Introduction
Protein-tyrosine phosphatase of regenerating liver-3
(PRL-3), also known as PTP4A3 (tyrosine phosphatase type IVA 3), is
a protein with relatively small molecular weight of 22 kDa. It
belongs to the protein tyrosine phosphatases superfamily (1), and contains three members, the PRL-1,
PRL-2, and PRL-3. Of them, PRL-3 is known to be highly expressed in
distant metastatic sites of colon cancers (2). High level of PRL-3 expression was
observed in other types of cancers, including breast (3), ovary (4,5),
liver (6), and stomach (7) tumors. Moreover, elevated PRL-3
expression correlates with cancer cell proliferation, motility,
invasiveness, and tumor angiogenesis (8–10) in
cancer cell line-based systems. Growing evidence has indicated that
high expression level of PRL-3 is an adverse prognostic factor
(11). Mechanistically, PRL-3
modulates multiple signaling pathways including Rho GTPase
(12), Src (13), and PI3K AKT (14) in different tumors. All these
observations indicate that PRL-3 plays an important role in various
cancer in progression and metastasis.
Although evidence links PRL-3 expression to
tumorigenesis and metastasis in tumor cells and tissues, expression
pattern of PRL-3 during development and consequence of its general
expression during embryogenesis is not known. PRL-3 expression in
adult tissues is detected PRL-3 in the heart and skeletal muscle
while moderate expression is detected in the pancreas (15,16).
Mouse PRL-3 is also detected in the villus epithelial cells of the
small intestine (17).
Importantly, PRL-3 protein has been detected in rat fetal heart and
developing blood vessels, but not in adult rat or human heart and
mature blood vessels (9). These
observations suggest that PRL-3 expression pattern is
developmentally regulated in mammals and PRL-3 may have potential
functions in cell proliferation.
To our knowledge, the expression pattern of
prl-3 during vertebrate development and consequence of its
aberrant upregulation has not been explored. In this study, we used
zebrafish as our vertebrate model and determined the expression
pattern of prl-3 during different stages of zebrafish
development by whole mount in situ hybridization.
Interestingly, overexpression of either zebrafish Prl-3 (zPrl-3) or
human PRL-3 (hPRL-3) led to notochord malformation reminiscent of
chordoma which we confirmed with chordoma-specific markers.
Relevance of the role of PRL-3 in chordoma is supported by
immunochemical detection of human PRL-3 in clinical chordoma
specimens.
Materials and methods
Fish lines and maintenance
Zebrafish (strain AB) embryos were collected from
the zebrafish model animal facility, institute of clinical and
translation research, Sun Yat-sen University. The fish was
maintained in a circulating rack system with alternate exposure of
14 h light and 10 h dark at 28.5°C, and fed three times daily.
Embryos were staged on hours of post fertilization (hpf) or days
post fertilization (dpf) at 28.5°C.
Whole mount in situ hybridization and
imaging
The zebrafish prl-3 (zprl-3, ptp4a2a
protein tyrosine phosphatase type IVA, member 2a, Gene ID: 449541)
probe was designed to include the-3′-UTR region based on database
(NM.001005583). Total mRNA isolated from zebrafish embryos was used
for reverse transcription of the first-strand cDNAs. Zebrafish
prl-3 (zprl-3) was amplified by PCR with a pair of
primers (zPRL3-931F and zPRL3-1659R, Table I), cloned into the PGEM -T Easy
Vector (Promega) and confirmed by sequencing. RNA probes of
zprl-3 were labeled with digoxigenin-dUTP (DIG, Roche, cat
no. 11277073910) by in vitro transcription and purified
according to the manufacturer's instructions. Shh (sonic
hedgehog) probe was used as described previously (18). Plasmid encoding ntl (no
tail) was a gift from Professor Vladimir Korzh, Institute of
Molecular and Cell Biology, Singapore. Zebrafish embryos were fixed
with 4% para-formaldehyde (PFA) in phosphate-buffered saline (PBS)
overnight, followed by washing with PBST (0.1% Tween in PBS).
Embryos were then empirically treated by protease K (Roche, cat no.
3115844001) for appropriate time, depending on stage of embryo
development. Embryos were post-fixed after digestion and washed in
PBST before pre-hybridization in HYB buffer (50% formamide, 5X
standard saline citrate, 0.1% Tween-20, 50 μg/ml heparin, 0.5 mg/ml
total yeast RNA, 9.2 mM citric acid) at 65°C for 2 h. Incubation
with respective digoxigenin-dUTP labeled probes (2 μg/ml) was
carried out at 65°C overnight. After removal of probes, embryos
were washed at 65°C with 100, 75, 50 and 25% formamide in 2X SSCT
(15 mM citrate, 150 mM NaCl, 0.1% Tween-20, pH 7.0). This was
followed by a 0.2X SSCT wash. Embryos were then incubated in MABT
buffer (150 mM maleic acid, 100 mM NaCl, 0.1% Tween-20, pH 7.5) and
blocked for 2 h with blocking reagent (Roche) dissolved in MABT.
Alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche, cat
no. 11093274910) diluted at 1:2,000 was added and incubated
overnight at 4°C. This was followed by 4 times PBST washes, and 1
wash with pH adjusted Tris-HCl staining buffer (0.1 M NaCl, 5 mM
MgCl2, 0.1 M Tris-HCl, 0.1% Tween-20, pH 9.2). Proper
staining was initiated with the addition of NBT and BCIP to the
staining buffer (54 μl of 50 mg/ml NBT and 42 μl of 75 mg/ml BCIP
in 10 ml Tris-HCl staining buffer) until the embryo was stained
with visually acceptable signal to noise ratio. Stained embryos
were imaged using a Leica DFC550 camera attached to a stereoscope
(LeicaM205FA). Image contrasts were processed by Photoshop CS.
 | Table IPrimers used in this study. |
Table I
Primers used in this study.
| Name | Sequence
(5′→3′) | Application |
|---|
| zPRL-3-931F |
5′-GAAATATCGGCCCAAACAGAGACT-3′ | For probe |
| zPRL-3-1659R |
5′-CAGATAAACACGCAGAAGAAACAT-3′ | For probe |
| zPRL-3-inner
(500R) |
5′-GTCGGTTCATGCGAGCCATA-3′ | For 5′RACE |
| zPRL-3-outer
(568R) |
5′-GGTGGAGTTTGTTGGGTTGT-3′ | For 5′RACE |
|
zPRL-3-470F-Cla1 |
5′-CCATCGATGGAAGCACAACTATGGCTCG-3′ | For mRNA |
|
zPRL-3-1010R-Xho1 |
5′-CCGCTCGAGTTCGCAGTCACATGATACAGCAC-3′ | For mRNA |
|
zPRL-3-mutation-F |
5′-GAAGCACAACTTAGGCTCGCTAGAACCGACCGG-3′ | For nonsense
mRNA |
|
zPRL-3-mutation-R |
5′-CCGGTCGGTTCTAGCGAGCCTAAGTTGTGCTTC-3′ | For nonsense
mRNA |
|
hPRL-3-845F-Cla1 |
5′-CCATCGATGCCACCAATGGCTCGGATGAACCG-3′ | For mRNA |
|
hPRL-3-1378R-Xho1 |
5′-CCGCTCGAGGAGCTACATAACGCAGCACCG-3′ | For mRNA |
|
hPRL-3-mutation-F |
5′-CATCGATGCCACCATAGGCTCGGTAGAACCGC-3′ | For nonsense
mRNA |
|
hPRL-3-mutation-R |
5′-GGTTCTACCGAGCCTATGGTGGCATCGATGGG-3′ | For nonsense
mRNA |
Northern blot hybridization
Total RNA was extracted from zebrafish embryos at
their indicated stages, using TRIzol (Life Technologies, cat no.
15596026). Ten μg RNA was ran by formaldehyde-based denaturing gel
and transferred onto nylon membrane (GE, cat. no. RPN303B) and
fixed with UV cross-linker. Northern blot hybridization was carried
out following the manufacturer's instructions (Roche, cat no.
12039672910) using the same digoxigenin-labeled zprl-3
antisense probes as used in zebrafish whole mount
hybridization.
5′RACE of zebrafish PRL-3 and
microinjection of PRL-3 mRNA and its plasmids into zebrafish
embryos
Total RNA was extracted from 2 dpf embryos. 5′-RACE
of zprl-3 was performed with Ambion's
FirstChoice® RLM RACE kit with indicated primers
(Table I) according to the
manufacturer's instructions. Encoding regions of both zebrafish and
human PRL-3 with their proximal part of 5′UTR fragments were
amplified by PCR using iProof High-Fidelity PCR kit (Bio-Rad, cat
no. 172-5331) and cloned into pCS2 plasmids for functional mRNA
synthesis. Negative controls used in the overexpression experiments
contain nonsense mutantions of zebrafish prl-3
(zprl-3) and human PRL-3 (hPRL-3) are obtained
using QuickChange Site-Directed Mutagenesis kit (Stratagene, cat
no. 200518). PCS2-zprl-3 and PCS2-hPRL-3 were used as
templates to produce the initiation codon point mutation (ATG→TAG)
of PRL-3 following the manufacturer's instructions. The indicated
primers for mutations are also listed in Table I. All above mentioned constructs
were validated by DNA sequencing. Functional zprl-3 and
hPRL-3 mRNAs and their nonsense mRNAs were synthesized using
the Message Machine kit (Ambion, cat no. AM1340). mRNAs (75–100 pg)
and injected into zebrafish embryos at one-cell stage with
Microinjector (Warner PLI-100A). hPRL-3 overexpression was achieved
by simultaneous co-injection of 100 pg plasmids encoding
PEGFP-PRL-3 (19) and PEGFP into
one-cell stage zebrafish embryos. Embryos injected with nonsense
mRNA were treated as normal controls.
Western blot analyses
Zebrafish embryos were collected, washed with
pre-cooled PBS and pipetted up and down to remove yolks. Fifteen
embryos were disintegrated in lysis buffer containing protease
inhibitors for 1 h on ice. Afterward, the lysates were centrifuged
at 4°C and the supernatants were collected. Equal amounts of
proteins were separated by SDS-PAGE and transferred onto a
polyvinylidene fluoride membrane (PVDF, Roche, cat. no.
03010040001). The transferred membranes were blocked in 5% skim
milk for 1 h and then incubated with primary antibodies at 4°C
overnight. After that, membrane was incubated with secondary
antibodies at room temperature for 1 h. Signals were detected using
the enhanced chemiluminescence system (ELC, Millipore, cat no.
WBKLS0500) according to the manufacturer's instructions. Because
zebrafish PRL-3 has a high identity to human PRL-3, the human PRL-3
monoclonal antibody (clones 318) was used to detect the
overexpression of zebrafish PRL-3 in this analysis as described
previously (4).
H&E staining
Embryos were PFA-fixed, paraffin-embedded and sliced
into 5-μm thickness with Leica Microtome (RM2135). This is followed
by dewaxing in fresh xylene twice for 15 min each. All slides were
then subjected to stepwise rehydration with 100, 95, 80 and 75%
ethanol and water followed by hematoxylin staining for 2 min.
Stained slides were subsequently rinsed in distilled water and
immersed briefly in 1% acid alcohol (1% HCl in 70% ethanol).
Treated slides were immediately stained with eosin solution for
10–30 sec, followed by stepwise dehydration in 75, 80, 95 and 100%
ethanol. Slides were then cleared with xylenne and mounted in
neutral balsam before microscopic imaging (Zeiss Axio Imager
Z1).
Immunohistochemistry (IHC)
IHC experiments were conducted using monoclonal
PRL-3 antibody (clone 318) (4) to
examine PRL-3 expression in clinical chordoma specimens collected
from the First Affiliated Hospital, Sun Yat-sen University. The
clinical chordoma specimens were formalin-fixed, paraffin-embedded
and sliced into 5-μm thickness. Slides were baked at 60°C for 1 h
and then dewaxed in xylene for 10 min. This step was repeated
twice. Treated slides were rehydrated with 100%, 95%, 80%, 75%
ethanol and PBS, followed by antigen retrieval in 0.01M sodium
citrate buffer at 95°C for 15 min. Slides were then cooled and
washed with PBS for 3 times, incubated with 3%
H2O2 for 10 min, and then washed several
times with PBS. Treated slides were blocked in 10% goat serum for 2
h and then incubated at 4°C overnight with PRL-3 antibodies diluted
at 1:200. After rinsing with PBS, slides were incubated with
secondary antibodies for 30 min at room temperature. Colorimetric
detection was achieved with 3, 3-diaminobenzidine (DAB). The
reaction was terminated with water and slides were mounted in
neutral balsam, observed and imaged using Zeiss Axio Imager Z1
microscope.
Results
The expression pattern of PRL-3 in
zebrafish
Evolutionally, the strength of sequence identity
between orthologous genes parallels their conservation of gene
functions. Database search identified zebrafish Prl-3 and found
that it shares 90% protein identity with its human orthologue
(Fig. 1A), suggesting similar
roles during vertebrate development. Therefore, understanding
dynamic expression patterns of prl-3 in zebrafish may help
to appreciate its physiological role in mammals. The expression
pattern of prl-3 during zebrafish development was examined
by whole mount in situ hybridization (ISH) using
zPrl-3 antisense dig labeled probes. Zebrafish prl-3
transcripts were detected as early as the 8-cell stage of embryonic
development (Fig. 1B), indicating
prl-3 as a maternally expressed transcript. Ubiquitous
prl-3 mRNA expression continues in the whole embryo from 12
hpf (Fig. 1C) to 24 hpf (Fig. 1D), suggesting generic importance of
prl-3 in the first 24 h of embryogenesis. Tissue restricted
expression was detected from 48 hpf where zebrafish prl-3
was enriched in the zebrafish brain (br), suggesting a role in
neurogenesis (Fig. 1E). Additional
sites where prl-3 transcripts are detected include the
digestive tract (dt), muscles (mu) and vessels (ve) (Fig. 1E). Increasingly restricted
expression continues from 72 to 96 hpf where prl-3 mRNA
expression is now concentrated in specific organs, including the
esophagus (es), notochord (no), vessels (ve), and intestine (in)
(Fig. 1F and G), suggesting its
potential role in their organogenesis. Continued detection of
prl-3 transcripts in the digestive tracts from 48 hpf
(Fig. 1E) to 96 hpf (Fig. 1F and G) suggest a significant role
in the development of the digestive system. By 96 hpf prl-3
transcripts are restricted to the endothelia of zebrafish intestine
and stomach (Fig. 1H–J). Trace
expression of prl-3 transcripts is also detected in the
liver (li) at 96 hpf (Fig. 1G).
Our ISH results here clearly document the dynamic expression of
prl-3 in developing tissues. These dynamic changes of
prl-3 mRNA expression from 24 to 96 hpf were additionally
verified using northern hybridization. Relative expression value at
tested developmental stages is numerically indicated under each
band. Computed results showed reduction of prl-3 transcripts
with progressive embryonic development (Fig. 1K), in line with that observed in
ISH.
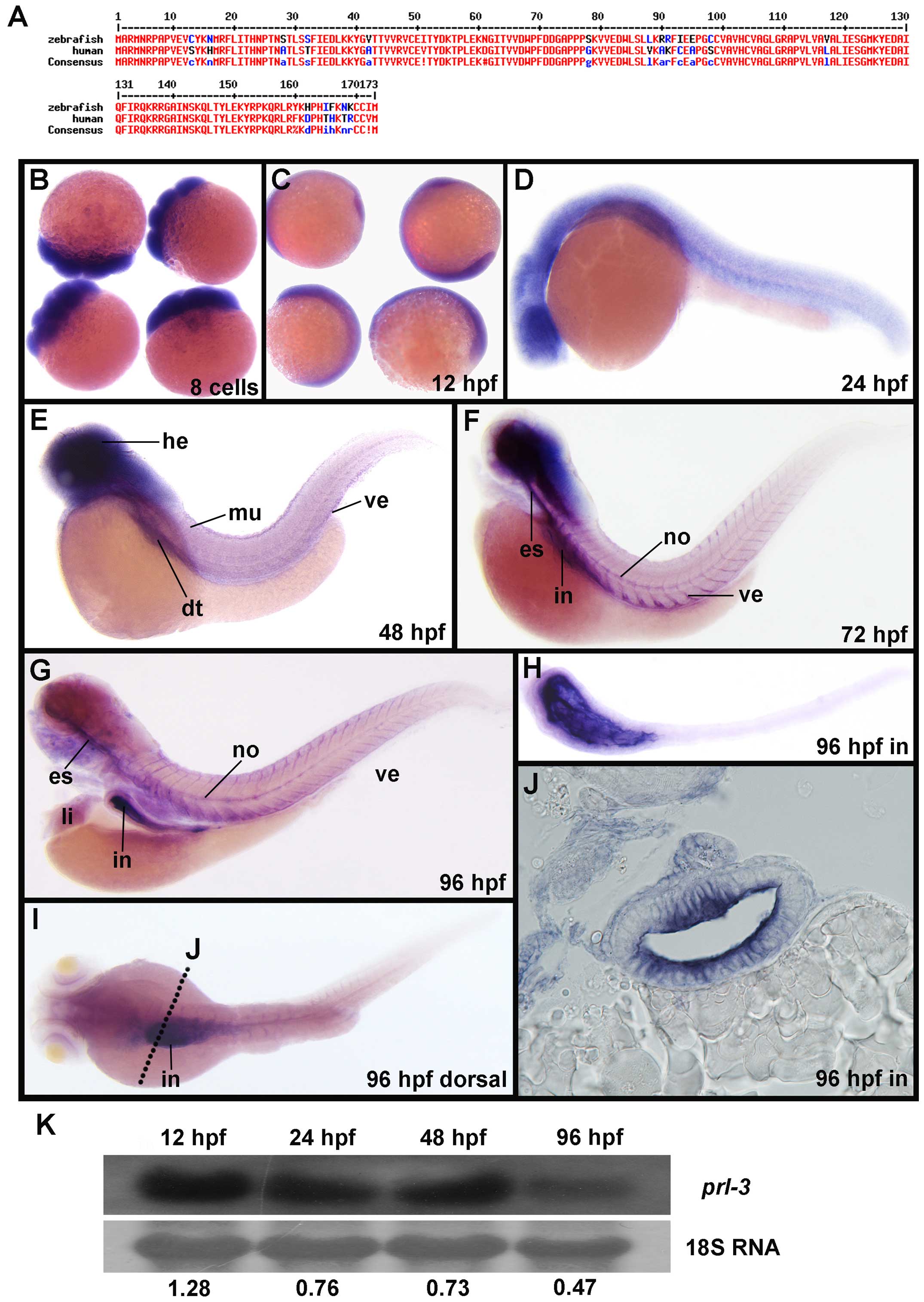 | Figure 1Dynamic expression pattern of
prl-3 transcripts in zebrafish embryo development. (A) Amino
acid sequence alignment of human and zebrafish PRL-3 protein. The
identical amino acids are highlighted in red. (B–G) Prl-3
mRNA expression pattern in zebrafish by whole mount in situ
hybridization of embryos with prl-3 specific antisense
probes. Typical lateral views of embryos at 8-cell stage (B), 12 h
post-fertilization (hpf) (C), 24 hpf (D), 48 hpf (E), 72 hpf (F),
and in 96 hpf (G) are presented. (H) Intestines peeled from embryos
at 96 hpf for in situ hybridization. (I) Dorsal view of
Prl-3 expression in the intestines of the whole embryos.
Dotted line indicates the transection of zebrafish larvea in (J).
(J) In situ hybridization of Prl-3 in the
transectioned intestines of embryos in 96 hpf. (K) Northern blot
analysis of prl-3 expression in WT embryos at 24, 48, 72 and
96 hpf. The ratio of prl-3 mRNA level versus 18S RNA level
is shown under each lane. Es, esophagus; dt, digestive tract; he,
head; in, intestine; li, liver; no, notochord; ve vessel. |
Overexpression of Zebrafish PRL-3 induces
notochord malformation
The dynamic expression pattern of prl-3 mRNA
suggests a regulated role in embryonic development. Overt
overexpression of prl-3 in zebrafish embryos will address
the consequence of perturbing prl-3 dosage during development. To
induce overt expression of endogenous prl-3 expression, we
microinjected zebrafish prl-3 mRNA into one cell stage
embryos and observe the impact on embryonic development. Control
population was microinjected with prl-3 nonsense mRNA (by
mutation of ATG→TAG) instead. Due to zebrafish PRL-3 having a high
identity to human PRL-3, the human antibody was used to detect this
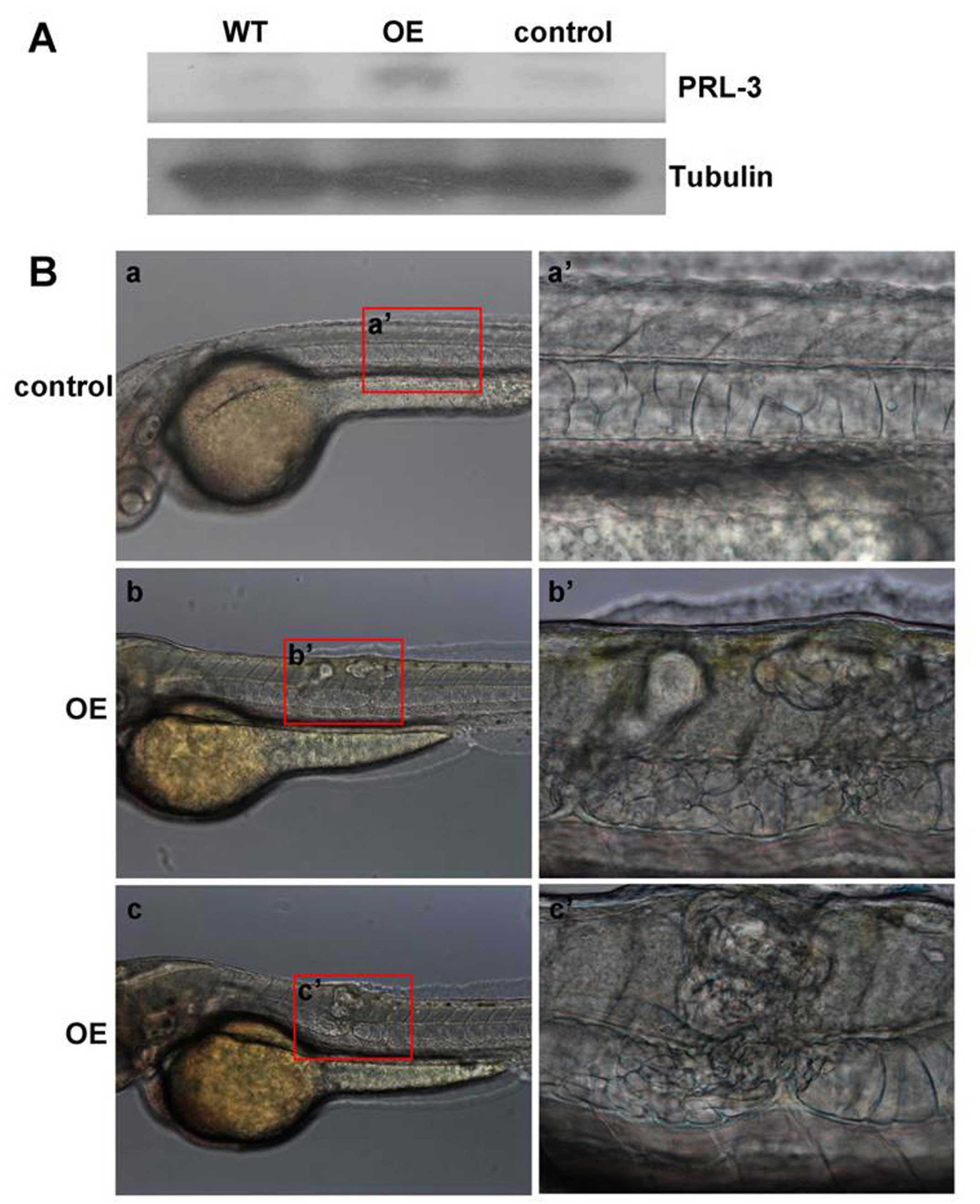
protein. Western blot analyses supported successful translation of
the microinjected prl-3 mRNA into PRL-3 protein as it
contains the highest PRL-3 protein level when compared to wild-type
or control population injected with the nonsense control mRNA
(Fig. 2A). Developmental
deformations were then tracked at 12, 24, 30, 50 and 96 hpf.
Notochord deformation characterized by aberrant cellular
proliferation in and around the notochord. Our results demonstrated
that the notochord cells of control embryos are neatly aligned into
a column (Figs. 2B-a and -a′, and
3A and A′), whereas consistent
defects of bulked lumps were observed in the PRL-3 overexpression
(OE) embryos (Figs. 2B-b, b′, c and
c′, and 3B, B′, C, C′, D and
D′). Statistical analysis confirmed that the incidence of
notochord abnormality between embryos with PRL-3 overexpression and
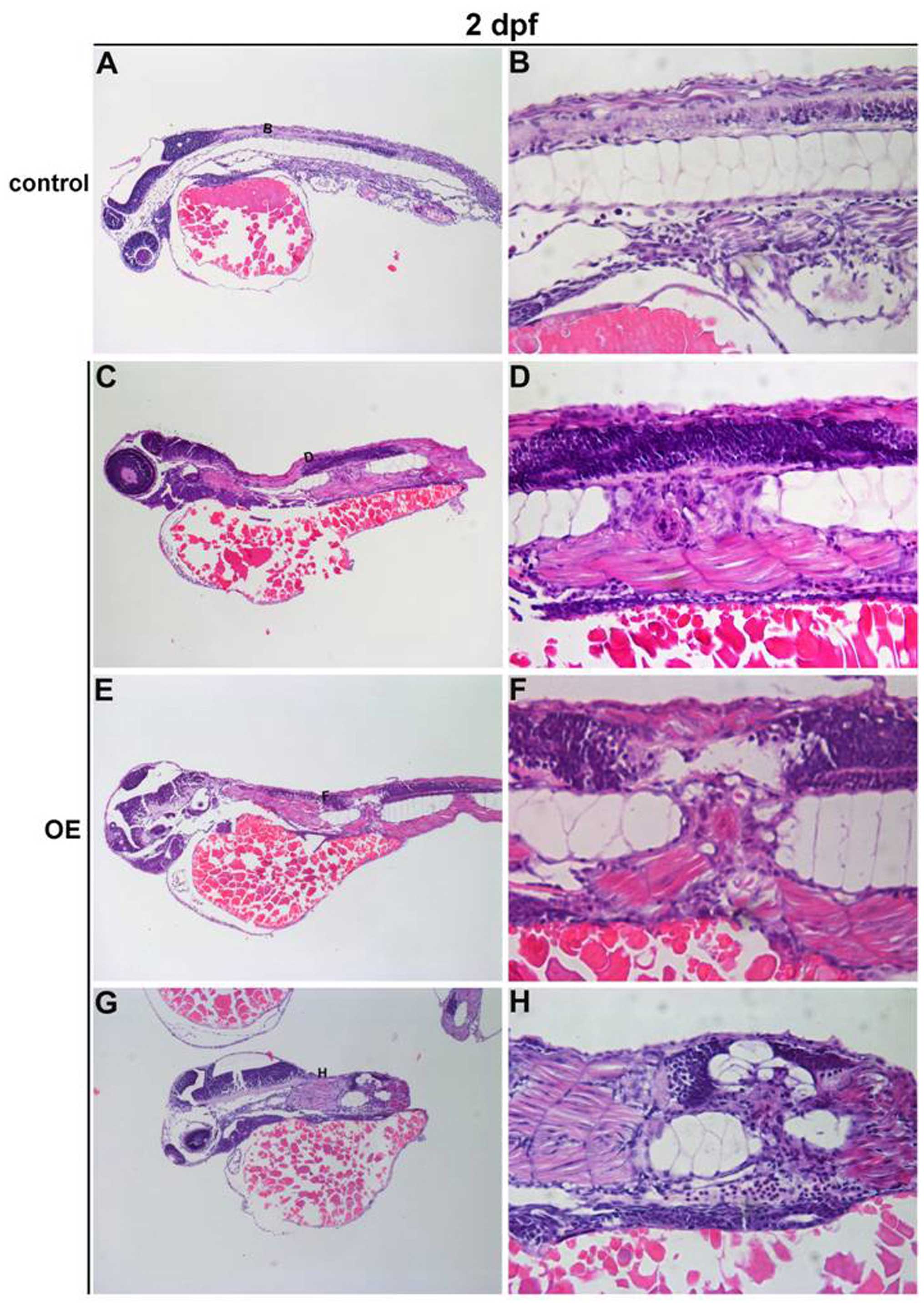
controls is statistically significant (Table II). Furthermore, H&E staining
of PRL-3 OE embryos at 2 dpf confirmed aberrant cellular
proliferation in the vicinity of the malformed notochord. The clear
difference from normal control notochord cells (Fig. 4A and B) are large vacuolated
epithelial cells, regularly aligned along a primary axis in PRL-3
OE embryos (Fig. 4C–H black box).
Taken together, our results indicated that the observed notochord
malformation could be the typical phenotype of chordoma-like
malignant tumor that arise from remnants of the embryonic
notochord, with its origin in the bones of the axial skeleton
(20).
 | Table IIStatistical data of notochordoma
occurrence rate. |
Table II
Statistical data of notochordoma
occurrence rate.
| Groups | Notochordoma
occurrence rate | P-value |
|---|
| zPRL-3 mRNA | 38% (47/125) | P<0.0001 |
| zPRL-3 nonsense
mRNA | 5% (48/1021) | |
| hPRL-3 mRNA | 24% (63/267) | P<0.0001 |
| hPRL-3 nonsense
mRNA | 3% (21/630) | |
The notochord abnormality induced by
PRL-3 overexpression is chordoma
The zebrafish notochord is an embryonic midline
structure that plays an structural role in vertebrate development
(21) and shh is used as a
molecular probe for notochord in early zebrafish embryo development
(22–25). To clarify whether the observed
notochord malformation is a result of notochord proliferation, ISH
was performed with antisense shh probes. Our results showed
that no ectopic shh mRNA expression was detected in control
population (Fig. 5A-a–c), however,
the enrichment of shh mRNA in domains of malformed notochord
was observed, which was consistently observed at various
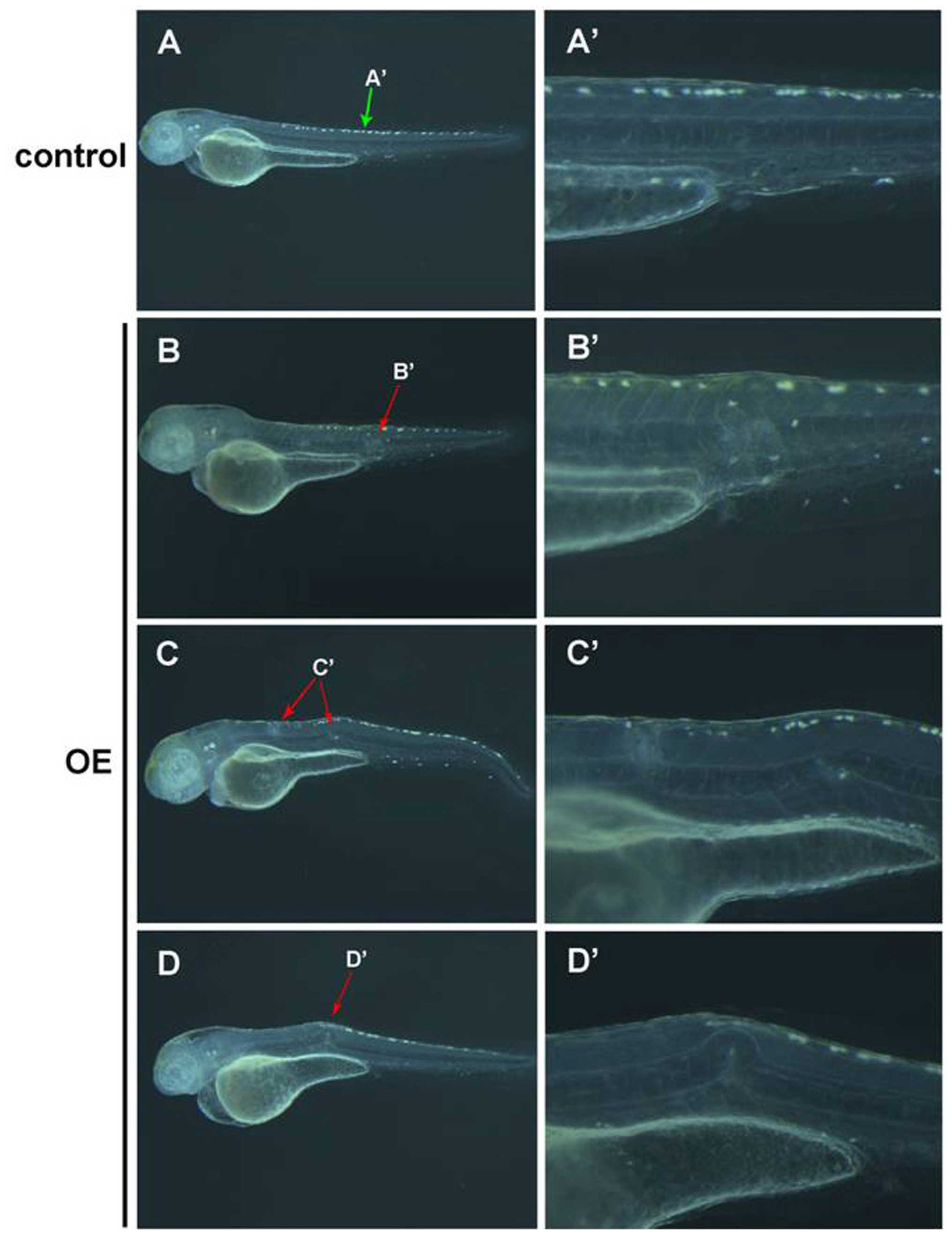
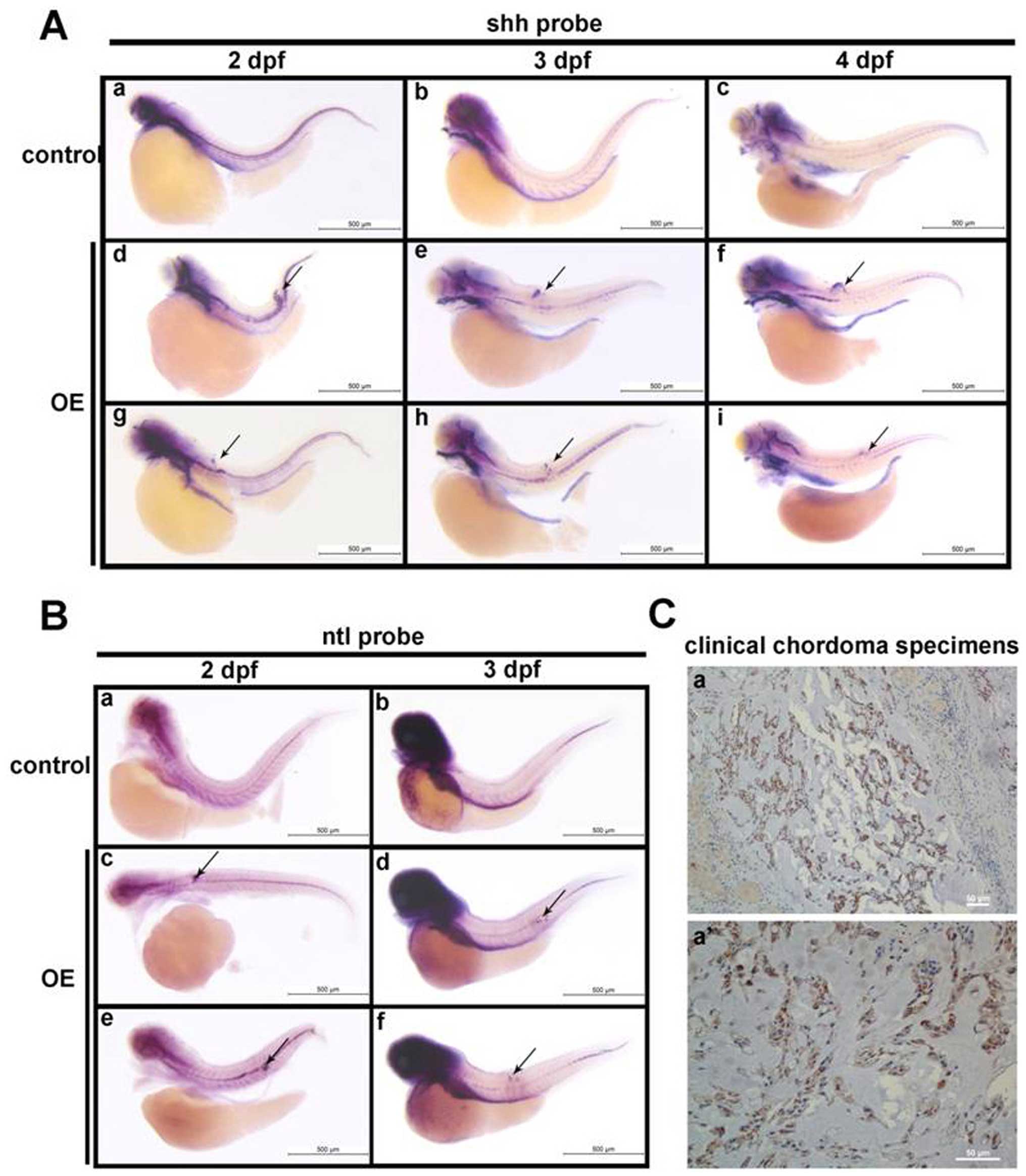
developmental stages at 2, 3 and 4 dpf (Fig. 5A-d–i). The above results suggest
the observed aberrant cellular proliferation was attributed to the
immature and undifferentiated notochord cells.
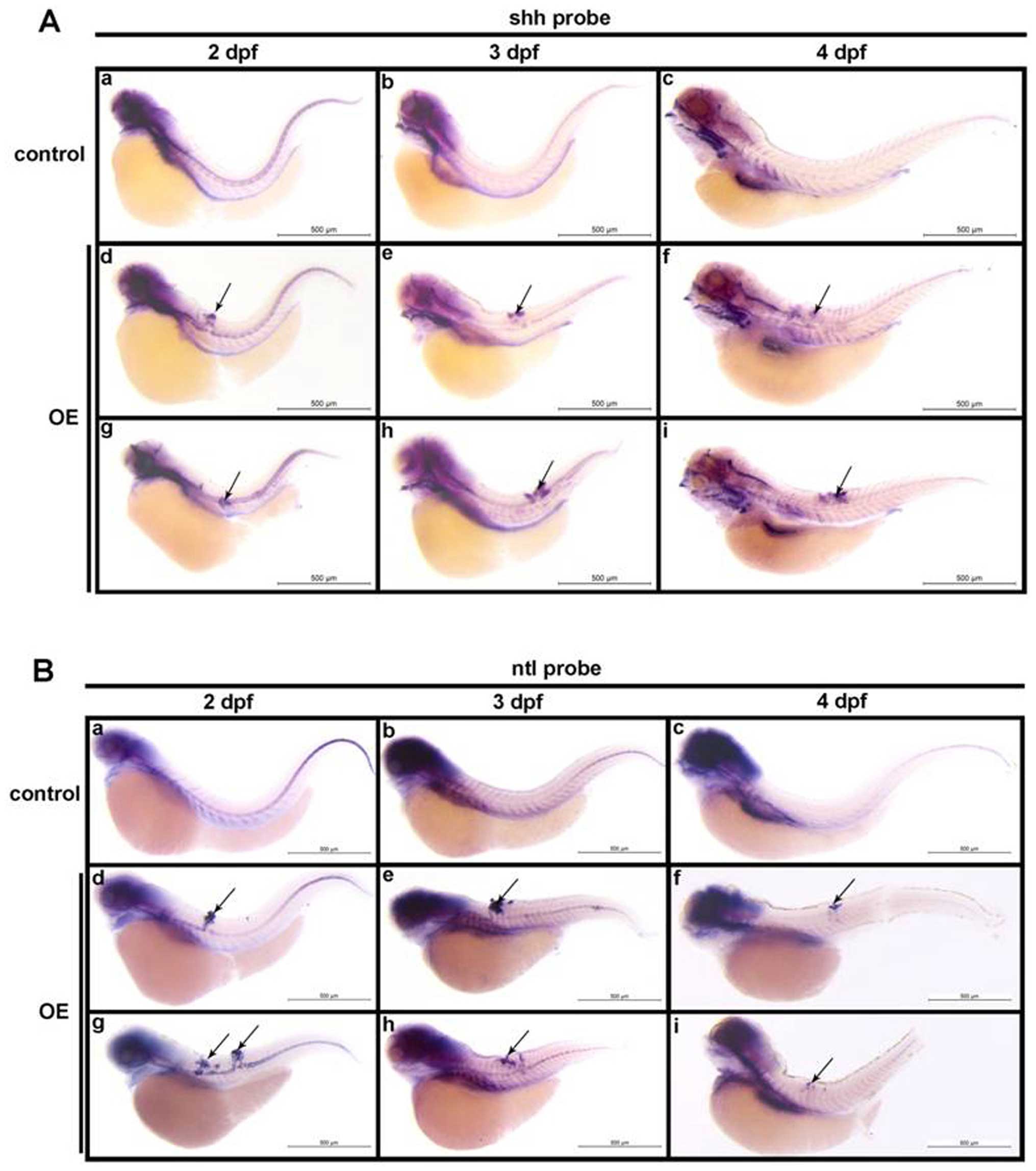 | Figure 5Identification of notochord
malformation as chordoma by in situ hybridization of
shh and ntl expression. (A) Whole mount in
situ hybridization of shh in control embyos at 2 dpf
(a), 3 dpf (b), and 4 dpf (c), compared with that in notochord
malformation induced by prl-3 upregulation (OE) at 2 dpf (d
and g), 3 dpf (e and h), and 4 dpf (f and i). The notochord
malformation areas with shh positive signals are indicated
with arrows. (B) Whole mount in situ hybridization of
chordoma-specific ntl in normal control embyos at 2 dpf (a),
3 dpf (b), and 4 dpf (c), compared with that in notochord
malformation induced by prl-3 upregulation (OE) at 2 dpf (d
and g), 3 dpf (e and h), and 4 dpf (f and i). The notochord
malformation areas with ntl-positive signals are indicated
with arrows. All bars are shown as 500 μm. |
Given that chordoma is arisen from notochord
remnants of the early embryos, it is recognized that accumulation
of shh positive cells in notochord is the phenotype of
chordoma. In zebrafish, Ntl (no tail), as a hallmarked
transcription factor, is typically expressed in the immature
notochord (26–28), and continuous Ntl expression
endorses the immature notochord expansion as neoplasia, which is
coincident to the clinical diagnosis of chordoma with brachyury as
a specific biomarker (29,30). As zebrafish ntl is
orthologous gene to brachyury, which has been evaluated in mice
(26,31,32),
we thus clarified whether the phenotype induced by prl-3
mRNA overexpression is chordoma by ISH with zebrafish ntl
probe. Our results revealed that as notochord cells become
vacuolated, the expression of ntl is extinguished in the
notochord of control embryos (Fig.
5B-a–c). In contrast, we observed the maintenance of ntl
expression in 3 dpf specimens with PRL-3 overexpression, and the
ectopic masses of cells detected in notochord malformation regions
are all ntl-positive till 4 dpi (Fig. 5B-d–i), supporting the chordoma
phenotype. Using shh and ntl probes, our results
confirmed that notochord abnormalities induced by PRL-3
overexpression during embryo development is a result of
chordoma.
Overexpression of orthologues human PRL-3
also leads to chordoma in zebrafish
We showed that zebrafish Prl-3 overexpression can
initiate chordoma in early zebrafish development. To check whether
overexpression of the human orthologue results in a similar
outcome, human PRL-3 mRNA was microinjected into one-cell
embryos. Endogenous PRL-3 level reflected in the control population
was injected with human PRL-3 nonsense mRNA. Additionally,
plasmids encoding eGFP-PRL-3 (EGFP-PRL-3) (19) and eGFP alone were also
microinjected into zebrafish embryos at one-cell stage to
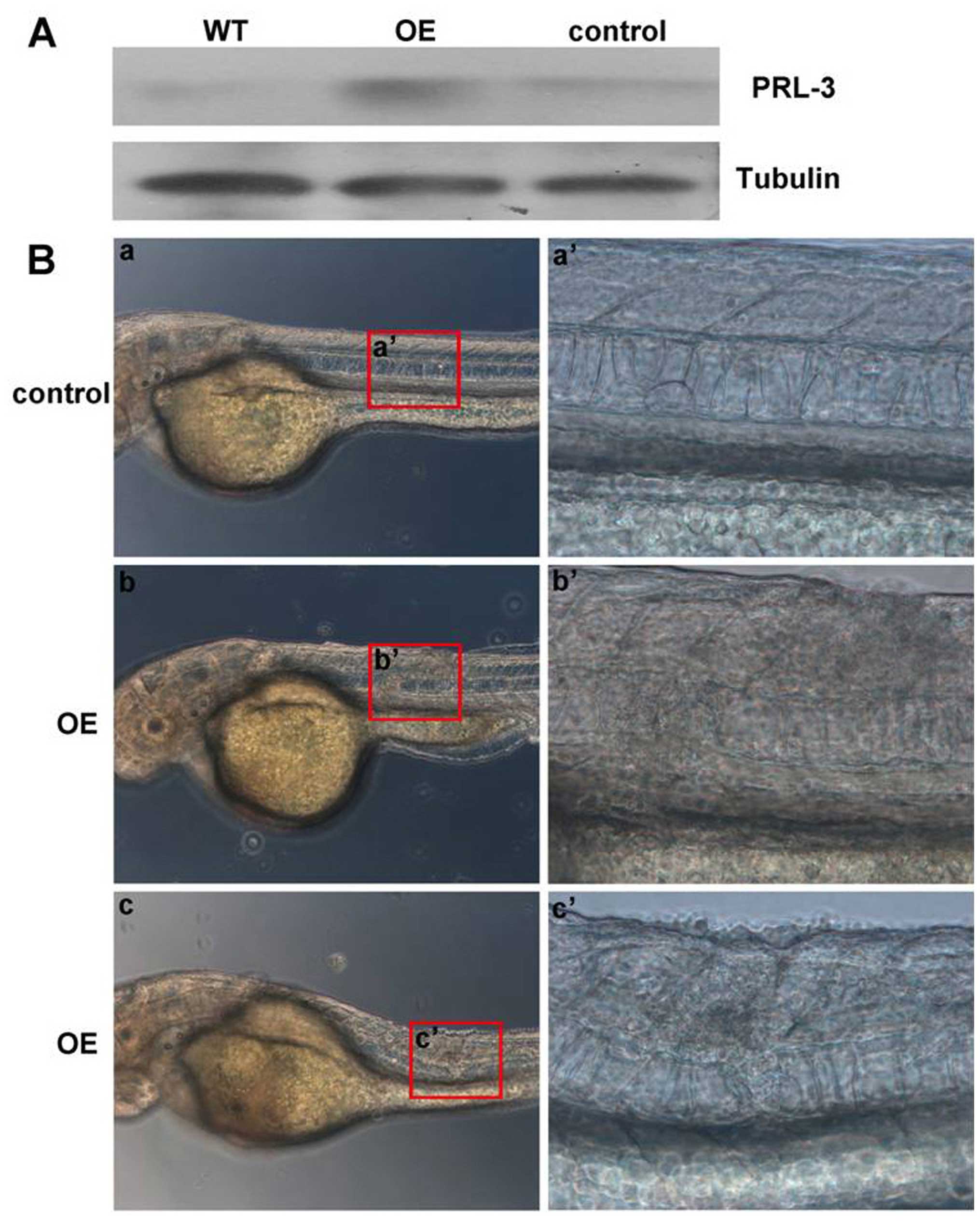
upregulate hPRL-3 expression. Western blot results confirmed
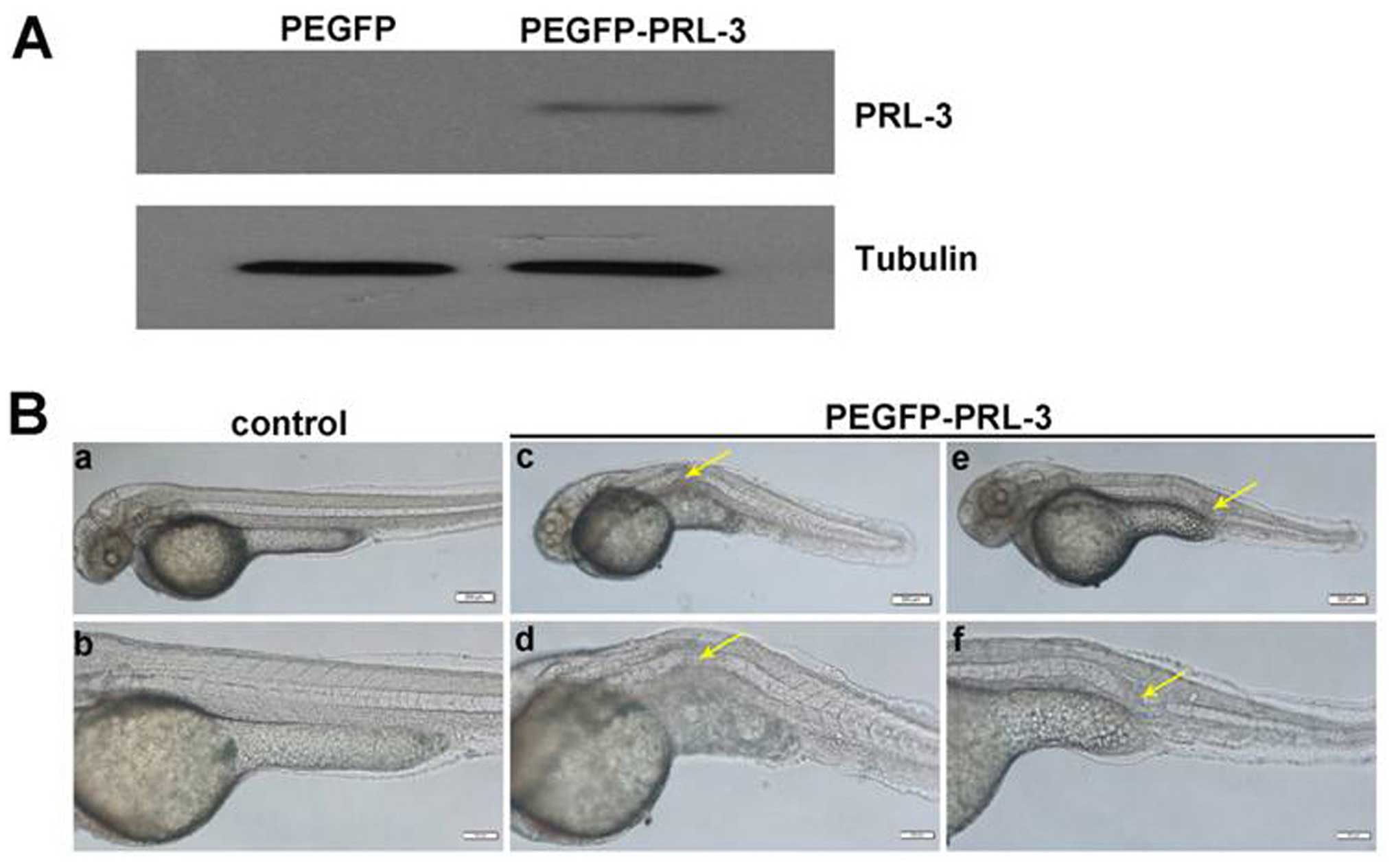
successful expression of the exogenous human PRL-3 RNA (Fig. 6A) and the GFP-PRL-3 (Fig. 7A) in injected zebrafish embryos
while level of PRL-3 in control is comparable to un-injected
wild-type. Embryos with notochord malformation were observed by 30
h post-injection and appearance of which is similar to those
induced by zPrl-3 overexpression. Examples of injected
embryos depicting the notochord deformation phenotype are
photographed at 48 h post-PRL-3 mRNA (Fig. 6B) or its GFP-PRL-3 plasmid
(Fig. 7B) injection. Statistical
analysis showed that overexpressing hPRL-3 by either PRL-3 mRNA
injection or its plasmids can significantly induce notochord
malformation, compared to the control groups (Table II). This phenomenon revealed
functional conservation of human Prl-3 and zebrafish PRL-3 in
inducing notochord developmental distortion. Furthermore, the
abnormal notochords caused by human PRL-3 overexpression were
confirmed by ISH as aberrant proliferation of notochord cells with
shh and ntl probes, defining features of chordoma
(Fig. 8A and B). Most importantly,
PRL-3 was detected in the few clinical chordoma specimens with
PRL-3 antibody by immunohistochemistry (IHC) analysis (Fig. 8C), further supporting PRL-3 as a
predictor and therapeutic target for chordoma.
Discussion
Previous reports show that PRL-3 is usually
expressed in heart, skeletal muscle and small intestine of mouse
tissues (15,16), and similarly expressed in human
fetal heart, skeletal muscle and pre-erythrocytes of bone marrow
(9,33). However, one recent report showed
that PRL-3 is mainly expressed in somites of zebrafish (34). To clarify these confusions, we
investigated the dynamic expression pattern of prl-3 in
zebrafish and showed that prl-3 is expressed maternally and
is ubiquitously expressed in early stages of zebrafish development
(Fig. 1B and C). Progressive
embryo development from 2 to 4 days post-fertilization results in
progressive decline in prl-3 expression but its expression
is retained in proliferative areas, including the anterior
intestine, esophagus, vessel and notochord (Fig. 1D–F). Hence prl-3 is
dynamically expressed during embryonic development.
Given that brachyury (ntl) is
expressed in the zebrafish notochord at the beginning of
gastrulation and eventually ntl and shh expression
are extinguished in notochord when notochord cells becomes
vacuolated (26–28), our results revealed that ntl
and shh were persistently expressed in the deformed
notochord regions in PRL-3-overexpressed zebrafishes, further
highlighting the impact of overt zebrafish and human PRL-3
expression in notochord malformation (Figs. 2B, 4 and 6B). We further demonstrated for the first
time that this notochord deformation is attributed to aberrant
proliferation of immature notochord cells (Figs. 5 and 8A and B). Despite the rapid onset of
chordoma in zebrafish embryos, developed tumors have similar
histological characteristics with that of human chordoma (20,35).
Taken together, our discoveries indicate PRL-3 may play an
important role in notochord development and aberrant expression of
PRL-3 is detrimental to zebrafish embryonic development due to
aberrant proliferation of notochord cells. This suggests a key role
of PRL-3 in chordoma formation.
Previous studies correlate enhanced PRL-3 expression
as a driver of cancer metastasis and a prognostic biomarker for
various human cancers (8,14,36).
The discovery of high PRL-3 protein expression in clinical
notochordoma samples (Fig. 8C)
additionally suggests the possible usage of PRL-3 as a specific
biomarker for chordoma diagnosis.
Acknowledgements
The authors thank Professor Jun Chen, Zhejiang
University, for providing the PCS2 plasmid as a gift. We also
appreciate Dr Cathleen Teh (IMCB, A*STAR, Singapore) for
her helpful editing of this manuscript. This study was supported by
National Science Foundation of China (no. 81472730) to W.H. and
Guangzhou Science Technology and Innovation Commission (no.
201510010144) to Y.S.
References
|
1
|
Stephens BJ, Han H, Gokhale V and Von Hoff
DD: PRL phosphatases as potential molecular targets in cancer. Mol
Cancer Ther. 4:1653–1661. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saha S, Bardelli A, Buckhaults P,
Velculescu VE, Rago C, St Croix B, Romans KE, Choti MA, Lengauer C,
Kinzler KW, et al: A phosphatase associated with metastasis of
colorectal cancer. Science. 294:1343–1346. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Radke I, Götte M, Kersting C, Mattsson B,
Kiesel L and Wülfing P: Expression and prognostic impact of the
protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast
cancer. Br J Cancer. 95:347–354. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Polato F, Codegoni A, Fruscio R, Perego P,
Mangioni C, Saha S, Bardelli A and Broggini M: PRL-3 phosphatase is
implicated in ovarian cancer growth. Clin Cancer Res. 11:6835–6839.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ren T, Jiang B, Xing X, Dong B, Peng L,
Meng L, Xu H and Shou C: Prognostic significance of phosphatase of
regenerating liver-3 expression in ovarian cancer. Pathol Oncol
Res. 15:555–560. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao WB, Li Y, Liu X, Zhang LY and Wang X:
Evaluation of PRL-3 expression, and its correlation with
angiogenesis and invasion in hepatocellular carcinoma. Int J Mol
Med. 22:187–192. 2008.PubMed/NCBI
|
|
7
|
Ooki A, Yamashita K, Kikuchi S, Sakuramoto
S, Katada N, Waraya M, Kawamata H, Nishimiya H, Nakamura K and
Watanabe M: Therapeutic potential of PRL-3 targeting and clinical
significance of PRL-3 genomic amplification in gastric cancer. BMC
Cancer. 11:1222011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Al-Aidaroos AQ and Zeng Q: PRL-3
phosphatase and cancer metastasis. J Cell Biochem. 111:1087–1098.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo K, Li J, Wang H, Osato M, Tang JP,
Quah SY, Gan BQ and Zeng Q: PRL-3 initiates tumor angiogenesis by
recruiting endothelial cells in vitro and in vivo. Cancer Res.
66:9625–9635. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu X, Zeng H, Zhang X, Zhao Y, Sha H, Ge
X, Zhang M, Gao X and Xu Q: Phosphatase of regenerating liver-3
promotes motility and metastasis of mouse melanoma cells. Am J
Pathol. 164:2039–2054. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guzińska-Ustymowicz K and Pryczynicz A:
PRL-3, an emerging marker of carcinogenesis, is strongly associated
with poor prognosis. Anticancer Agents Med Chem. 11:99–108. 2011.
View Article : Google Scholar
|
|
12
|
Fiordalisi JJ, Keller PJ and Cox AD: PRL
tyrosine phosphatases regulate rho family GTPases to promote
invasion and motility. Cancer Res. 66:3153–3161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang F, Liang J, Wang WQ, Sun JP, Udho E
and Zhang ZY: PRL3 promotes cell invasion and proliferation by
down-regulation of Csk leading to Src activation. J Biol Chem.
282:5413–5419. 2007. View Article : Google Scholar
|
|
14
|
Wang H, Quah SY, Dong JM, Manser E, Tang
JP and Zeng Q: PRL-3 down-regulates PTEN expression and signals
through PI3K to promote epithelial-mesenchymal transition. Cancer
Res. 67:2922–2926. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matter WF, Estridge T, Zhang C, Belagaje
R, Stancato L, Dixon J, Johnson B, Bloem L, Pickard T, Donaghue M,
et al: Role of PRL-3, a human muscle-specific tyrosine phosphatase,
in angiotensin-II signaling. Biochem Biophys Res Commun.
283:1061–1068. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng Q, Hong W and Tan YH: Mouse PRL-2 and
PRL-3, two potentially prenylated protein tyrosine phosphatases
homologous to PRL-1. Biochem Biophys Res Commun. 244:421–427. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng Q, Si X, Horstmann H, Xu Y, Hong W
and Pallen CJ: Prenylation-dependent association of
protein-tyrosine phosphatases PRL-1, -2, and -3 with the plasma
membrane and the early endosome. J Biol Chem. 275:21444–21452.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang SL, Aw SS, Chang C, Korzh S, Korzh V
and Peng J: Depletion of Bhmt elevates sonic hedgehog transcript
level and increases β-cell number in zebrafish. Endocrinology.
152:4706–4717. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng Q, Dong JM, Guo K, Li J, Tan HX, Koh
V, Pallen CJ, Manser E and Hong W: PRL-3 and PRL-1 promote cell
migration, invasion, and metastasis. Cancer Res. 63:2716–2722.
2003.PubMed/NCBI
|
|
20
|
Burger A, Vasilyev A, Tomar R, Selig MK,
Nielsen GP, Peterson RT, Drummond IA and Haber DA: A zebrafish
model of chordoma initiated by notochord-driven expression of
HRASV12. Dis Model Mech. 7:907–913. 2014. View Article : Google Scholar :
|
|
21
|
Stemple DL: Structure and function of the
notochord: An essential organ for chordate development.
Development. 132:2503–2512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krauss S, Concordet JP and Ingham PW: A
functionally conserved homolog of the Drosophila segment polarity
gene hh is expressed in tissues with polarizing activity in
zebrafish embryos. Cell. 75:1431–1444. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roelink H, Augsburger A, Heemskerk J,
Korzh V, Norlin S, Ruiz i Altaba A, Tanabe Y, Placzek M, Edlund T,
Jessell TM, et al: Floor plate and motor neuron induction by vhh-1,
a vertebrate homolog of hedgehog expressed by the notochord. Cell.
76:761–775. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan YL, Hatta K, Riggleman B and
Postlethwait JH: Expression of a type II collagen gene in the
zebrafish embryonic axis. Dev Dyn. 203:363–376. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Corallo D, Trapani V and Bonaldo P: The
notochord: Structure and functions. Cell Mol Life Sci.
72:2989–3008. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Halpern ME, Ho RK, Walker C and Kimmel CB:
Induction of muscle pioneers and floor plate is distinguished by
the zebrafish no tail mutation. Cell. 75:99–111. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kispert A and Hermann BG: The Brachyury
gene encodes a novel DNA binding protein. EMBO J. 12:4898–4899.
1993.PubMed/NCBI
|
|
28
|
Schulte-Merker S, Ho RK, Herrmann BG and
Nüsslein-Volhard C: The protein product of the zebrafish homologue
of the mouse T gene is expressed in nuclei of the germ ring and the
notochord of the early embryo. Development. 116:1021–1032.
1992.PubMed/NCBI
|
|
29
|
Barresi V, Ieni A, Branca G and Tuccari G:
Brachyury: A diagnostic marker for the differential diagnosis of
chordoma and hemangioblastoma versus neoplastic histological
mimickers. Dis Markers. 2014:5147532014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nibu Y, José-Edwards DS and Di Gregorio A:
From notochord formation to hereditary chordoma: The many roles of
Brachyury. BioMed Res Int. 2013:8264352013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schulte-Merker S, van Eeden FJ, Halpern
ME, Kimmel CB and Nüsslein-Volhard C: no tail (ntl) is the
zebrafish homologue of the mouse T (Brachyury) gene. Development.
120:1009–1015. 1994.PubMed/NCBI
|
|
32
|
Kispert A and Herrmann BG:
Immunohistochemical analysis of the Brachyury protein in wild-type
and mutant mouse embryos. Dev Biol. 161:179–193. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dumaual CM, Sandusky GE, Crowell PL and
Randall SK: Cellular localization of PRL-1 and PRL-2 gene
expression in normal adult human tissues. J Histochem Cytochem.
54:1401–1412. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin MD, Lee HT, Wang SC, Li HR, Hsien HL,
Cheng KW, Chang YD, Huang ML, Yu JK and Chen YH: Expression of
phosphatase of regenerating liver family genes during
embryogenesis: An evolutionary developmental analysis among
Drosophila, amphioxus, and zebrafish. BMC Dev Biol. 13:182013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ferrari L, Pistocchi A, Libera L, Boari N,
Mortini P, Bellipanni G, Giordano A, Cotelli F and Riva P: FAS/FASL
are dysregulated in chordoma and their loss-of-function impairs
zebrafish notochord formation. Oncotarget. 5:5712–5724. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peng L, Ning J, Meng L and Shou C: The
association of the expression level of protein tyrosine phosphatase
PRL-3 protein with liver metastasis and prognosis of patients with
colorectal cancer. J Cancer Res Clin Oncol. 130:521–526. 2004.
View Article : Google Scholar : PubMed/NCBI
|