents the G0/G1 progression of CRC cells
To investigate the biological function of
UCA1, we conducted UCA1 knockdown assays using RNA
interference in CRC cells. Firstly, we confirmed that UCA1
expression in HCT116 and DLD1 cells transfected with
UCA1-specific siRNA was significantly lower than that in
cells transfected with the negative control siRNA: the expression
levels of UCA1 were reduced by 90% and 80%, as detected by
qPCR. Real-time cellular analysis (RTCA) indicated that cell
proliferation was reduced in CRC cells when UCA1 was knocked
down. For HCT116 cells, we observed a 42.8% reduction in the cell
proliferation rate when UCA1 was silenced (0.28 ±0.07 for
the siRNA-UCA1 group vs. 0.49±0.08 for the siControl group,
P=0.028) (Fig. 2A). For DLD1
cells, we observed a 21.9% reduction in the cell proliferation rate
when UCA1 was silenced (0.25±0.03 for the siRNA-UCA1 group
vs. 0.32±0.02 for the siControl group, P=0.033) (Fig. 2B). In addition,
fluorescence-activated cell sorting (FACS) analysis was conducted
to analyze the effect of UCA1 on cell cycle progression.
Knockdown of UCA1 increased the proportion of cells in G0/G1
phases (HCT116 cell line: 73.9±8.57% for the siRNA-UCA1 group vs.
52.1±10.3% for the siControl group, P=0.0074; DLD1 cell line:
70.1±8.92% for the siRNA-UCA1 group vs. 54.7±10.0% for the
siControl group, P=0.029, respectively) (Fig. 2C and D). Collectively, these data
suggested that silencing of UCA1 contributed to
proliferation inhibition via cell cycle arrest.
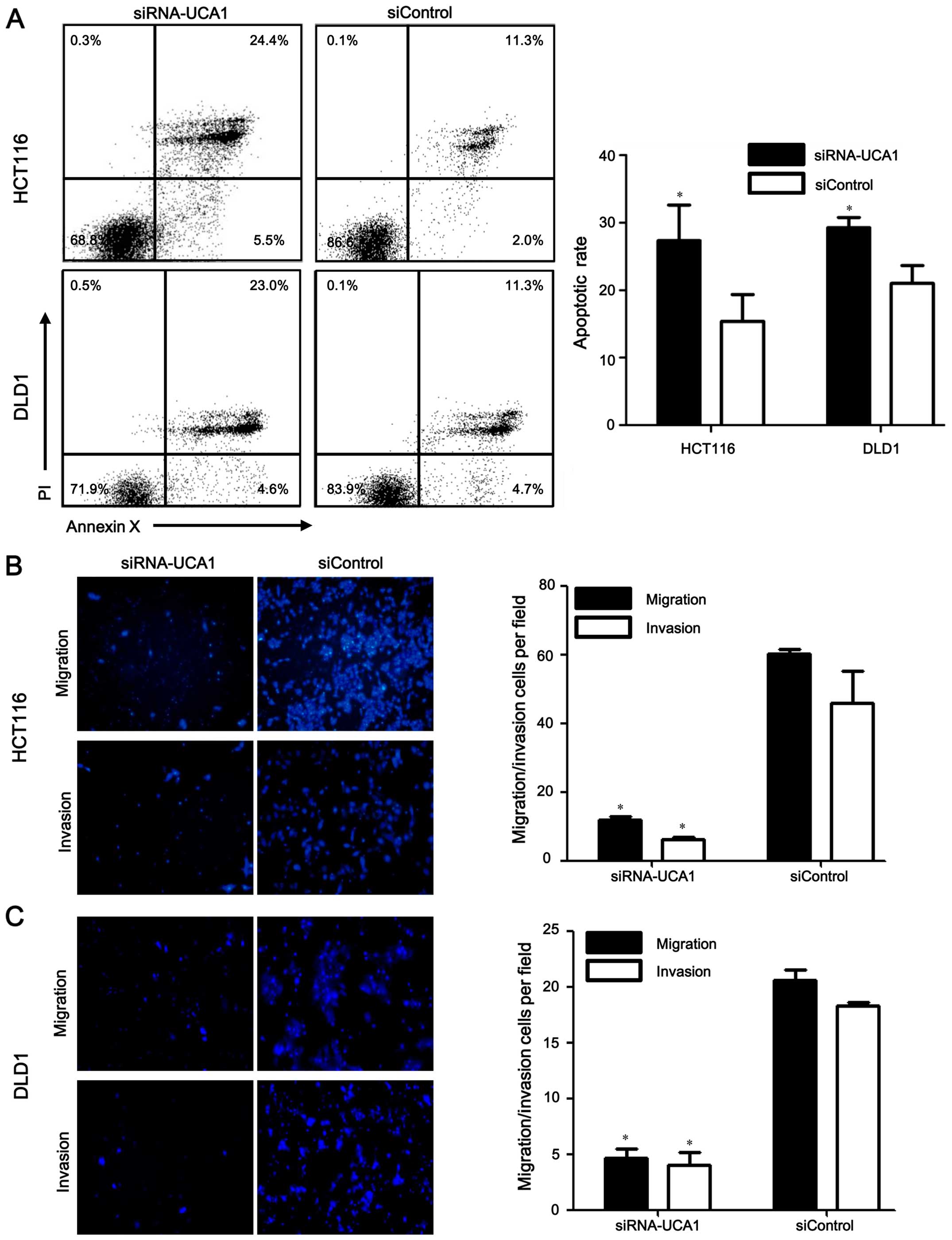
Silencing UCA1 promotes apoptosis,
inhibits migration and invasion in CRC cells
Consistent with decreased cell proliferation, there
was a significant increase in the apoptosis of UCA1
knockdown cells relative to that of the negative control cells
(HCT116 cell line: 25.7±8.8% for the siRNA-UCA1 group vs.
15.2±7.0% for the siControl group, P=0.020; DLD1 cell line:
28.9±3.0% for the siRNA-UCA1 group vs. 21.7±3.3% for the siControl
group, P=0.009, respectively) (Fig.
3A). Therefore, the data suggested that UCA1 could
inhibit apoptosis of CRC cells. We further analyzed whether
UCA1 knockdown affected cell migration and invasion of CRC
cells using Transwell migration assays. The results indicated that
UCA1-silenced HCT116 and DLD1 cells showed less potential of
migration and invasion ability compared with the negative control
cell lines (all P<0.01) (Fig. 3B
and C). These data suggested that UCA1 induced migration
and invasion in CRC cells.
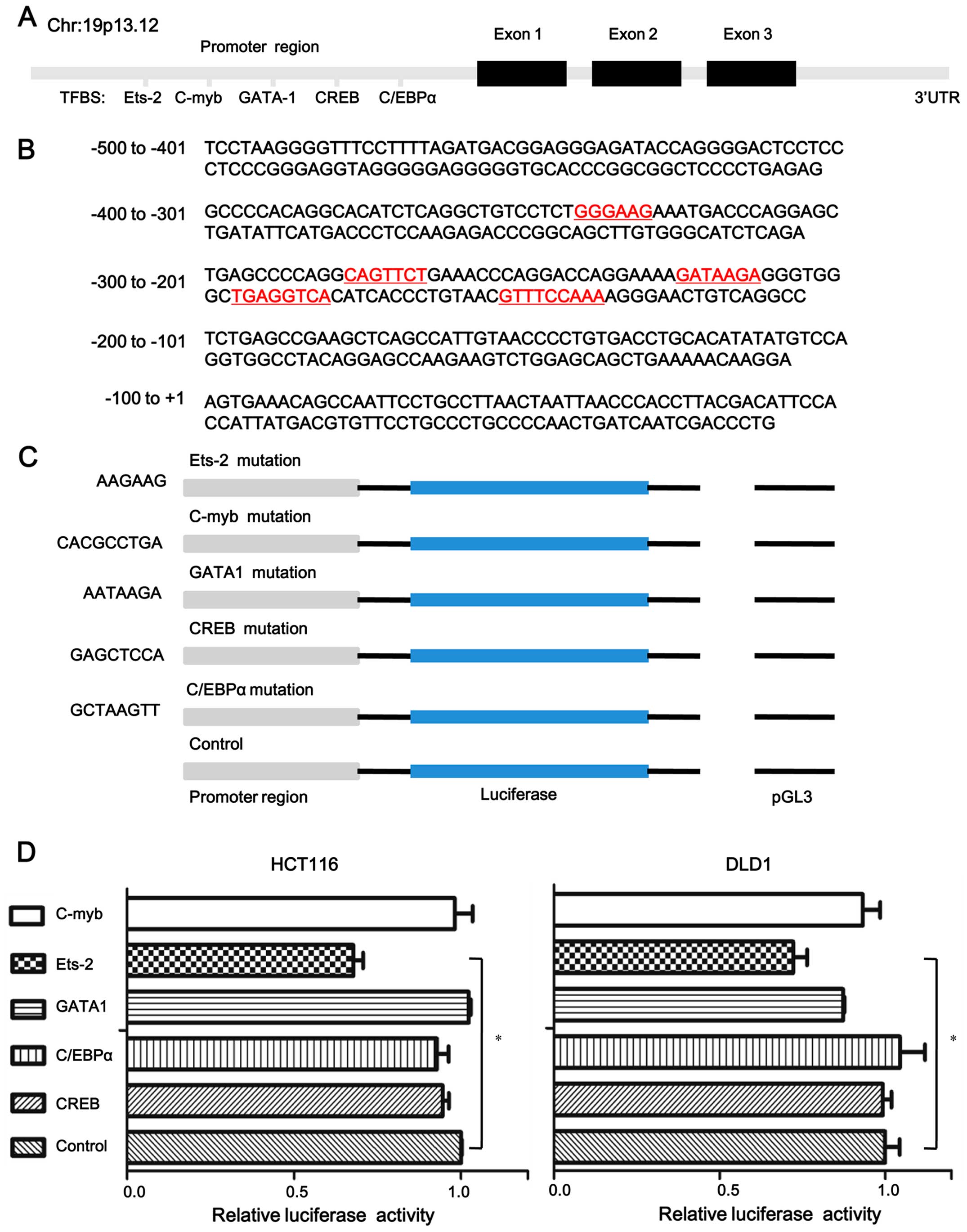
Ets-2 is critical for UCA1 promoter
activity
To obtain insight into the potential regulators of
UCA1, we analyzed the core promoter of UCA1 (from
nucleotides −500 to 200) using several bioinformatics software
programs and predicted five potential transcription factor binding
sites (Ets-2, C/EBPα, c-myb, GATA-1 and CREB) (Fig. 4A and B). To investigate which
transcription factor is critical for UCA1 promoter activity,
we replaced the five transcription factor binding sites to mutated
sites in pGL3-UCA1-700-mut and the mutated sites are shown
in Fig. 4C. Compared with the
wild-type pGL-3-UCA1-700 construct, HCT116 and DLD1 cells
yielded a lower promoter activity when transcription factor Ets-2
binding site was mutated, as assessed by luciferase reporter assays
(0.677±0.06 for Ets-2 vs. 1 for control, P=0.0084, 0.725±0.08 for
Ets-2 vs. 1 for control, P=0.019, respectively) (Fig. 4D). The results suggested that Ets-2
binding sites play an essential role in the regulation of the
UCA1 promoter activity.
Discussion
CRC is the process whereby benign polyps develop
into adenoma and then into tumors; thus, early screening and
diagnosis are important to decrease the death rate (21). Numerous genetic and epigenetic
alterations, such as DNA methylation and microRNAs, have been
studied as potential biomarkers for screening or diagnosis in CRC.
However, few such markers can be applied for clinical diagnosis and
treatment. Thus, it is essential to screen out new effective
biomarkers.
LncRNAs have attracted increased interest from
researchers in recent years. A growing number of studies (22–24)
suggest that abnormal expressions of lncRNAs are closely related to
tumor initiation and progression. Unlike protein-coding mRNAs,
which are expressed in multiple tissue types, most lncRNAs show
tissue-specific expression patterns. For example, Yang et al
(25) found that CCAT1 was
upregulated in gastric cancer and might be a potential therapeutic
marker. Qi et al (26) also
suggested that low expression of lncRNA loc285194 was related to
poor prognosis in CRC. There is no doubt that lncRNAs have
significant roles in cancer progression and development. However,
only a limited number of lncRNAs have been investigated in detail
(27–29) and the functional roles of lncRNAs
in CRC are not yet well elucidated.
In the present study, we observed that UCA1 was
highly expressed in CRC tissues, suggesting a positive role for
UCA1 in CRC tumourigenesis. In addition, the expression of
UCA1 was associated with lymph mode metastasis (P=0.04),
distant metastasis (P=0.043) and tumor stage (P=0.01). In the
survival analysis, high UCA1 expression levels were related
with poor prognosis, indicating that UCA1 could be a
valuable prognostic biomarker. In fact, UCA1 has been
extensively studied in the context of cancer (30–32),
especially in bladder cancer (33). Fang et al (34) have reported that a significantly
elevated expression of UCA1 in tongue squamous cell
carcinomas was found and there was higher expression of UCA1
in lymph node metastases than in paired primary tumors. Previous
studies indicated that other lncRNAs were also considered as
molecular biomarkers in CRC. CCAT1 (35), an upregulated lncRNA in CRC, was
explored for early screening and detection. Elevated levels of
lincRNA-p21 were significantly associated with CRC disease state.
In this regard, UCA1 could be used as a potential prognostic
indicator in CRC.
We identified the biological functions of
UCA1 in CRC cells. Proliferative activity and the ability to
metastasis were significantly suppressed in vitro after
silencing of UCA1. Besides, knockdown of UCA1 induced
G0/G1 phase arrest and promoted apoptosis in CRC cells. Similar to
these findings, Fan et al (20) reported that ectopic UCA1 expression
enhances the tumorigenic potential and increases invasion. Some of
our results were similar to those of Han et al (36), who also observed that elevated
UCA1 expression in CRC could influence cancer cell
proliferation and apoptosis. However, our results differ in terms
of the metastatic phenotype. Our results showed that high
expression of UCA1 could significantly enhance migration and
invasion of CRC cell lines, whereas those of Han et al did
not. The contradictory results may be due in part to the different
cells used. The results suggest that UCA1 acts as an
oncogene in CRC.
Recent studies put forward that the transcription
factors regulate lncRNAs via binding with the promoters of lncRNAs,
which has been proved to play a pivotal role in tumor progression
(37–39). In the present study, we found that
Ets-2 could bind to the UCA1 core promoter and stimulated
UCA1 transcriptional activation in CRC cells. Similar to
these findings, Wu et al (40) have also demonstrated that Ets-2
bound to the UCA1 promoter region and regulated cell
apoptosis via Akt pathway in bladder cancer. Our data support that
Ets-2 takes part in CRC development, but the exact regulatory
mechanisms of UCA1 by Ets-2 in CRC require further
investigation.
Some studies have reported that UCA1 may act
as a switch to regulate gene expression in different cell signal
pathways in cancer. Yang et al (38) observed that UCA1 regulated
the cell cycle through CREB via the PI3K-AKT pathway in bladder
carcinoma. In another study, UCA1 was observed to regulate
bladder cancer cell glucose metabolism through the cascade of
mTOR-STAT3/miR143-HK2 (41). These
results highlighted the effect of UCA1 on signal
transduction pathways. Besides, the report that UCA1 could
be regulated by hsa-miR-1 in bladder cancer showed that lncRNAs may
act as a novel set of targets for microRNA (42). Other regulatory factors such as
histone modification and DNA methylation that influence UCA1
expression in CRC need to be further studied.
Our results showed that UCA1 expression was
significantly increased in CRC tissues and cell lines. Elevated
levels of UCA1 were statistically correlated with lymph mode
metastasis, distant metastasis and tumor stage, and predicted poor
prognosis in CRC. In vitro, we demonstrated that UCA1
was associated with tumor migration, invasion and proliferation of
CRC cells. These findings provide important insight into exploring
new biomarkers for the diagnosis and therapy of CRC. The results
indicate that UCA1 may be a promising target for future
therapy of CRC.
Acknowledgements
The present study was partly supported by the
National Natural Scientific Foundation of China (grant nos.
81201581 to D.C. and 81372566 to L.W.), the Guangdong Provincial
Scientific Research Foundation (grant no. S2013010013478 to D.C.),
the Sun Yat-Sen University Young Teacher Training Program (grant
no. 88000-3126201 to D.C.), National Key Technology Support Program
(grant no. 2014BAI09B06) and Science and Technology Plan Project of
Guangzhou (grant no. 2013J4500045). We thank Z. Wang for his
assistance in recruiting the subjects and X. Fu for his laboratory
assistance.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
UCA1
|
urothelial carcer associated 1
|
|
LncRNA
|
long non-coding RNA
|
|
qPCR
|
quantitative real-time polymerase
chain reaction
|
|
sncRNA
|
small non-coding RNAs
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
RTCA
|
real-time cellular analysis
|
|
FACS
|
fluorescence-activated cell
sorting
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar
|
|
3
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Øster B, Linnet L, Christensen LL, Thorsen
K, Ongen H, Dermitzakis ET, Sandoval J, Moran S, Esteller M, Hansen
TF, et al: COLOFOL steering group: Non-CpG island promoter
hypomethylation and miR-149 regulate the expression of SRPX2 in
colorectal cancer. Int J Cancer. 132:2303–2315. 2013. View Article : Google Scholar
|
|
5
|
Doberstein K, Steinmeyer N, Hartmetz AK,
Eberhardt W, Mittelbronn M, Harter PN, Juengel E, Blaheta R,
Pfeilschifter J and Gutwein P: MicroRNA-145 targets the
metalloprotease ADAM17 and is suppressed in renal cell carcinoma
patients. Neoplasia. 15:218–230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponjavic J, Ponting CP and Lunter G:
Functionality or transcriptional noise? Evidence for selection
within long noncoding RNAs. Genome Res. 17:556–565. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar
|
|
10
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: A long non-coding RNA and its important 3′ end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI
|
|
12
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xue Y, Ma G, Gu D, Zhu L, Hua Q, Du M, Chu
H, Tong N, Chen J, Zhang Z, et al: Genome-wide analysis of long
noncoding RNA signature in human colorectal cancer. Gene.
556:227–234. 2015. View Article : Google Scholar
|
|
14
|
Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu
Y, Li X, Cai G and Cai S: Low expression of novel lncRNA
RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal
cancer. Med Oncol. 31:312014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhai H, Fesler A, Schee K, Fodstad O,
Flatmark K and Ju J: Clinical significance of long intergenic
noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer.
12:261–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Br J Cancer.
110:164–171. 2014. View Article : Google Scholar :
|
|
17
|
Deng Q, He B, Gao T, Pan Y, Sun H, Xu Y,
Li R, Ying H, Wang F, Liu X, et al: Up-regulation of 91H promotes
tumor metastasis and predicts poor prognosis for patients with
colorectal cancer. PLoS One. 9:e1030222014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang F, Li X, Xie X, Zhao L and Chen W:
UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma
and embryo, influencing cell growth and promoting invasion. FEBS
Lett. 582:1919–1927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gellad ZF and Provenzale D: Colorectal
cancer: National and international perspective on the burden of
disease and public health impact. Gastroenterology. 138:2177–2190.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Chen Z, Tian L, Zhou C, He MY, Gao
Y, Wang S, Zhou F, Shi S, Feng X, et al: LncRNA profile study
reveals a three-lncRNA signature associated with the survival of
patients with oesophageal squamous cell carcinoma. Gut.
63:1700–1710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y
and Fang G: Long noncoding RNA CCAT1, which could be activated by
c-Myc, promotes the progression of gastric carcinoma. J Cancer Res
Clin Oncol. 139:437–445. 2013. View Article : Google Scholar
|
|
26
|
Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C,
Zhou XY and Du X: Low expression of LOC285194 is associated with
poor prognosis in colorectal cancer. J Transl Med. 11:1222013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pandey GK, Mitra S, Subhash S, Hertwig F,
Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S,
et al: The risk-associated long noncoding RNA NBAT-1 controls
neuroblastoma progression by regulating cell proliferation and
neuronal differentiation. Cancer Cell. 26:722–737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan SX, Tao QF, Wang J, Yang F, Liu L,
Wang LL, Zhang J, Yang Y, Liu H, Wang F, et al: Antisense long
non-coding RNA PCNA-AS1 promotes tumor growth by regulating
proliferating cell nuclear antigen in hepatocellular carcinoma.
Cancer Lett. 349:87–94. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hiemer SE, Szymaniak AD and Varelas X: The
transcriptional regulators TAZ and YAP direct transforming growth
factor β-induced tumorigenic phenotypes in breast cancer cells. J
Biol Chem. 289:13461–13474. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu SP, Yang JX, Cao DY and Shen K:
Identification of differentially expressed long non-coding RNAs in
human ovarian cancer cells with different metastatic potentials.
Cancer Biol Med. 10:138–141. 2013.
|
|
32
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xue M, Li X, Li Z and Chen W: Urothelial
carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted
long noncoding RNA that enhances hypoxic bladder cancer cell
proliferation, migration, and invasion. Tumour Biol. 35:6901–6912.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fang Z, Wu L, Wang L, Yang Y, Meng Y and
Yang H: Increased expression of the long non-coding RNA UCA1 in
tongue squamous cell carcinomas: A possible correlation with cancer
metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 117:89–95.
2014. View Article : Google Scholar
|
|
35
|
Nissan A, Stojadinovic A,
Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, Bochem A,
Dayanc BE, Ritter G, Gomceli I, et al: Colon cancer associated
transcript-1: A novel RNA expressed in malignant and pre-malignant
human tissues. Int J Cancer. 130:1598–1606. 2012. View Article : Google Scholar
|
|
36
|
Han Y, Yang YN, Yuan HH, Zhang TT, Sui H,
Wei XL, Liu L, Huang P, Zhang WJ and Bai YX: UCA1, a long
non-coding RNA up-regulated in colorectal cancer influences cell
proliferation, apoptosis and cell cycle distribution. Pathology.
46:396–401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xue M, Li X, Wu W, Zhang S, Wu S, Li Z and
Chen W: Upregulation of long non-coding RNA urothelial carcinoma
associated 1 by CCAAT/enhancer binding protein α contributes to
bladder cancer cell growth and reduced apoptosis. Oncol Rep.
31:1993–2000. 2014.PubMed/NCBI
|
|
38
|
Yang C, Li X, Wang Y, Zhao L and Chen W:
Long non-coding RNA UCA1 regulated cell cycle distribution via CREB
through PI3-K dependent pathway in bladder carcinoma cells. Gene.
496:8–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kabbout M, Garcia MM, Fujimoto J, Liu DD,
Woods D, Chow CW, Mendoza G, Momin AA, James BP, Solis L, et al:
ETS2 mediated tumor suppressive function and MET oncogene
inhibition in human non-small cell lung cancer. Clin Cancer Res.
19:3383–3395. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu W, Zhang S, Li X, Xue M, Cao S and Chen
W: Ets-2 regulates cell apoptosis via the Akt pathway, through the
regulation of urothelial cancer associated 1, a long non-coding
RNA, in bladder cancer cells. PLoS One. 8:e739202013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Z, Li X, Wu S, Xue M and Chen W: Long
non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase
2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci.
105:951–955. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang T, Yuan J, Feng N, Li Y, Lin Z, Jiang
Z and Gui Y: Hsa-miR-1 downregulates long non-coding RNA urothelial
cancer associated 1 in bladder cancer. Tumour Biol. 35:10075–10084.
2014. View Article : Google Scholar : PubMed/NCBI
|