MicroRNAs (miRNAs or miRs) are a family of small
(19–25 nucleotides in length) non-coding RNAs that have a key role
in the regulation of gene expression through the inhibition or the
reduction of protein synthesis following mRNA complementary
sequence base pairing (1–4). A single or multiple mRNAs can be
targeted at the 3′ untranslated region (3′UTR), coding sequence
(CDS) or 5′ untranslated region (5′UTR) sequence, and it is
calculated that >60% of human mRNAs are recognized by miRNAs
(1–4). The miRNA/mRNA interaction occurs at
the level of the RNA-induced silencing complex (RISC) and causes
translational repression or mRNA degradation, depending on the
degree of complementarity with target mRNA sequences (5–8).
Since their discovery and first characterization, the number of
human miRNAs identified and deposited in the miRBase databases
(miRBase v.21, www.mirbase.org) has increaed (it is
>2,500) (9–16) and the research studies on miRNAs
have confirmed the very high complexity of the networks constituted
by miRNAs and RNA targets (17–22).
Alterations in miRNA expression have been
demonstrated to be associated with different human pathologies, and
guided alterations of specific miRNAs have been suggested as novel
approaches for the development of innovative therapeutic protocols
(23,24). Studies have conclusively
demonstrated that miRNAs are deeply involved in tumor onset and
progression, either behaving as tumor-promoting miRNAs (oncomiRNAs
and metastamiRNAs) or as tumor suppressor miRNAs (25,26).
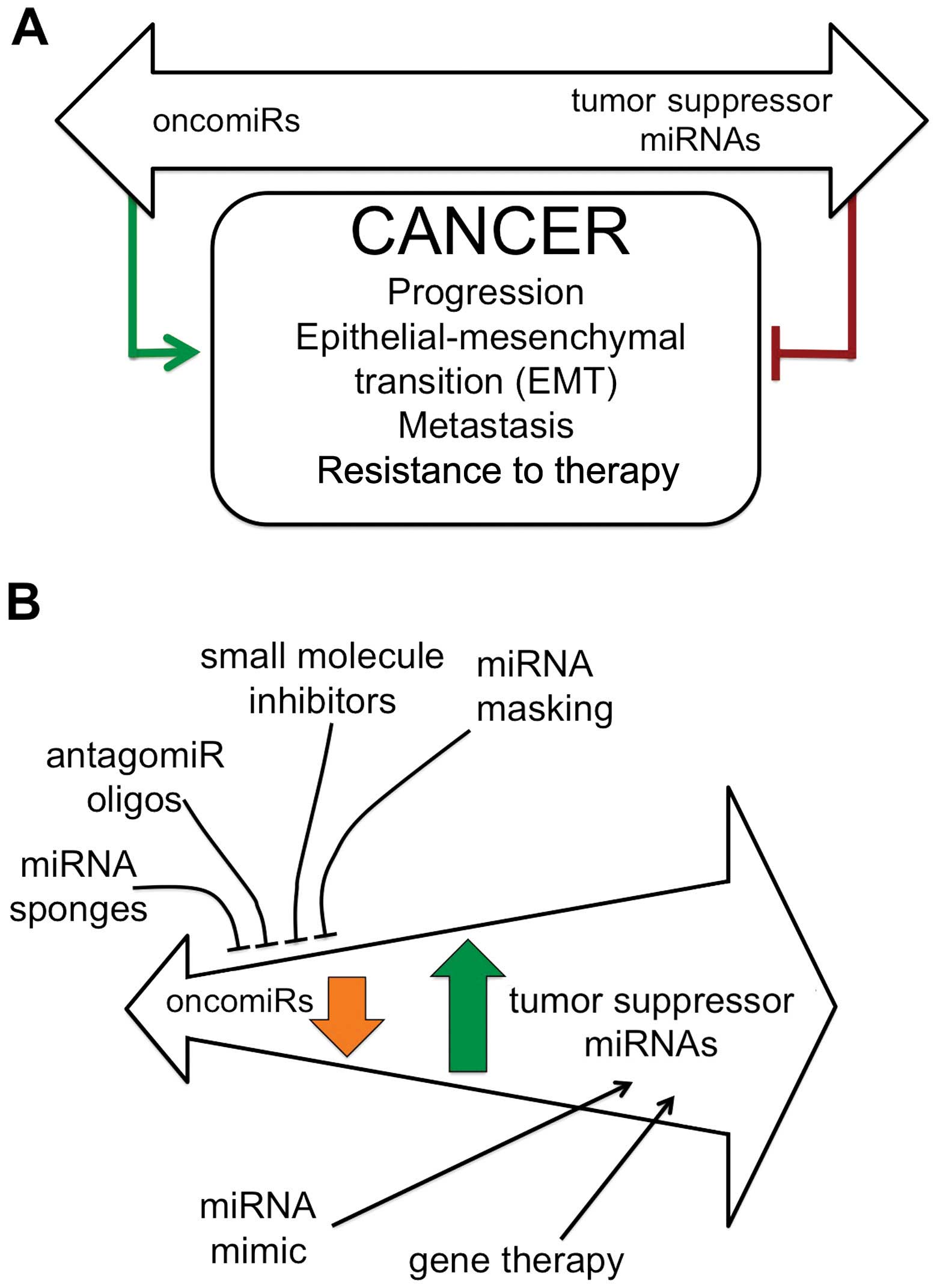
In general, miRNAs able to promote cancer target mRNAs coding for
tumor suppressor proteins, whereas miRNAs exhibiting tumor
suppressor properties usually target mRNAs coding oncoproteins (see
the scheme depicted in Fig. 1A).
This has a very important implication in diagnosis and/or
prognosis, including the recent discovery that the pattern of
circulating cell-free miRNAs in serum allows us to perform
molecular analyses on these non-invasive liquid biopsies with deep
diagnostic and prognostic implications. This research field has
confirmed that cancer-specific miRNAs are present in extracellular
body fluids, and may play a very important role in the crosstalk
between cancer cells and surrounding normal cells (27–32).
Interestingly, the evidence of the presence of
miRNAs in serum, plasma and saliva supports their potential as an
additional set of biomarkers for cancer. The extracellular miRNAs
are protected by exosome-like structures, small intraluminal
vesicles shed from a variety of cells (including cancer cells),
with a biogenesis connected with endosomal sorting complex required
for transport machinery in multivesicular bodies (29). For instance, miR-141 and
miR-221/222 are predicted biomarkers in liquid biopsies from
patients with colon cancer (33,34).
On the other hand, tumor-associated miRNAs are
suitable targets for intervention therapeutics, as previously
reported (35–44) and summarized in Fig. 1B. The inhibition of miRNA activity
can be readily achieved by the use of miRNA inhibitors and
oligomers, including RNA, DNA and DNA analogues (miRNA antisense
therapy) (45–47), small molecule inhibitors, locked
nucleic acids (LNAs) (48–53), peptide nucleic acids (PNAs)
(54–57), morpholinos (58–60),
miRNA sponges (61–67), mowers (68) or through miRNA masking that
inhibits miRNA function by masking the miRNA binding site of a
target mRNA using a modified single-stranded RNA complementary to
the target sequence (69–75). On the contrary, the enhancement of
miRNA function (miRNA replacement therapy) can be achieved by the
use of modified miRNA mimetics, either synthetic, or produced by
plasmid or lentiviral vectors carrying miRNA sequences (76–81).
Several miRNAs exhibit onco-suppressor properties by
targeting mRNAs coding oncoproteins (82–105). Therefore, these onco-suppressor
miRNAs have been found to be often downregulated in tumors. For
instance, Fernandez et al (106) recently described the intriguing
tumor suppressor activity of miR-340, showing the miR-340-mediated
inhibition of multiple negative regulators of p27, a protein
involved in apoptosis and cell cycle progression. These
interactions with oncoprotein-coding mRNA targets determine the
inhibition of cell cycle progression, the induction of apoptosis
and growth inhibition. The miR-340-mediated downregulation of three
post-transcriptional regulators [Pumilio RNA-binding family member
(PUM)1, PUM2 and S-phase kinase-associated protein 2 (SKP2)]
correlates with the upregulation of p27. PUM1 and PUM2 inhibit p27
at the translational level, by rendering the p27 transcript
available to interact with two oncomiRs (miR-221 and miR-222),
while the oncoprotein SKP2 inhibits the CDK inhibitor at the
post-translational level by triggering the proteasomal degradation
of p27, showing that miR-340 affected not only the synthesis but
also the decay of p27. Moreover their data confirm the recent
identification of transcripts encoding several pro-invasive
proteins such as c-Met, implicated in breast cancer cell migration,
RhoA and Rock1, implicated in the control of the migration and
invasion of osteosarcoma cells, and E-cadherin mRNA, involved in
the miR-340-induced loss of intercellular adhesion (106 and refs within).
Recently, miR-18a was demonstrated to play a
protective role in colorectal carcinoma (CRC) by inhibiting the
proliferation, invasion and migration of CRC cells by directly
targeting the TBP-like 1 (TBPL1) gene. The onco-suppressor activity
of miR-18a in CRC tissues and cell lines was supported by the
finding that the content of this mRNA is markedly lower in tumor
cells with respect to normal control tissues and cells (107). In addition Xishan et al
(108) found that miR-320a acts
as a novel tumor suppressor gene in chronic myelogenous leukemia
(CML) and can decrease the migratory, invasive, proliferative and
apoptotic behavior of CML cells, as well as epithelial-mesenchymal
transition (EMT), by attenuating the expression of the BCR/ABL
oncogene. Furthermore Zhao et al (109) demonstrated that miR-449a
functions as a tumor suppressor in neuroblastoma by inducing cell
differentiation and cell cycle arrest. Finally, Kalinowski et
al (110) and Gu et al
(111) demonstrated the
significant role of miR-7 in cancer which functions by directly
targeting and inhibiting key oncogenic signaling molecules involved
in cell cycle progression, proliferation, invasion and metastasis.
A partial list of onco-suppressor miRNAs is presented in Table I.
miRNAs can act as oncogenes and have been
demonstrated to play a causal role in the onset and progression of
human cancer (oncomiRNAs) (224–233). Recent findings have nevertheless
identified a subclass of miRNAs whose expression is highly
associated with the acquisition of metastatic phenotypes and are
referred to as miRs endowed with either metastasis-promoting or
tumor suppressor inhibitory activities (213,234,235).
Recent data have revealed that miR-25 may act as an
onco-miRNA in osteosarcoma, negatively regulating the protein
expression of the cell cycle inhibitor, p27. In agreement with this
hypothesis restoring the p27 level in miR-25-over-expressing cells
was shown to reverse the enhancing effect of miR-25 on Saos-2 and
U2OS cell proliferation (236).
In addition a recent study published by Siu et al (237), describes miR-96 as a potential
target of therapeutics for metastatic prostate cancer,
demonstrating the enhanced effects in cellular growth and
invasiveness of miR-96 in cell lines (AC1, AC3 and SC1) derived
from prostate-specific, Pten/Tp53 double knockout mice and
confirmed in tissue samples from prostate cancer patients. miR-96
acts as an oncomiR and metastamiR through TGF-β/mTOR signaling,
promoting bone metastasis and contributing to a reduced survival
rate in prostate cancer (237).
Furthermore Xia et al (238) demonstrated that the
overexpression of miR-1908 significantly decreased the expression
of PTEN in glioblastoma cells, one of the most frequently mutated
tumor suppressors in human cancer, resulting in an increase in
proliferation, migration and invasion. Finally Sachdeva et
al (239), found that miR-182
targets multiple genes in lung metastasis and regulates
intravasation, thus increasing the number of circulating tumor
cells (CTCs). Only the simultaneous restoration of miR-182 target
genes decreased the number of metastases in vivo,
demonstrating that a single miRNA can regulate the metastasis of
primary tumors in vivo by the coordinated regulation of
multiple genes. Selected examples of oncomiRNAs and metastamiRNAs
are presented in Tables II and
III. All these miRNAs act by
inhibiting tumor suppressor pathways.
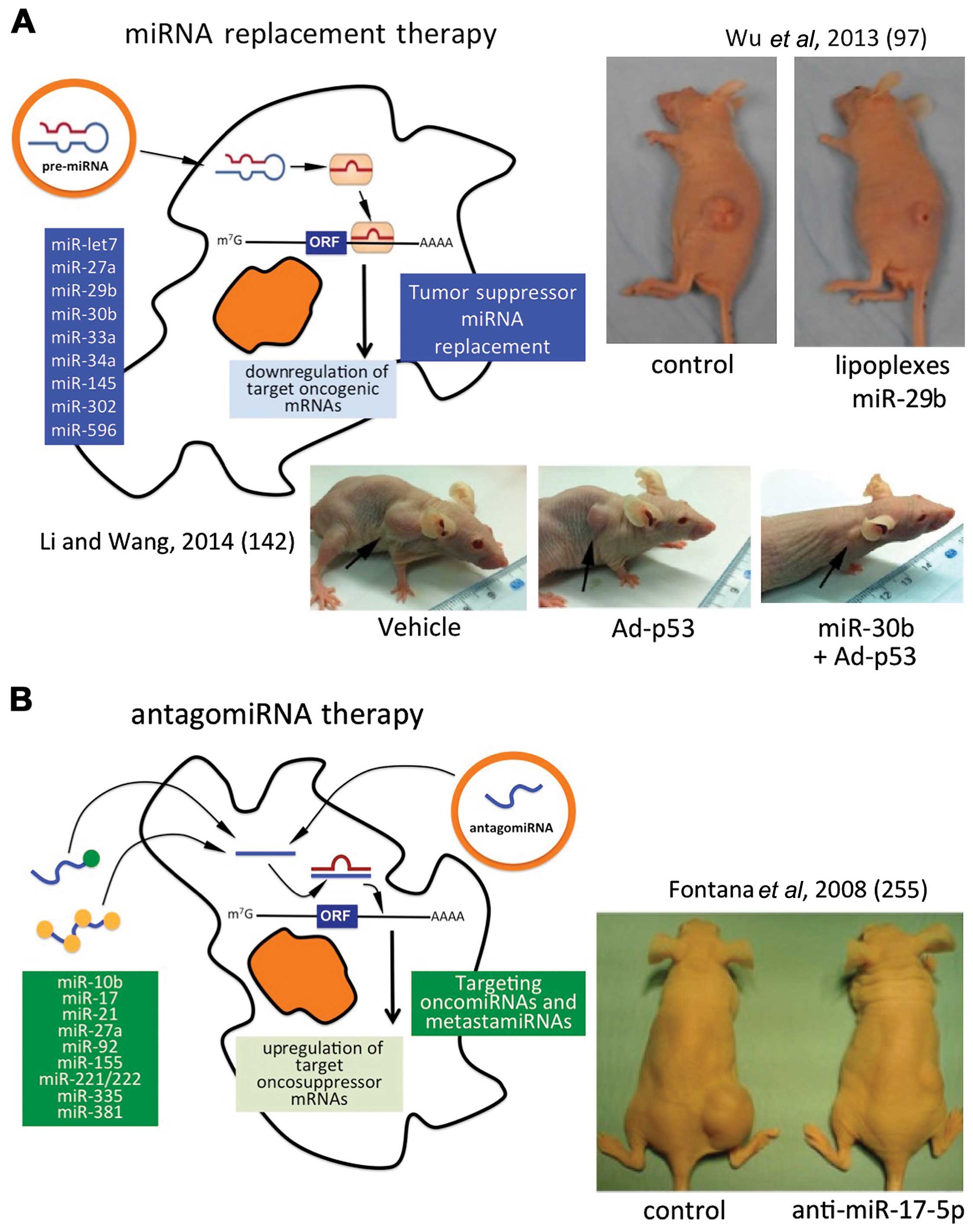
Using the development of anticancer therapies as a
representative field of investigation, the therapeutic strategy
based on miRNA replacement is targeted to pathological cells which
downregulate onco-suppressor miRNAs playing a role in controlling
the expression of mRNAs encoding key oncoproteins. The
downregulation of these oncogene-targeting miRNAs is clearly the
key step for oncogene upregulation leading to tumor onset and
progression. Table IV presents
selected examples of miRNA replacement therapy in cancer research
and treatment (90–92,94–97,99).
The effects of therapeutic molecules against miRNAs
have been the object of very recent studies, in part summarized in
Table V (309–316). Of course, the endpoint of the
treatment of target cells with molecules against selected miRNAs is
the alteration of miRNA-regulated genes. As a first example,
Wagenaar et al (317)
developed potent and specific single-stranded oligonucleotide
inhibitors of miR-21 and used them to verify dependency on miR-21
in a panel of liver cancer cell lines. Treatment with anti-miR-21,
but not with a mismatch control anti-miRNA, resulted in the
significant derepression of direct targets of miR-21 and led to the
loss of viability in the majority of HCC cell lines tested. The
robust induction of caspase activity, apoptosis and necrosis was
noted in the anti-miR-21-treated HCC cells. Furthermore, the
ablation of miR-21 activity resulted in the inhibition of HCC cell
migration and in the suppression of clonogenic growth (317).
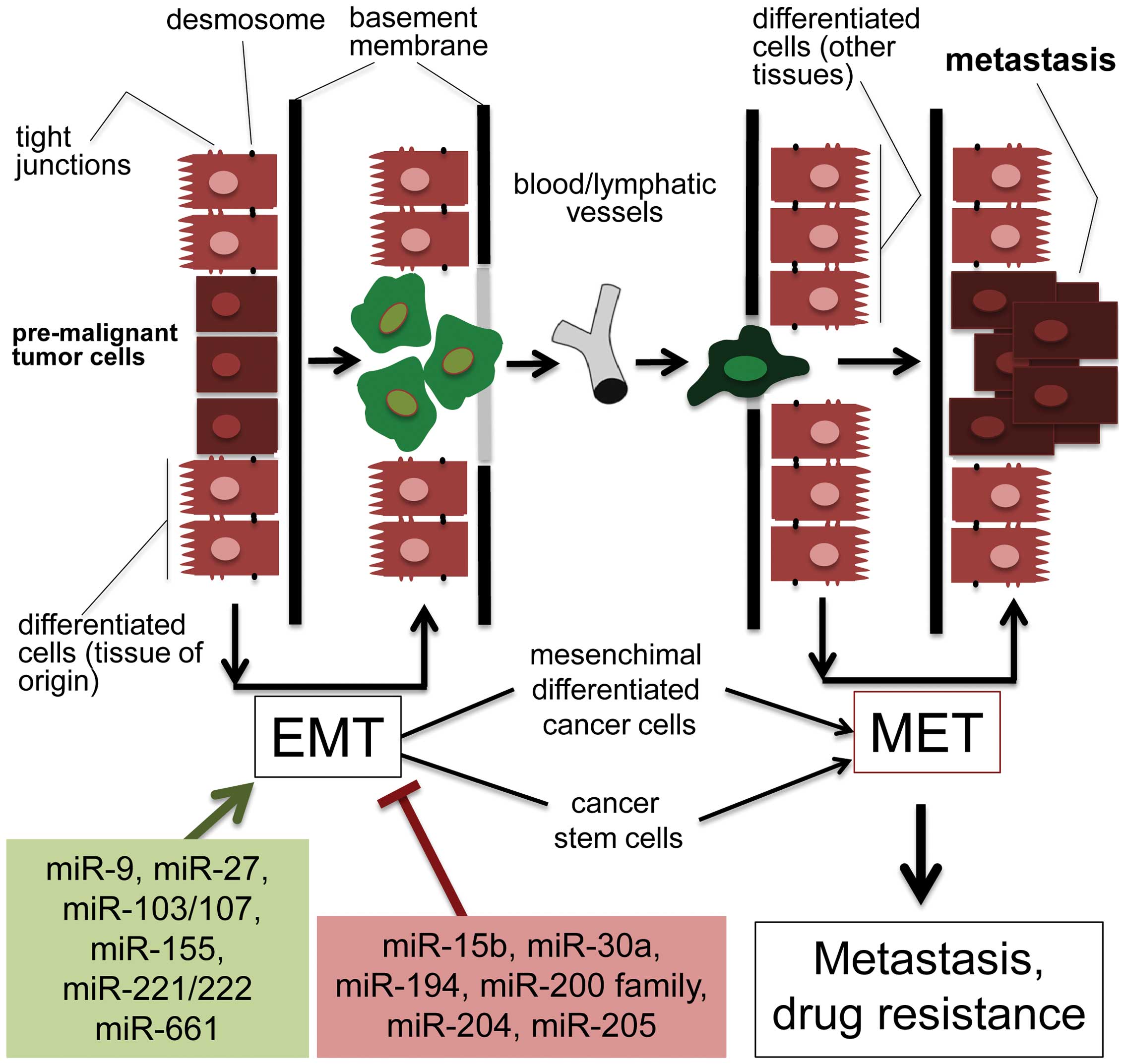
EMT is a powerful process in tumor invasion,
metastasis and tumorigenesis, and describes the molecular
reprogramming and phenotypic changes that are characterized by a
transition from polarized immotile epithelial cells to motile
mesenchymal cells (Fig. 3). This
process is characterized by the loss of polarity and cell-cell
contacts by the differentiated epithelial cells, with deep
alterations occurring at the level of tight junctions and
desmosomes. The breach of the basement membrane is a following
step, leading to the invasion of blood and/or lymphatic vessels by
these mesenchymal differentiated cancer cells, which at the end of
the process, causes migration, often accompanied by drug resistance
(Fig. 3). It is now well-known
that several miRNAs are important regulators of EMT. Some of these
are miR-7, miR-17/20, miR-22, miR-30, miR-200 and its family
members. Most of these miRNAs potentiate EMT, while some
well-characterized miRNAs play a suppressive role in EMT. For
instance, the metastasis suppressor role of the miR-200 members is
strongly associated with the inhibition of EMT. This is well
described in the published review by Zhang and Ma (321), and in the studies by Zaravinos
et al (322) and Kiesslich
et al (323), showing the
most recent advances regarding the influence of miRNAs in EMT and
the regulatory effects they exert on major signaling pathways in
various types of cancer (Fig. 3).
In Caski cervical cancer cells, the oncomiR-155 acts as a tumor
suppressor and suppresses EGF-induced EMT, decreasing
migration/invasion capacities, inhibiting cell proliferation and
enhancing the chemosensitivity to DDP in humans (324). Chang et al (279) demonstrated that the
overexpression of miR-20a in gallbladder carcinoma cells induced
EMT and promoted metastasis via the direct inhibition of Smad7,
correlating this miRNA with local invasion, distant metastasis and
a poor prognosis in patients with gallbladder carcinoma.
In the ovarian surface epithelium, EMT is considered
the key regulator of the post-ovulatory repair process and it can
be triggered by a range of environmental stimuli. The aberrant
expression of the miR-200 family (miR-200a, miR-200b, miR-200c,
miR-141 and miR-429) in ovarian cancer, and its involvement in the
initiation and progression of ovarian cancer have been well
demonstrated. The miR-200 family members seem to be strongly
associated with EMT and to have a metastasis suppressor role. miRNA
signatures can accurately distinguish ovarian cancer from the
normal ovary and can be used as diagnostic tools to predict the
clinical response to chemotherapy. Recent evidence suggests a
growing list of novel miRNAs (miR-187, miR-34a, miR-506, miRNA-138,
miR-30c, miR-30d, miR-30e-3p, miR-370 and miR-106a, among others)
that are also implicated in ovarian cancer-associated EMT, either
enhancing or suppressing it. MicroRNA-based gene therapy provides a
prospective antitumor approach for integrated cancer therapy
(325).
As regards the molecular targets of EMT-regulating
miRNAs, several are known and validated. Among these, transcription
factors play a very important role. For instance, Gao et al
(326) identified SOX2 as a key
player in EMT, by examining the effects of its overexpression. They
demonstrated that SOX2-overexpressing Eca-109 cells exhibited an
enhanced cell migration/invasion capacity. Moreover, these cells
exhibited characteristics of EMT, that is, a significantly
suppressed expression of the epithelial cell marker with a
concomitant enhancement in the expression of mesenchymal markers.
An increased expression of Slug in SOX2-overexpressing cells
suggested the involvement of this transcription factor in
SOX2-regulated metastasis. Finally, the expression levels of
STAT3/HIF-1α were found to be upregulated in SOX2-expressing cells,
and the blockade of these transcription factors resulted in the
inhibition of Slug expression at both the protein and mRNA
level.
Of interest, is also the finding that miR-221/222,
which are involved in EMT as positive regulators, can be
transcriptionally controlled by Slug. This was demonstrated by
Lambertini et al (327),
who showed that Slug silencing significantly decreased the level of
miR-221, strongly suggesting that miR-221 is a Slug target gene.
This was further confirmed by the characterization of a specific
region of the miR-221 promoter that is transcriptionally active and
is bound by the transcription factor Slug in vivo.
On the other hand, various miRNAs have been reported
to directly target EMT-promoting transcription factors. For
instance Qiu et al (328)
found that miR-139-5p functions as a suppressor of EMT in HCC and
metastasis by targeting ZEB1 and ZEB2, and that it may be a
therapeutic target for metastatic HCC. In conclusion, miRNAs
targeting and miRNA mimicking strategies are both expected to be
suitable for the control of EMT.
A very important step in tumor dissemination and
metastasis is neoangiogenesis. This is a very complex process in
which several proteins and protein networks participate, for
instance interleukin (IL)-8, vascular endothelial growth factor
(VEGF), basic fibroblast growth factor (bFGF), angiopoietins and
matrix metalloproteinases (MMPs). As far as the expression of the
IL-8 gene is concerned, the increase in IL-8 gene expression from
the healthy brain to low-grade glioma (LGG) can be explained by
alterations in the regulatory networks associated with IL-8 gene
transcription. Among these, the nuclear factor-κB (NF-κB) network
should be proposed, since i) NF-κB is one of the major
transcription factors involved in IL-8 gene regulation (329); ii) NF-κB is a marker of glioma
onset and progression (330–333); iii) miR-16 inhibits glioma cell
growth through the suppression of the NF-κB signaling pathway
(334). In addition to
transcription factors, miRNAs can directly modulate pro-angiogenic
factors. For instance, the increased IL-8 gene expression in
high-grade glioma (HGG; with respect to LGG) may be associated with
decrease of its inhibitory miRNA, miR-93, at least in a subset of
HGG patients. The decrease in miR-93 expression in these HGG
patients, in addition to IL-8, may lead to the post-transcriptional
upregulation of VEGF, monocyte chemoattractant protein-1 (MCP-1)
and platelet-derived growth factor (PDGF)-bb, well recognized
markers of the late tumor stages of gliomas (335–337). However, it should be mentioned
that HGG samples are highly heterogeneous with respect to miR-93
levels, suggesting the involvement of multiple regulatory pathways
in controlling the level of IL-8 gene expression.
One of the better described examples of tumor
suppressor miRNAs is miR-124. This miRNA has been found to play a
significant role in several types of cancer (168–173,338). Specifically, miR-124 expression
is reportedly downregulated in the cells and tissues of esophageal
cancer (339), breast cancer
(340), renal cell carcinoma
(341) and CRC (172). Accordingly, the ectopic
expression of miR-124 by target tumor cells inhibits tumor-related
parameters in experimental model systems mimicking prostate cancer,
medulloblastoma, hepatocellular carcinoma, gastric cancer, glioma,
osteosarcoma and CRC.
Furthermore, they found that miR-124a directly
targeted and suppressed IQ motif containing GTPase activating
protein 1 (IQGAP1), a well-known regulator of actin dynamics and
cell motility (342). Taken
together all these data clearly demonstrate that miR-124a is an
important tumor suppressor miRNA which is downregulated in cancer
cells; accordingly antitumor effects can be achieved following the
administration of miR-124, pre-miR-124 or a variety of miR-124
mimics to cancer cells.
Finally, the translational relevance of the role of
miR-124 in antitumor drug sensitivity is suggested by the finding
that the increased miR-124 expression correlates with an improved
breast cancer prognosis, specifically in patients receiving
chemotherapy. This finding suggests that miR-124 may potentially be
used as a therapeutic agent to improve the efficacy of
chemotherapy, including that based on DNA-damaging agents via ATM
interactor (ATMIN)- and poly(ADP-ribose) polymerase 1
(PARP1)-mediated mechanisms (343).
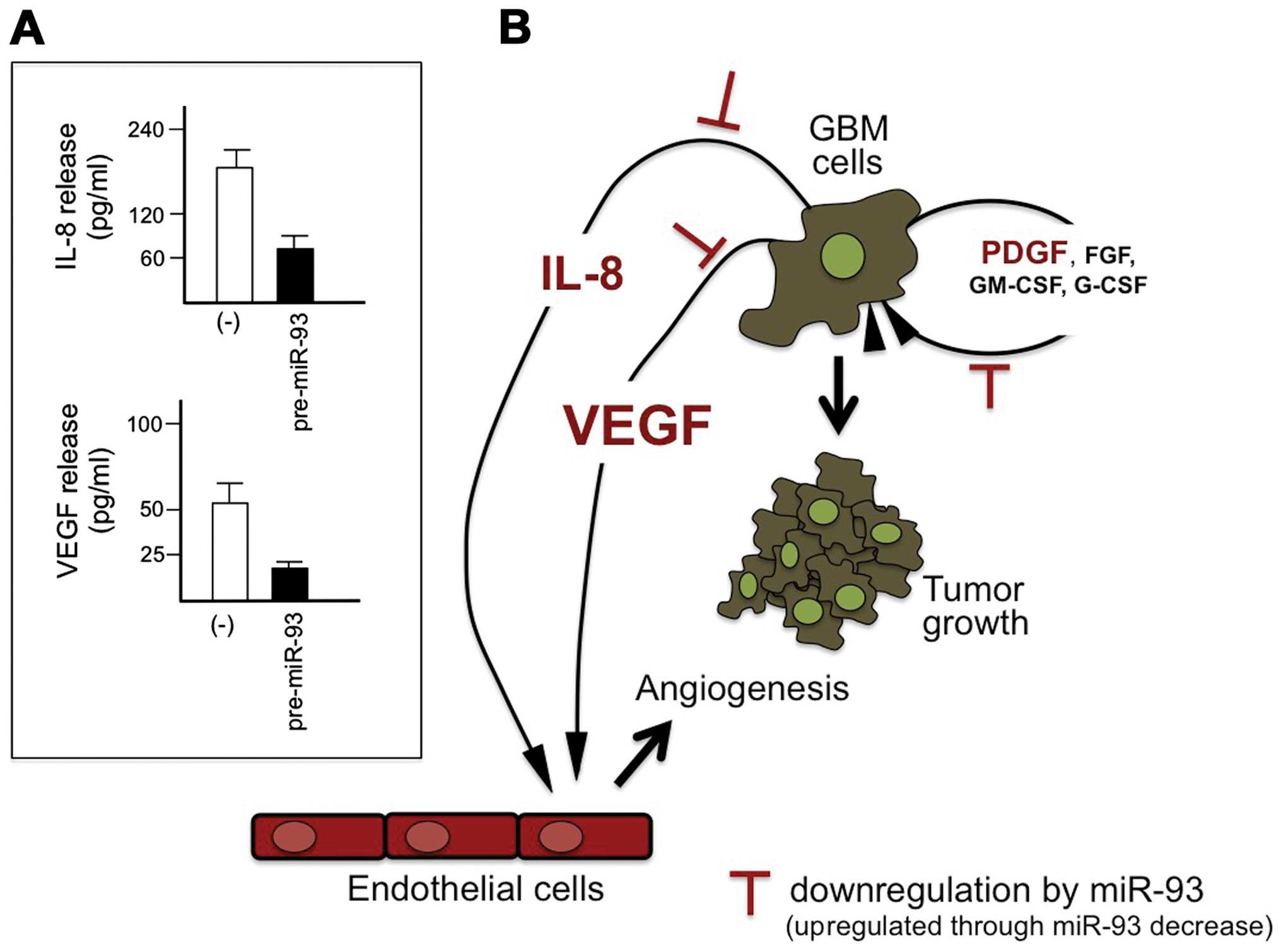
A second example of possible miRNA replacement
therapy is based on the inhibition of IL-8 and VEGF by the
transfection of tumor target cells with pre-miR-93. This was
performed in human glioma cell lines (U251 and T98G), as well as on
the SK-N-AS neuroblastoma cell line.
The first conclusion of this research activity is
that the miRNA, miR-93, is involved in the control of the
expression of the IL-8 gene in the glioma U251 and in the
neuroblastoma SK-N-AS cell lines (344,345). The effects of these treatments
were analyzed by RT-qPCR (looking at the IL-8 mRNA content) or by
Bio-plex analysis (looking at IL-8 protein secretion). In addition,
Fabbri et al (344) found
that the transfection of target cells with pre-miR-93 led to the
downregulation of VEGF (see the results depicted in Fig. 4A), suggesting that, as shown in
Fig. 4B, miR-93 has effects on the
growth of gliomas [by interfering with growth factors, including
PDGF, fibroblast growth factor (FGF), granulocyte-macrophage
colony-stimulating factor (GM-CSF) and granulocyte-colony
stimulating factor(G-CSF)], as well as on neoangiogenesis.
Gliomas, as other tumors, express miR-221 at high
levels, promoting malignant progression through activation of the
Akt pathway and the inhibition of p27Kip1 (346–349). In addition miR-221 mediates the
downregulation of other genes, such as PUMA (258), intercellular adhesion molecule 1
(ICAM-1) (350), TIMP
metallopeptidase inhibitor 3 (TIMP3) (351) and phosphatase and tensin homolog
(PTEN) (352), and may thus be
associated with cancer onset and progression (353). Therefore, miR-221 appears to be a
specific target for the treatment of gliomas (354,355). Zhang et al (354) reported that the co-suppression of
the miR-221/222 cluster suppressed human glioma cell growth by
affecting p27Kip1 expression in vitro and in
vivo. In our own laboratory, we have also examined the effects
of a PNA against miR-221 and showed that it is able to induce a
sharp decrease in miR-221 biological activity. The employed PNA
carryed an Arg(8) peptide to
facilitate PNA uptake by target cells. Two studies were published
on this specific issue. In the first study by Brognara et al
(319), we demonstrated that
targeting miR-221 induced a sharp increase in the expression of the
miR-221 target p27Kip1 mRNA in a breast cancer cell line
(319). In a more recent study of
ours, Brognara et al (56)
demonstrated that the PNA against miR-221 can be internalized by
glioma cells when linked to a Arg(8) tail (R8), leading to the inhibition of
miR-221 functions, associated with the increased expression of
p27Kip1 in U251 and T98G cells. In addition, the
expression of another miR-221 target gene, TIMP3, was upregulated
following treatment of the T98G cells with R8-PNA-a221. These data
support the concept that targeting miR-221 with antagomiR molecules
may provide novel options for developing protocols for the
treatment of gliomas. This is supported by the finding that the
treatment of all the glioma cells lines with R8-PNA-a221 induced
the activation of the early apoptotic pathway (56).
Several tumors express upregulated levels of several
miRNAs, suggesting that a possible limit to anti-mRNA therapeutics
may be the requirement of the co-targeting of several miRNAs to
obtain the programmed biological effects. Moreover, an important
anti-miRNA strategy may be associated with the obvious need for the
co-targeting of different miRNAs belonging to the same miRNA
family.
The co-treatment of target cells with antagomiR
molecules selective for different miRNAs has been recently
described. For instance, Lee et al (357) investigated the role of miRNAs
targeting runt related transcription factor 3 (RUNX3) in early
tumorigenesis. Under hypoxic conditions, miR-130a and miR-495 are
upregulated and target RUNX3 by binding to its 3′-UTR in gastric
cancer cells. Using matrigel plug assay, they found that antagomiRs
specific for miR-130a and miR-495 significantly reduced
angiogenesis in vivo and hypothesized that the co-targeting
of miR-130a and miR-495 may prove to be a potential therapeutic
strategy with which to recover RUNX3 expression under hypoxic
conditions and in early tumorigenesis (357).
In our own laboratory, we have approached the same
issue using PNAs. We have previously reported that a PNA targeting
miR-221 can be internalized by glioma cells and exert biological
effects on miR-221-dependent functions when it is linked to an
octaarginine tail (R8) (56). The
major results of the more recent study by Brognara et al
(358) are the following: i)
R8-conjugated PNAs against miR-221 (R8-PNA-a221) and miR-222
(R8-PNA-a222) exhibit selective biological activity on miR-221 and
miR-222; ii) when R8-PNA-a221 and R8-PNA-a222 are singularly
administered to glioma cells, the specific inhibition of
hybridization to miR-221 and miR-222 is obtained following RT-qPCR
analysis; iii) both R8-PNA-a221 and R8-PNA-a222 induce the
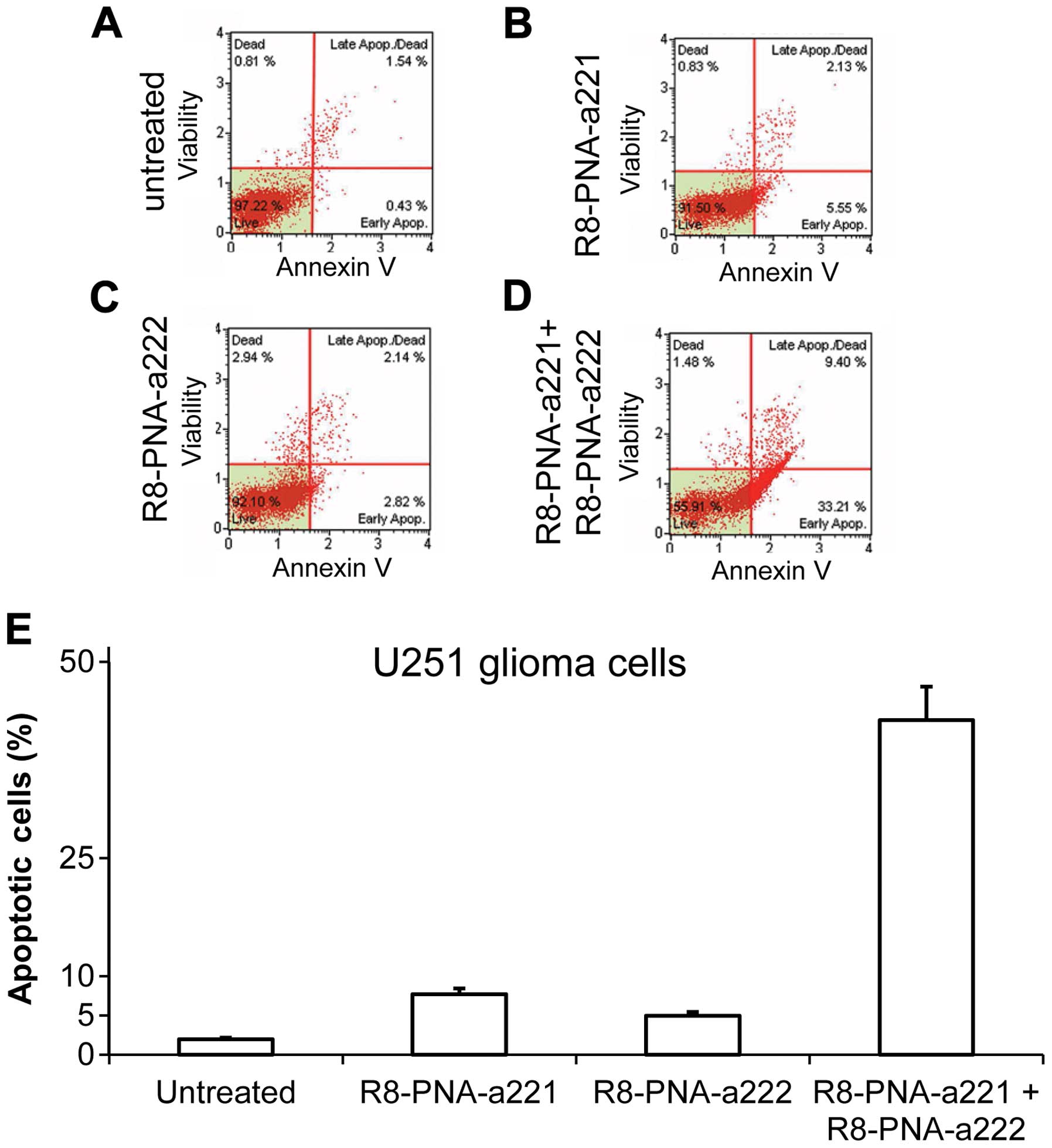
apoptosis of U251, U373 and T98G glioma cells. Finally, the
co-administration of R8-PNA-a221 and R8-PNA-a222 was associated
with the most prominent effects of this treatment in inducing
apoptosis (see the representative experimental results shown in
Fig. 5) (358).
One of the most interesting results obtained to date
using miRNA therapeutics is the formal demonstration that, when
used in combination with antitumor drugs, satisfactory therapeutic
effects may be achieved (359).
This has been demonstrated using both miRNA mimicking approaches,
as well as anti-miRNA molecules.
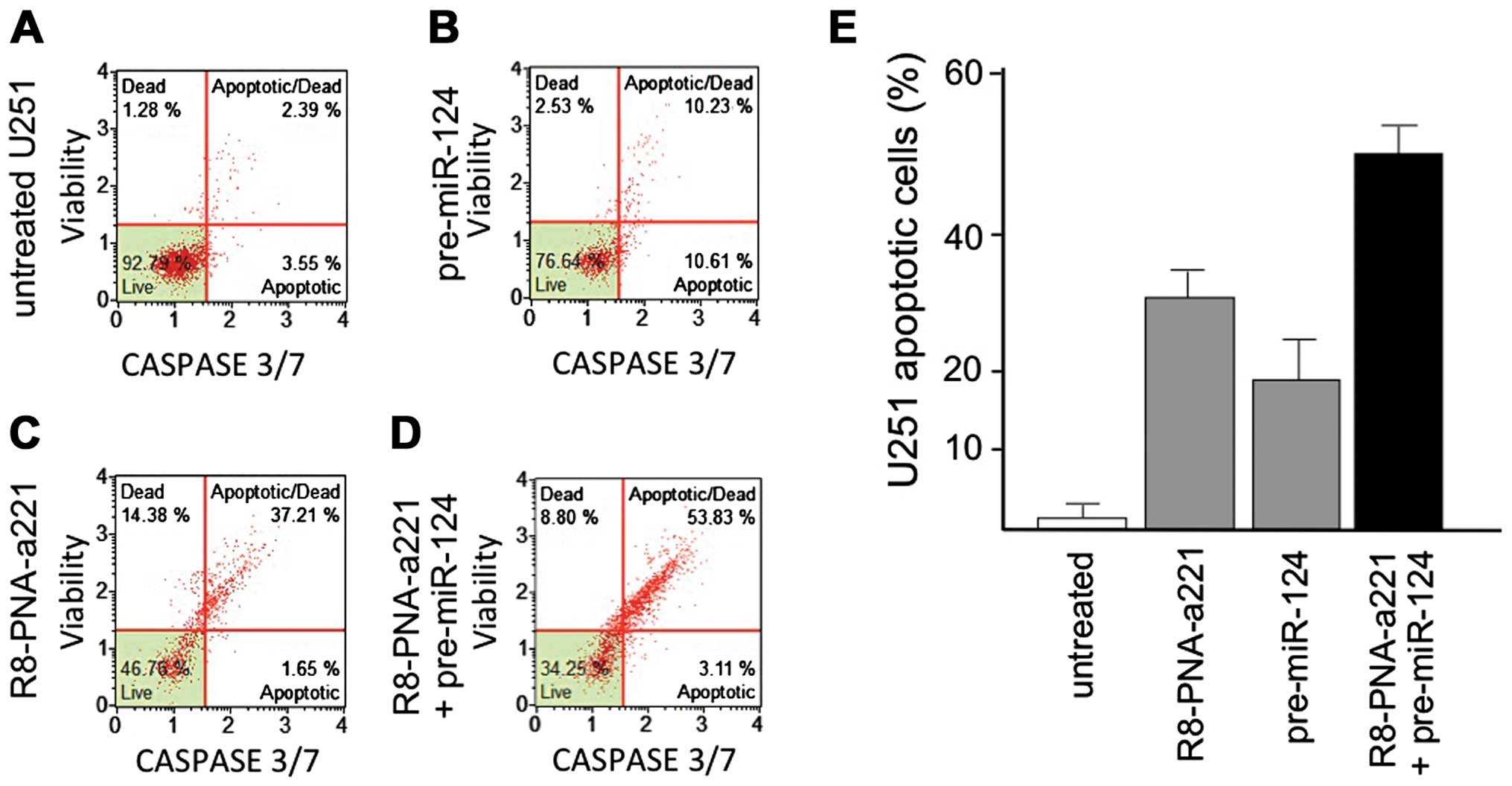
In our own laboratory, we further analyzed the
possible co-admistration of temozolomide (TMZ) and the tumor
suppressor pre-miR-124. This was investigated in one neuroblastoma
and two glioma cell lines. For miRNA replacement, we employed
transfection with pre-miR-124, since miR-124 is a powerful tumor
suppressor pro-apoptotic miRNA. In order to demonstrate the
activity of the combined treatment, the anti-proliferative and
pro-apoptotic effects were analyzed. This set of data confirm that
miRNA therapeutics can be successfully combined with chemical
treatments to obtain greater effects with low doses of reagents. In
conclusion, our data showed that, in addition to the combinations
between antitumor drugs and antagomiR-based protocols, interesting
results can be obtained by the combination of drugs with miRNA
replacement agents (Fabbri et al, unpublished data).
A recent study reported the co-delivery of
antagomiR-10b and PTX by a liposomal delivery and showed that it
efficiently inhibited tumor growth and reduced the incidence of
lung metastasis. In fact, antagomiR-10b impeded the migration of 4
T1 cells in vitro, silencing miR-10b and upregulating Hoxd10
both in vitro and in vivo, while PTX elicited potent
tumor cell inhibitory effects (368). The same antitumor efficacy and
delivery to the tumor site may be achieved by the dual loading of
miR-218 mimic (bio-drug) and temozolomide (chemo-drug) using a new
delivery nanogel system approach (369).
A further example in this very exciting field of
investigation was reported by Hu et al (372), studying Bcl-2, a prominent member
of the Bcl-2 family of proteins that regulate the induction of
apoptosis. They investigated the effect of Bcl-2 siRNAs combined
with miR-15a oligonucleotides on the growth of Raji cells.
Following transfection of these combined reagents, the protein and
mRNA levels of Bcl-2 were markedly decreased. The growth of the
cells was significantly inhibited compared with the cells
transfected with Bcl-2 siRNA or miR-15a alone and the apoptotic
rate significantly increased. These results suggest that the
combination of Bcl-2 siRNA and miR-15a oligonucleotides increases
the apoptosis of Raji cells, and strongly support the concept that
the combination of Bcl-2 siRNA and miR-15a may be a useful approach
in the treatment of lymphoma.
MicroRNA therapeutics in cancer are based on
targeting or mimicking miRNAs involved in cancer onset,
progression, angiogenesis, EMT and metastasis. This strategy has
been proposed several years ago and is based on the well-recognized
fact that miRNAs play a key role in the post-transcriptional
control of gene expression by the sequence-selective targeting of
mRNAs and are key players in several biological functions and
pathological processes, including cancer. In this respect, several
studies have conclusively demonstrated that miRNAs are deeply
involved in tumor onset and progression, either behaving as
tumor-promoting miRNAs (oncomiRNAs and metastamiRNAs) or as tumor
suppressor miRNAs. In general, miRNAs able to promote cancer target
mRNAs coding for tumor suppressor proteins, whereas miRNAs
exhibiting tumor suppressor properties usually target mRNAs coding
oncoproteins. This has a very important implication in diagnosis
and/or prognosis, including the recent discovery that the pattern
of circulating cell-free miRNAs in serum allows us to perform
molecular analyses on these non-invasive liquid biopsies. This
research field has confirmed that cancer-specific miRNAs are
present in extracellular body fluids, and may play a very important
role in the crosstalk between cancer cells and surrounding normal
cells. Interestingly, the evidence of the presence of miRNAs in
serum, plasma and saliva supports their potential as an additional
set of biomarkers for cancer.
This review has focused on the most promising
examples potentially leading to the development of anticancer,
miRNA-based therapeutic protocols. The inhibition of miRNA activity
can be readily achieved by the use of miRNA inhibitors and
oligomers, including RNA, DNA, DNA analogues (miRNA antisense
therapy), small molecule inhibitors, miRNA sponges or through miRNA
masking. On the contrary, the enhancement of miRNA function (miRNA
replacement therapy) can be achieved by the use of modified miRNA
mimetics and plasmids or lentiviral vectors carrying miRNA
sequences. However, we should carefully consider that a single
miRNA can target several mRNAs (not only tumor-associated mRNAs)
and a single mRNA may contain in the 3′UTR sequence several signals
for miRNA recognition. In this case, antagomiRNA-based therapy
should be designed to target multiple miRNAs. MicroRNA targeting
and mimicking is further complicated by the facts that, since their
discovery and first characterization, the number of miRNA sequences
deposited in the miRBase databases is increasing, and research
studies on miRNAs in cancer have confirmed the very high complexity
of the networks constituted by miRNAs and RNA targets.
One possible approach includes the combination
strategies based on the co-administration of anticancer agents, as
shown by the observation that i) the combined administration of
different antagomiR molecules induces greater antitumor effects and
ii) some anti-miR molecules can sensitize drug-resistant tumor cell
lines to drug treatment. In this review, we approached two
additional issues: i) the combination of miRNA replacement therapy
with drug administration and ii) the combination of antagomiR and
miRNA replacement therapy. One of the solid results emerging from
different independent studies is the demonstration that miRNA
replacement therapy can enhance the antitumor effects of the
antitumor drugs.
The second important conclusion of the reviewed
studies is that the combination of anti-miRNA and miRNA replacement
strategies may lead to excellent results, in terms of antitumor
effects. This possible combined strategy is in its infancy and very
few studies are available in the literature. Proof-of-principle
data are presented as examples of possible combined treatments in
Fig. 6. Our data indicate that the
co-treatment of U251 glioblastoma cells with PNAs targeting miR-221
or miR-222 in the presence of pre-miR-124 transfection leads to a
much higher level of apoptosis as opposed to singularly
administered reagents. These data further extend the possible
combined antitumor treatment based on antitumor drugs and
antagomiR-molecules, and present the very novel possibility of
combining antagomiR and miRNA replacement therapies.
This study was funded by CIB, by COFIN-2009 and by
AIRC (IG 13575: peptide nucleic acids targeting oncomiR and
tumor-suppressor miRNAs: cancer diagnosis and therapy). EB is
supported by an Umberto Veronesi fellowship. We would like to thank
the Horizon-2020 ULTRAPLACAD (ULTRA sensitive PLAsmonic devices for
early Cancer Diagnosis) n.633937 project for supporting the
research on circulating miRNAs as diagnostic tools.
|
1
|
Mazière P and Enright AJ: Prediction of
microRNA targets. Drug Discov Today. 12:452–458. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Witkos TM, Koscianska E and Krzyzosiak WJ:
Practical aspects of microRNA target prediction. Curr Mol Med.
11:93–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ghelani HS, Rachchh MA and Gokani RH:
MicroRNAs as newer therapeutic targets: A big hope from a tiny
player. J Pharmacol Pharmacother. 3:217–227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
5
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chekulaeva M and Filipowicz W: Mechanisms
of miRNA-mediated post-transcriptional regulation in animal cells.
Curr Opin Cell Biol. 21:452–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cammaerts S, Strazisar M, De Rijk P and
Del Favero J: Genetic variants in microRNA genes: Impact on
microRNA expression, function, and disease. Front Genet. 6:1862015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Friedländer MR, Lizano E, Houben AJS,
Bezdan D, Báñez-Coronel M, Kudla G, Mateu-Huertas E, Kagerbauer B,
González J, Chen KC, et al: Evidence for the biogenesis of more
than 1,000 novel human microRNAs. Genome Biol. 15:R572014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng WC, Chung IF, Tsai CF, Huang TS,
Chen CY, Wang SC, Chang TY, Sun HJ, Chao JY, Cheng CC, et al:
YM500v2: a small RNA sequencing (smRNA-seq) database for human
cancer miRNome research. Nucleic Acids Res. 43:D862–D867. 2015.
View Article : Google Scholar :
|
|
11
|
Londin E, Loher P, Telonis AG, Quann K,
Clark P, Jing Y, Hatzimichael E, Kirino Y, Honda S, Lally M, et al:
Analysis of 13 cell types reveals evidence for the expression of
numerous novel primate- and tissue-specific microRNAs. Proc Natl
Acad Sci USA. 112:E1106–E1115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar :
|
|
13
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar :
|
|
14
|
Taccioli C, Fabbri E, Visone R, Volinia S,
Calin GA, Fong LY, Gambari R, Bottoni A, Acunzo M, Hagan J, et al:
UCbase and miRfunc: A database of ultraconserved sequences and
microRNA function. Nucleic Acids Res. 37:D41–D48. 2009. View Article : Google Scholar
|
|
15
|
Witwer KW: Data submission and quality in
microarray-based microRNA profiling. Clin Chem. 59:392–400. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
A microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peter ME: Targeting of mRNAs by multiple
miRNAs: The next step. Oncogene. 29:2161–2164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bianchi N, Finotti A, Ferracin M,
Lampronti I, Zuccato C, Breveglieri G, Brognara E, Fabbri E,
Borgatti M, Negrini M, et al: Increase of microRNA-210, decrease of
raptor gene expression and alteration of mammalian target of
rapamycin regulated proteins following mithramycin treatment of
human erythroid cells. PLoS One. 10:e01215672015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Subramanian S and Steer CJ: MicroRNAs as
gatekeepers of apoptosis. J Cell Physiol. 223:289–298.
2010.PubMed/NCBI
|
|
21
|
Wang Y and Blelloch R: Cell cycle
regulation by MicroRNAs in embryonic stem cells. Cancer Res.
69:4093–4096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fabbri E, Borgatti M, Montagner G, Bianchi
N, Finotti A, Lampronti I, Bezzerri V, Dechecchi MC, Cabrini G and
Gambari R: Expression of microRNA-93 and Interleukin-8 during
Pseudomonas aeruginosa-mediated induction of proinflammatory
responses. Am J Respir Cell Mol Biol. 50:1144–1155. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Faruq O and Vecchione A: microRNA:
Diagnostic perspective. Front Med Lausanne. 2:512015.PubMed/NCBI
|
|
24
|
Shalaby T, Fiaschetti G, Baumgartner M and
Grotzer MA: Significance and therapeutic value of miRNAs in
embryonal neural tumors. Molecules. 19:5821–5862. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Palmero EI, de Campos SG, Campos M, de
Souza NC, Guerreiro ID, Carvalho AL and Marques MM: Mechanisms and
role of microRNA deregulation in cancer onset and progression.
Genet Mol Biol. 34:363–370. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weber JA, Baxter DH, Zhang S, Huang DY,
Huang KH, Lee MJ, Galas DJ and Wang K: The microRNA spectrum in 12
body fluids. Clin Chem. 56:1733–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fayyad-Kazan H, Bitar N, Najar M, Lewalle
P, Fayyad-Kazan M, Badran R, Hamade E, Daher A, Hussein N, ElDirani
R, et al: Circulating miR-150 and miR-342 in plasma are novel
potential biomarkers for acute myeloid leukemia. J Transl Med.
11:312013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Neviani P and Fabbri M: Exosomic microRNAs
in the tumor microenvironment. Front Med Lausanne.
2:472015.PubMed/NCBI
|
|
30
|
Köberle V, Kronenberger B, Pleli T, Trojan
J, Imelmann E, Peveling-Oberhag J, Welker MW, Elhendawy M, Zeuzem
S, Piiper A, et al: Serum microRNA-1 and microRNA-122 are
prognostic markers in patients with hepatocellular carcinoma. Eur J
Cancer. 49:3442–3449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He Y, Lin J, Kong D, Huang M, Xu C, Kim
TK, Etheridge A, Luo Y, Ding Y and Wang K: Current state of
circulating MicroRNAs as cancer biomarkers. Clin Chem.
61:1138–1155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Westphal M and Lamszus K: Circulating
biomarkers for gliomas. Nat Rev Neurol. 11:556–566. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yau TO, Wu CW, Dong Y, Tang CM, Ng SS,
Chan FK, Sung JJ and Yu J: microRNA-221 and microRNA-18a
identification in stool as potential biomarkers for the
non-invasive diagnosis of colorectal carcinoma. Br J Cancer.
111:1765–1771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Czech MP: MicroRNAs as therapeutic
targets. N Engl J Med. 354:1194–1195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brown BD and Naldini L: Exploiting and
antagonizing microRNA regulation for therapeutic and experimental
applications. Nat Rev Genet. 10:578–585. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kota SK and Balasubramanian S: Cancer
therapy via modulation of micro RNA levels: A promising future.
Drug Discov Today. 15:733–740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bader AG and Lammers P: The Therapeutic
Potential of microRNAs. Discovery Technology. 2011.
|
|
40
|
Rothschild SI: microRNA therapies in
cancer. Mol Cell Ther. 2:72014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
van Rooij E and Kauppinen S: Development
of microRNA therapeutics is coming of age. EMBO Mol Med. 6:851–864.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Orellana EA and Kasinski AL: MicroRNAs in
cancer: A historical perspective on the path from discovery to
therapy. Cancers (Basel). 7:1388–1405. 2015. View Article : Google Scholar
|
|
43
|
Berindan-Neagoe I, Monroig PC, Pasculli B
and Calin GA: MicroRNAome genome: A treasure for cancer diagnosis
and therapy. CA Cancer J Clin. 64:311–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bernardo BC, Ooi JY, Lin RC and McMullen
JR: miRNA therapeutics: A new class of drugs with potential
therapeutic applications in the heart. Future Med Chem.
7:1771–1792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Weiler J, Hunziker J and Hall J:
Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs
implicated in human disease? Gene Ther. 13:496–502. 2006.
View Article : Google Scholar
|
|
46
|
Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z
and Yang B: A single anti-microRNA antisense
oligodeoxyribonucleotide (AMO) targeting multiple microRNAs offers
an improved approach for microRNA interference. Nucleic Acids Res.
37:e242009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lennox KA and Behlke MA: Chemical
modification and design of anti-miRNA oligonucleotides. Gene Ther.
18:1111–1120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Obad S, dos Santos CO, Petri A, Heidenblad
M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al:
Silencing of microRNA families by seed-targeting tiny LNAs. Nat
Genet. 43:371–378. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
49
|
Elmén J, Lindow M, Schütz S, Lawrence M,
Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et
al: LNA-mediated microRNA silencing in non-human primates. Nature.
452:896–899. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Stenvang J, Silahtaroglu AN, Lindow M,
Elmen J and Kauppinen S: The utility of LNA in microRNA-based
cancer diagnostics and therapeutics. Semin Cancer Biol. 18:89–102.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chabot S, Teissié J and Golzio M: Targeted
electro-delivery of oligonucleotides for RNA interference: siRNA
and antimiR. Adv Drug Deliv Rev. 81:161–168. 2015. View Article : Google Scholar
|
|
52
|
Lundin KE, Højland T, Hansen BR, Persson
R, Bramsen JB, Kjems J, Koch T, Wengel J and Smith CI: Biological
activity and biotechnological aspects of locked nucleic acids. Adv
Genet. 82:47–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Staedel C, Varon C, Nguyen PH, Vialet B,
Chambonnier L, Rousseau B, Soubeyran I, Evrard S, Couillaud F and
Darfeuille F: Inhibition of gastric tumor cell growth using
seed-targeting LNA as specific, long-lasting MicroRNA inhibitors.
Mol Ther Nucleic Acids. 4:e2462015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Avitabile C, Accardo A, Ringhieri P,
Morelli G, Saviano M, Montagner G, Fabbri E, Gallerani E, Gambari R
and Romanelli A: Incorporation of naked peptide nucleic acids into
liposomes leads to fast and efficient delivery. Bioconjug Chem.
26:1533–1541. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fabbri E, Manicardi A, Tedeschi T, Sforza
S, Bianchi N, Brognara E, Finotti A, Breveglieri G, Borgatti M,
Corradini R, et al: Modulation of the biological activity of
microRNA-210 with peptide nucleic acids (PNAs). ChemMedChem.
6:2192–2202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Brognara E, Fabbri E, Bazzoli E, Montagner
G, Ghimenton C, Eccher A, Cantù C, Manicardi A, Bianchi N, Finotti
A, et al: Uptake by human glioma cell lines and biological effects
of a peptide-nucleic acids targeting miR-221. J Neurooncol.
118:19–28. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar
|
|
58
|
Morris JK, Chomyk A, Song P, Parker N,
Deckard S, Trapp BD, Pimplikar SW and Dutta R: Decrease in levels
of the evolutionarily conserved microRNA miR-124 affects
oligodendrocyte numbers in Zebrafish, Danio rerio. Invert Neurosci.
15:42015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Conte I, Hadfield KD, Barbato S, Carrella
S, Pizzo M, Bhat RS, Carissimo A, Karali M, Porter LF, Urquhart J,
et al: MiR-204 is responsible for inherited retinal dystrophy
associated with ocular coloboma. Proc Natl Acad Sci USA.
112:E3236–E3245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ristori E, Lopez-Ramirez MA, Narayanan A,
Hill-Teran G, Moro A, Calvo CF, Thomas JL and Nicoli S: A
Dicer-miR-107 interaction regulates biogenesis of specific miRNAs
crucial for neurogenesis. Dev Cell. 32:546–560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ebert MS and Sharp PA: MicroRNA sponges:
Progress and possibilities. RNA. 16:2043–2050. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kluiver J, Gibcus JH, Hettinga C, Adema A,
Richter MK, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen BJ and
van den Berg A: Rapid generation of microRNA sponges for microRNA
inhibition. PLoS One. 7:e292752012a. View Article : Google Scholar
|
|
64
|
Kluiver J, Slezak-Prochazka I,
Smigielska-Czepiel K, Halsema N, Kroesen BJ and van den Berg A:
Generation of miRNA sponge constructs. Methods. 58:113–117. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li KC, Chang YH, Yeh CL and Hu YC: Healing
of osteoporotic bone defects by baculovirus-engineered bone
marrow-derived MSCs expressing MicroRNA sponges. Biomaterials.
74:155–166. 2016. View Article : Google Scholar
|
|
66
|
de Melo Maia B, Ling H, Monroig P, Ciccone
M, Soares FA, Calin GA and Rocha RM: Design of a miRNA sponge for
the miR-17 miRNA family as a therapeutic strategy against vulvar
carcinoma. Mol Cell Probes. 29:420–426. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tay FC, Lim JK, Zhu H, Hin LC and Wang S:
Using artificial microRNA sponges to achieve microRNA
loss-of-function in cancer cells. Adv Drug Deliv Rev. 81:117–127.
2015. View Article : Google Scholar
|
|
68
|
Liu Y, Han Y, Zhang H, Nie L, Jiang Z, Fa
P, Gui Y and Cai Z: Synthetic miRNA-mowers targeting miR-183-96-182
cluster or miR-210 inhibit growth and migration and induce
apoptosis in bladder cancer cells. PLoS One. 7:e522802012.
View Article : Google Scholar
|
|
69
|
Choi WY, Giraldez AJ and Schier AF: Target
protectors reveal dampening and balancing of Nodal agonist and
antagonist by miR-430. Science. 318:271–274. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Haraguchi T, Ozaki Y and Iba H: Vectors
expressing efficient RNA decoys achieve the long-term suppression
of specific microRNA activity in mammalian cells. Nucleic Acids
Res. 37:e432009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Krol J, Busskamp V, Markiewicz I, Stadler
MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D,
et al: Characterizing light-regulated retinal microRNAs reveals
rapid turnover as a common property of neuronal microRNAs. Cell.
141:618–631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cassidy JJ, Straughan AJ and Carthew RW:
Differential masking of natural genetic variation by miR-9a in
Drosophila. Genetics. 202:675–687. 2016. View Article : Google Scholar
|
|
73
|
Wang Z: The principles of MiRNA-masking
antisense oligonucleotides technology. Methods Mol Biol. 676:43–49.
2011. View Article : Google Scholar
|
|
74
|
Bak RO, Hollensen AK and Mikkelsen JG:
Managing microRNAs with vector-encoded decoy-type inhibitors. Mol
Ther. 21:1478–1485. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Murakami K and Miyagishi M: Tiny masking
locked nucleic acids effectively bind to mRNA and inhibit binding
of microRNAs in relation to thermodynamic stability. Biomed Rep.
2:509–512. 2014.PubMed/NCBI
|
|
76
|
Shin KJ, Wall EA, Zavzavadjian JR, Santat
LA, Liu J, Hwang JI, Rebres R, Roach T, Seaman W, Simon MI, et al:
A single lentiviral vector platform for microRNA-based conditional
RNA interference and coordinated transgene expression. Proc Natl
Acad Sci USA. 103:13759–13764. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Askou AL, Aagaard L, Kostic C, Arsenijevic
Y, Hollensen AK, Bek T, Jensen TG, Mikkelsen JG and Corydon TJ:
Multigenic lentiviral vectors for combined and tissue-specific
expression of miRNA- and protein-based antiangiogenic factors. Mol
Ther Methods Clin Dev. 2:140642015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Winbanks CE, Beyer C, Hagg A, Qian H,
Sepulveda PV and Gregorevic P: miR-206 represses hypertrophy of
myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS
One. 8:e735892013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Montgomery RL, Yu G, Latimer PA, Stack C,
Robinson K, Dalby CM, Kaminski N and van Rooij E: MicroRNA mimicry
blocks pulmonary fibrosis. EMBO Mol Med. 6:1347–1356. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Bader AG: miR-34 - a microRNA replacement
therapy is headed to the clinic. Front Genet. 3:1202012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kwekkeboom RF, Lei Z, Doevendans PA,
Musters RJ and Sluijter JP: Targeted delivery of miRNA therapeutics
for cardiovascular diseases: Opportunities and challenges. Clin Sci
(Lond). 127:351–365. 2014. View Article : Google Scholar
|
|
82
|
Sherr CJ: Principles of tumor suppression.
Cell. 116:235–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Park SM, Shell S, Radjabi AR, Schickel R,
Feig C, Boyerinas B, Dinulescu DM, Lengyel E and Peter ME: Let-7
prevents early cancer progression by suppressing expression of the
embryonic gene HMGA2. Cell Cycle. 6:2585–2590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Müller DW and Bosserhoff AK: Integrin beta
3 expression is regulated by let-7a miRNA in malignant melanoma.
Oncogene. 27:6698–6706. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Peng Y, Laser J, Shi G, Mittal K, Melamed
J, Lee P and Wei JJ: Antiproliferative effects by Let-7 repression
of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res.
6:663–673. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Bader AG, Brown D and Winkler M: The
promise of microRNA replacement therapy. Cancer Res. 70:7027–7030.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wiggins JF, Ruffino L, Kelnar K, Omotola
M, Patrawala L, Brown D and Bader AG: Development of a lung cancer
therapeutic based on the tumor suppressor microRNA-34. Cancer Res.
70:5923–5930. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Ibrahim AF, Weirauch U, Thomas M,
Grünweller A, Hartmann RK and Aigner A: MicroRNA replacement
therapy for miR-145 and miR-33a is efficacious in a model of colon
carcinoma. Cancer Res. 71:5214–5224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Trang P, Wiggins JF, Daige CL, Cho C,
Omotola M, Brown D, Weidhaas JB, Bader AG and Slack FJ: Systemic
delivery of tumor suppressor microRNA mimics using a neutral lipid
emulsion inhibits lung tumors in mice. Mol Ther. 19:1116–1122.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Buechner J, Tømte E, Haug BH, Henriksen
JR, Løkke C, Flægstad T and Einvik C: Tumour-suppressor microRNAs
let-7 and miR-101 target the proto-oncogene MYCN and inhibit cell
proliferation in MYCN-amplified neuroblastoma. Br J Cancer.
105:296–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Scheibner KA, Teaboldt B, Hauer MC, Chen
X, Cherukuri S, Guo Y, Kelley SM, Liu Z, Baer MR, Heimfeld S, et
al: MiR-27a functions as a tumor suppressor in acute leukemia by
regulating 14-3-3θ. PLoS One. 7:e508952012. View Article : Google Scholar
|
|
95
|
Thomas M, Lange-Grünweller K, Weirauch U,
Gutsch D, Aigner A, Grünweller A and Hartmann RK: The
proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 31:918–928.
2012. View Article : Google Scholar
|
|
96
|
Endo H, Muramatsu T, Furuta M, Uzawa N,
Pimkhaokham A, Amagasa T, Inazawa J and Kozaki K: Potential of
tumor-suppressive miR-596 targeting LGALS3BP as a therapeutic agent
in oral cancer. Carcinogenesis. 34:560–569. 2013. View Article : Google Scholar
|
|
97
|
Wu Y, Crawford M, Mao Y, Lee RJ, Davis IC,
Elton TS, Lee LJ and Nana-Sinkam SP: Therapeutic delivery of
microRNA-29b by cationic lipoplexes for lung cancer. Mol Ther
Nucleic Acids. 2:e842013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Huang X, Schwind S, Yu B, Santhanam R,
Wang H, Hoellerbauer P, Mims A, Klisovic R, Walker AR, Chan KK, et
al: Targeted delivery of microRNA-29b by transferrin-conjugated
anionic lipopolyplex nanoparticles: A novel therapeutic strategy in
acute myeloid leukemia. Clin Cancer Res. 19:2355–2367.
2013.PubMed/NCBI
|
|
99
|
Liang Z, Ahn J, Guo D, Votaw JR and Shim
H: MicroRNA-302 replacement therapy sensitizes breast cancer cells
to ionizing radiation. Pharm Res. 30:1008–1016. 2013. View Article : Google Scholar :
|
|
100
|
Møller HG, Rasmussen AP, Andersen HH,
Johnsen KB, Henriksen M and Duroux M: A systematic review of
microRNA in glioblastoma multiforme: Micro-modulators in the
mesenchymal mode of migration and invasion. Mol Neurobiol.
47:131–144. 2013. View Article : Google Scholar :
|
|
101
|
Hershkovitz-Rokah O, Modai S,
Pasmanik-Chor M, Toren A, Shomron N, Raanani P, Shpilberg O and
Granot G: Restoration of miR-424 suppresses BCR-ABL activity and
sensitizes CML cells to imatinib treatment. Cancer Lett.
360:245–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Lee YM, Lee JY, Ho CC, Hong QS, Yu SL,
Tzeng CR, Yang PC and Chen HW: miRNA-34b as a tumor suppressor in
estrogen-dependent growth of breast cancer cells. Breast Cancer
Res. 13:R1162011. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Huang P, Ye B, Yang Y, Shi J and Zhao H:
MicroRNA-181 functions as a tumor suppressor in non-small cell lung
cancer (NSCLC) by targeting Bcl-2. Tumour Biol. 36:3381–3387. 2015.
View Article : Google Scholar
|
|
104
|
Su R, Lin HS, Zhang XH, Yin XL, Ning HM,
Liu B, Zhai PF, Gong JN, Shen C, Song L, et al: MiR-181 family:
Regulators of myeloid differentiation and acute myeloid leukemia as
well as potential therapeutic targets. Oncogene. 34:3226–3239.
2015. View Article : Google Scholar
|
|
105
|
Bachetti T, Di Zanni E, Ravazzolo R and
Ceccherini I: miR-204 mediates post-transcriptional down-regulation
of PHOX2B gene expression in neuroblastoma cells. Biochim Biophys
Acta. 1849.1057–1065. 2015.
|
|
106
|
Fernandez S, Risolino M, Mandia N, Talotta
F, Soini Y, Incoronato M, Condorelli G, Banfi S and Verde P:
miR-340 inhibits tumor cell proliferation and induces apoptosis by
targeting multiple negative regulators of p27 in non-small cell
lung cancer. Oncogene. 34:3240–3250. 2015. View Article : Google Scholar
|
|
107
|
Liu G, Liu Y, Yang Z, Wang J, Li D and
Zhang X: Tumor suppressor microRNA-18a regulates tumor
proliferation and invasion by targeting TBPL1 in colorectal cancer
cells. Mol Med Rep. 12:7643–7648. 2015.PubMed/NCBI
|
|
108
|
Xishan Z, Ziying L, Jing D and Gang L:
MicroRNA-320a acts as a tumor suppressor by targeting BCR/ABL
oncogene in chronic myeloid leukemia. Sci Rep. 5:124602015.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhao Z, Ma X, Sung D, Li M, Kosti A, Lin
G, Chen Y, Pertsemlidis A, Hsiao TH and Du L: microRNA-449a
functions as a tumor suppressor in neuroblastoma through inducing
cell differentiation and cell cycle arrest. RNA Biol. 12:538–554.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kalinowski FC, Brown RA, Ganda C, Giles
KM, Epis MR, Horsham J and Leedman PJ: microRNA-7: A tumor
suppressor miRNA with therapeutic potential. Int J Biochem Cell
Biol. 54:312–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Gu DN, Huang Q and Tian L: The molecular
mechanisms and therapeutic potential of microRNA-7 in cancer.
Expert Opin Ther Targets. 19:415–426. 2015. View Article : Google Scholar
|
|
112
|
Nohata N, Hanazawa T, Enokida H and Seki
N: microRNA-1/133a and microRNA-206/133b clusters: Dysregulation
and functional roles in human cancers. Oncotarget. 3:9–21.
2012.PubMed/NCBI
|
|
113
|
Hudson RS, Yi M, Esposito D, Watkins SK,
Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB,
et al: MicroRNA-1 is a candidate tumor suppressor and prognostic
marker in human prostate cancer. Nucleic Acids Res. 40:3689–3703.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chang YS, Chen WY, Yin JJ,
Sheppard-Tillman H, Huang J and Liu YN: EGF receptor pomotes
prostate cancer bone metastasis by downregulating miR-1 and
activating TWIST1. Cancer Res. 75:3077–3086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zhang H, Cai K, Wang J, Wang X, Cheng K,
Shi F, Jiang L, Zhang Y and Dou J: MiR-7, inhibited indirectly by
lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of
breast cancer stem cells by downregulating the STAT3 pathway. Stem
Cells. 32:2858–2868. 2014a. View Article : Google Scholar
|
|
116
|
Okuda H, Xing F, Pandey PR, Sharma S,
Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y, et al:
miR-7 suppresses brain metastasis of breast cancer stem-like cells
by modulating KLF4. Cancer Res. 73:1434–1444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang
W, Di W and Qiu L: MicroRNA-7 inhibits tumor metastasis and
reverses epithelial-mesenchymal transition through AKT/ERK1/2
inactivation by targeting EGFR in epithelial ovarian cancer. PLoS
One. 9:e967182014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Dangi-Garimella S, Yun J, Eves EM, Newman
M, Erkeland SJ, Hammond SM, Minn AJ and Rosner MR: Raf kinase
inhibitory protein suppresses a metastasis signalling cascade
involving LIN28 and let-7. EMBO J. 28:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Shi XB, Tepper CG and deVere White RW:
Cancerous miRNAs and their regulation. Cell Cycle. 7:1529–1538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zheng L, Qi T, Yang D, Qi M, Li D, Xiang
X, Huang K and Tong Q: microRNA-9 suppresses the proliferation,
invasion and metastasis of gastric cancer cells through targeting
cyclin D1 and Ets1. PLoS One. 8:e557192013. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: Discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar
|
|
124
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar
|
|
125
|
Pekarsky Y and Croce CM: Role of miR-15/16
in CLL. Cell Death Differ. 22:6–11. 2015. View Article : Google Scholar
|
|
126
|
Bonci D, Coppola V, Musumeci M, Addario A,
Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C,
et al: The miR-15a-miR-16-1 cluster controls prostate cancer by
targeting multiple oncogenic activities. Nat Med. 14:1271–1277.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Kang W, Tong JH, Lung RW, Dong Y, Zhao J,
Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al: Targeting of YAP1
by microRNA-15a and microRNA-16-1 exerts tumor suppressor function
in gastric adenocarcinoma. Mol Cancer. 14:522015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chen F, Chen L, He H, Huang W, Zhang R, Li
P, et al: Up-regulation of microRNA-16 in glioblastoma inhibits the
function of endothelial cells and tumor angiogenesis by targeting
Bmi-1. Anticancer Agents Med Chem. 2015.
|
|
129
|
Humphreys KJ, McKinnon RA and Michael MZ:
miR-18a inhibits CDC42 and plays a tumour suppressor role in
colorectal cancer cells. PLoS One. 9:e1122882014. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zoni E, van der Horst G, van de Merbel AF,
Chen L, Rane JK, Pelger RC, Collins AT, Visakorpi T, Snaar-Jagalska
BE, Maitland NJ, et al: miR-25 modulates invasiveness and
dissemination of human prostate cancer cells via regulation of αv-
and α6 integrin expression. Cancer Res. 75:2326–2336. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Sengupta S, den Boon JA, Chen IH, Newton
MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B and
Ahlquist P: MicroRNA 29c is down-regulated in nasopharyngeal
carcinomas, up-regulating mRNAs encoding extracellular matrix
proteins. Proc Natl Acad Sci USA. 105:5874–5878. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Ugalde AP, Ramsay AJ, de la Rosa J, Varela
I, Mariño G, Cadiñanos J, Lu J, Freije JM and López-Otín C: Aging
and chronic DNA damage response activate a regulatory pathway
involving miR-29 and p53. EMBO J. 30:2219–2232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Garzon R, Heaphy CE, Havelange V, Fabbri
M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA,
et al: MicroRNA 29b functions in acute myeloid leukemia. Blood.
114:5331–5341. 2009a. View Article : Google Scholar
|
|
134
|
Garzon R, Liu S, Fabbri M, Liu Z, Heaphy
CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al:
MicroRNA-29b induces global DNA hypomethylation and tumor
suppressor gene reexpression in acute myeloid leukemia by targeting
directly DNMT3A and 3B and indirectly DNMT1. Blood. 113:6411–6418.
2009b. View Article : Google Scholar
|
|
135
|
Kapinas K, Kessler CB and Delany AM:
miR-29 suppression of osteonectin in osteoblasts: Regulation during
differentiation and by canonical Wnt signaling. J Cell Biochem.
108:216–224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Mott JL, Kobayashi S, Bronk SF and Gores
GJ: miR-29 regulates Mcl-1 protein expression and apoptosis.
Oncogene. 26:6133–6140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y,
Jia WH and Zhuang SM: Effects of microRNA-29 on apoptosis,
tumorigenicity, and prognosis of hepatocellular carcinoma.
Hepatology. 51:836–845. 2010.
|
|
139
|
Filkowski JN, Ilnytskyy Y, Tamminga J,
Koturbash I, Golubov A, Bagnyukova T, Pogribny IP and Kovalchuk O:
Hypomethylation and genome instability in the germline of exposed
parents and their progeny is associated with altered miRNA
expression. Carcinogenesis. 31:1110–1115. 2010. View Article : Google Scholar
|
|
140
|
Wang Y, Zhang X, Li H, Yu J and Ren X: The
role of miRNA-29 family in cancer. Eur J Cell Biol. 92:123–128.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Hu W, Dooley J, Chung SS, Chandramohan D,
Cimmino L, Mukherjee S, Mason CE, de Strooper B, Liston A and Park
CY: miR-29a maintains mouse hematopoietic stem cell self-renewal by
regulating Dnmt3a. Blood. 125:2206–2216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Li L and Wang B: Overexpression of
microRNA-30b improves adenovirus-mediated p53 cancer gene therapy
for laryngeal carcinoma. Int J Mol Sci. 15:19729–19740. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Hou C, Sun B, Jiang Y, Zheng J, Yang N, Ji
C, Liang Z, Shi J, Zhang R, Liu Y, et al: MicroRNA-31 inhibits lung
adenocarcinoma stem-like cells via down-regulation of MET-PI3K-Akt
signaling pathway. Anticancer Agents Med Chem. 16:501–518. 2016.
View Article : Google Scholar
|
|
144
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Sossey-Alaoui K, Downs-Kelly E, Das M,
Izem L, Tubbs R and Plow EF: WAVE3, an actin remodeling protein, is
regulated by the metastasis suppressor microRNA, miR-31, during the
invasion-metastasis cascade. Int J Cancer. 129:1331–1343. 2011.
View Article : Google Scholar :
|
|
146
|
Lin Y, Liu AY, Fan C, Zheng H, Li Y, Zhang
C, Wu S, Yu D, Huang Z, Liu F, et al: MicroRNA-33b inhibits breast
cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci Rep.
5:99952015. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Xu N, Li Z, Yu Z, Yan F, Liu Y, Lu X and
Yang W: MicroRNA-33b suppresses migration and invasion by targeting
c-Myc in osteosarcoma cells. PLoS One. 9:e1153002014. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Bommer GT, Gerin I, Feng Y, Kaczorowski
AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al:
p53-mediated activation of miRNA34 candidate tumorsuppressor genes.
Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Fujita Y, Kojima K, Hamada N, Ohhashi R,
Akao Y, Nozawa Y, Deguchi T and Ito M: Effects of miR-34a on cell
growth and chemoresistance in prostate cancer PC3 cells. Biochem
Biophys Res Commun. 377:114–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Leucci E, Cocco M, Onnis A, De Falco G,
van Cleef P, Bellan C, van Rijk A, Nyagol J, Byakika B, Lazzi S, et
al: MYC translocation-negative classical Burkitt lymphoma cases: An
alternative pathogenetic mechanism involving miRNA deregulation. J
Pathol. 216:440–450. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Saito Y, Nakaoka T and Saito H:
microRNA-34a as a therapeutic agent against human cancer. J Clin
Med. 4:1951–1959. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Wei JS, Song YK, Durinck S, Chen QR, Cheuk
AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J, et al: The MYCN
oncogene is a direct target of miR-34a. Oncogene. 27:5204–5213.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Lodygin D, Tarasov V, Epanchintsev A,
Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J and Hermeking
H: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A,
Chen H, Ding F, Wang X and Liu Z: MicroRNA-34 suppresses breast
cancer invasion and metastasis by directly targeting Fra-1.
Oncogene. 32:4294–4303. 2013. View Article : Google Scholar
|
|
157
|
Yang P, Li QJ, Feng Y, Zhang Y, Markowitz
GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, et al:
TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment
promotes venous metastases of HBV-positive hepatocellular
carcinoma. Cancer Cell. 22:291–303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Krzeszinski JY, Wei W, Huynh H, Jin Z,
Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, et
al: miR-34a blocks osteoporosis and bone metastasis by inhibiting
osteoclastogenesis and Tgif2. Nature. 512:431–435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Wang LG, Ni Y, Su BH, Mu XR, Shen HC and
Du JJ: MicroRNA-34b functions as a tumor suppressor and acts as a
nodal point in the feedback loop with Met. Int J Oncol. 42:957–962.
2013.PubMed/NCBI
|
|
161
|
Yu Z, Kim J, He L, Creighton CJ, Gunaratne
PH, Hawkins SM and Matzuk MM: Functional analysis of miR-34c as a
putative tumor suppressor in high-grade serous ovarian cancer. Biol
Reprod. 91:1132014. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Liu XY, Liu ZJ, He H, Zhang C and Wang YL:
MicroRNA-101-3p suppresses cell proliferation, invasion and
enhances chemotherapeutic sensitivity in salivary gland adenoid
cystic carcinoma by targeting Pim-1. Am J Cancer Res. 5:3015–3029.
2015.PubMed/NCBI
|
|
163
|
Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ,
Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, et al: MicroRNA-122
plays a critical role in liver homeostasis and
hepatocarcinogenesis. J Clin Invest. 122:2884–2897. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Taniguchi K, Sugito N, Kumazaki M,
Shinohara H, Yamada N, Nakagawa Y, Ito Y, Otsuki Y, Uno B, Uchiyama
K, et al: MicroRNA-124 inhibits cancer cell growth through
PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett.
363:17–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Huang TC, Chang HY, Chen CY, Wu PY, Lee H,
Liao YF, Hsu WM, Huang HC and Juan HF: Silencing of miR-124 induces
neuroblastoma SK-N-SH cell differentiation, cell cycle arrest and
apoptosis through promoting AHR. FEBS Lett. 585:3582–3586. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Kato T, Enomoto A, Watanabe T, Haga H,
Ishida S, Kondo Y, Furukawa K, Urano T, Mii S, Weng L, et al:
TRIM27/MRTF-B-dependent integrin β1 expression defines leading
cells in cancer cell collectives. Cell Rep. 7:1156–1167. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH,
Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al: The putative
tumour suppressor microRNA-124 modulates hepatocellular carcinoma
cell aggressiveness by repressing ROCK2 and EZH2. Gut. 61:278–289.
2012. View Article : Google Scholar
|
|
168
|
Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y,
Gu X and Meng F: miR-124 exerts tumor suppressive functions on the
cell proliferation, motility and angiogenesis of bladder cancer by
fine-tuning UHRF1. FEBS J. 282:4376–4388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Zhang C, Hu Y, Wan J and He H:
MicroRNA-124 suppresses the migration and invasion of osteosarcoma
cells via targeting ROR2-mediated non-canonical Wnt signaling.
Oncol Rep. 34:2195–2201. 2015.PubMed/NCBI
|
|
170
|
Sun Y, Ai X, Shen S and Lu S:
NF-κB-mediated miR-124 suppresses metastasis of non-small-cell lung
cancer by targeting MYO10. Oncotarget. 6:8244–8254. 2015a.
View Article : Google Scholar
|
|
171
|
Sun Y, Luo ZM, Guo XM, Su DF and Liu X: An
updated role of microRNA-124 in central nervous system disorders: A
review. Front Cell Neurosci. 9:1932015b. View Article : Google Scholar
|
|
172
|
Chen Z, Liu S, Tian L, Wu M, Ai F, Tang W,
Zhao L, Ding J, Zhang L and Tang A: miR-124 and miR-506 inhibit
colorectal cancer progression by targeting DNMT3B and DNMT1.
Oncotarget. 6:38139–38150. 2015.PubMed/NCBI
|
|
173
|
Zhang Y, Li H, Han J and Zhang Y:
Down-regulation of microRNA-124 is correlated with tumor metastasis
and poor prognosis in patients with lung cancer. Int J Clin Exp
Pathol. 8:1967–1972. 2015.PubMed/NCBI
|
|
174
|
Cowden Dahl KD, Dahl R, Kruichak JN and
Hudson LG: The epidermal growth factor receptor responsive miR-125a
represses mesenchymal morphology in ovarian cancer cells.
Neoplasia. 11:1208–1215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang
L, Han B, Meng J, Yan Z, Yan X, et al: MiR-125a suppresses tumor
growth, invasion and metastasis in cervical cancer by targeting
STAT3. Oncotarget. 6:25266–25280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Sun Y, Bai Y, Zhang F, Wang Y, Guo Y and
Guo L: miR-126 inhibits non-small cell lung cancer cells
proliferation by targeting EGFL7. Biochem Biophys Res Commun.
391:1483–1489. 2010. View Article : Google Scholar
|
|
177
|
Xiong Y, Kotian S, Zeiger MA, Zhang L and
Kebebew E: miR-126-3p inhibits thyroid cancer cell growth and
metastasis, and is associated with aggressive thyroid cancer. PLoS
One. 10:e01304962015. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Wang CZ, Yuan P and Li Y: MiR-126
regulated breast cancer cell invasion by targeting ADAM9. Int J
Clin Exp Pathol. 8:6547–6553. 2015.PubMed/NCBI
|
|
179
|
Wen Q, Zhao J, Bai L, Wang T, Zhang H and
Ma Q: miR-126 inhibits papillary thyroid carcinoma growth by
targeting LRP6. Oncol Rep. 34:2202–2210. 2015.PubMed/NCBI
|
|
180
|
Jiang L, He A, Zhang Q and Tao C: miR-126
inhibits cell growth, invasion, and migration of osteosarcoma cells
by downregulating ADAM-9. Tumour Biol. 35:12645–12654. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie
H, Zhou L, Wu J and Zheng S: MiR-126-3p suppresses tumor metastasis
and angiogenesis of hepatocellular carcinoma by targeting LRP6 and
PIK3R2. J Transl Med. 12:2592014. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Zhang Y, Wang X, Xu B, Wang B, Wang Z,
Liang Y, Zhou J, Hu J and Jiang B: Epigenetic silencing of miR-126
contributes to tumor invasion and angiogenesis in colorectal
cancer. Oncol Rep. 30:1976–1984. 2013.PubMed/NCBI
|
|
183
|
Png KJ, Halberg N, Yoshida M and Tavazoie
SF: A microRNA regulon that mediates endothelial recruitment and
metastasis by cancer cells. Nature. 481:190–194. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian
X, Yin Y, Zhao P, Wang YY, Wang XF, et al: MiR-128 inhibits tumor
growth and angiogenesis by targeting p70S6K1. PLoS One.
7:e327092012. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Wuchty S, Arjona D, Li A, Kotliarov Y,
Walling J, Ahn S, Zhang A, Maric D, Anolik R, Zenklusen JC, et al:
Prediction of associations between microRNAs and gene expression in
glioma biology. PLoS One. 6:e146812011. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan
X, Gong Y, Yin B, Liu W, Qiang B, et al: MicroRNA-128 inhibits
glioma cells proliferation by targeting transcription factor E2F3a.
J Mol Med Berl. 87:43–51. 2009. View Article : Google Scholar
|
|
187
|
Huang CY, Huang XP, Zhu JY, Chen ZG, Li
XJ, Zhang XH, Huang S, He JB, Lian F, Zhao YN, et al: miR-128-3p
suppresses hepatocellular carcinoma proliferation by regulating
PIK3R1 and is correlated with the prognosis of HCC patients. Oncol
Rep. 33:2889–2898. 2015.PubMed/NCBI
|
|
188
|
Kano M, Seki N, Kikkawa N, Fujimura L,
Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M and
Matsubara H: miR-145, miR-133a and miR-133b: Tumor-suppressive
miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J
Cancer. 127:2804–2814. 2010. View Article : Google Scholar
|
|
189
|
Kroiss A, Vincent S, Decaussin-Petrucci M,
Meugnier E, Viallet J, Ruffion A, Chalmel F, Samarut J and Allioli
N: Androgen-regulated microRNA-135a decreases prostate cancer cell
migration and invasion through downregulating ROCK1 and ROCK2.
Oncogene. 34:2846–2855. 2015. View Article : Google Scholar
|
|
190
|
Liang L, Li X, Zhang X, Lv Z, He G, Zhao
W, Ren X, Li Y, Bian X, Liao W, et al: MicroRNA-137, an HMGA1
target, suppresses colorectal cancer cell invasion and metastasis
in mice by directly targeting FMNL2. Gastroenterology.
144:624–635.e4. 2013. View Article : Google Scholar
|
|
191
|
Xia H, Sun S, Wang B, Wang T, Liang C, Li
G, Huang C, Qi D and Chu X: miR-143 inhibits NSCLC cell growth and
metastasis by targeting Limk1. Int J Mol Sci. 15:11973–11983. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Gao P, Xing AY, Zhou GY, Zhang TG, Zhang
JP, Gao C, Li H and Shi DB: The molecular mechanism of microRNA-145
to suppress invasion-metastasis cascade in gastric cancer.
Oncogene. 32:491–501. 2013. View Article : Google Scholar
|
|
193
|
Zhang H, Pu J, Qi T, Qi M, Yang C, Li S,
Huang K, Zheng L and Tong Q: MicroRNA-145 inhibits the growth,
invasion, metastasis and angiogenesis of neuroblastoma cells
through targeting hypoxia-inducible factor 2 alpha. Oncogene.
33:387–397. 2014. View Article : Google Scholar
|
|
194
|
Bhaumik D, Scott GK, Schokrpur S, Patil
CK, Campisi J and Benz CC: Expression of microRNA-146 suppresses
NF-kappaB activity with reduction of metastatic potential in breast
cancer cells. Oncogene. 27:5643–5647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Lin SL, Chiang A, Chang D and Ying SY:
Loss of miR-146a function in hormone-refractory prostate cancer.
RNA. 14:417–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Zhang JP, Zeng C, Xu L, Gong J, Fang JH
and Zhuang SM: MicroRNA-148a suppresses the epithelial-mesenchymal
transition and metastasis of hepatoma cells by targeting Met/Snail
signaling. Oncogene. 33:4069–4076. 2014. View Article : Google Scholar
|
|
197
|
Cimino D, De Pittà C, Orso F, Zampini M,
Casara S, Penna E, Quaglino E, Forni M, Damasco C, Pinatel E, et
al: miR148b is a major coordinator of breast cancer progression in
a relapse-associated microRNA signature by targeting ITGA5, ROCK1,
PIK3CA, NRAS, and CSF1. FASEB J. 27:1223–1235. 2013. View Article : Google Scholar
|
|
198
|
Bischoff A, Huck B, Keller B, Strotbek M,
Schmid S, Boerries M, Busch H, Müller D and Olayioye MA: miR149
functions as a tumor suppressor by controlling breast epithelial
cell migration and invasion. Cancer Res. 74:5256–5265. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Visone R, Veronese A, Rassenti LZ, Balatti
V, Pearl DK, Acunzo M, Volinia S, Taccioli C, Kipps TJ and Croce
CM: miR-181b is a biomarker of disease progression in chronic
lymphocytic leukemia. Blood. 118:3072–3079. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Kouri FM, Hurley LA, Daniel WL, Day ES,
Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C, et al:
miR-182 integrates apoptosis, growth, and differentiation programs
in glioblastoma. Genes Dev. 29:732–745. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Leivonen SK, Rokka A, Ostling P, Kohonen
P, Corthals GL, Kallioniemi O and Perälä M: Identification of
miR-193b targets in breast cancer cells and systems biological
analysis of their functional impact. Mol Cell Proteomics.
10:M110.0053222011. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Yang H, Liu P, Zhang J, Peng X, Lu Z, Yu
S, Meng Y, Tong WM and Chen J: Long noncoding RNA MIR31HG exhibits
oncogenic property in pancreatic ductal adenocarcinoma and is
negatively regulated by miR-193b. Oncogene. Nov 9–2015.(Epub ahead
of print). View Article : Google Scholar
|
|
203
|
Tan S, Li R, Ding K, Lobie PE and Zhu T:
miR-198 inhibits migration and invasion of hepatocellular carcinoma
cells by targeting the HGF/c-MET pathway. FEBS Lett. 585:2229–2234.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Bao W, Wang HH, Tian FJ, He XY, Qiu MT,
Wang JY, Zhang HJ, Wang LH and Wan XP: A TrkB-STAT3-miR-204-5p
regulatory circuitry controls proliferation and invasion of
endometrial carcinoma cells. Mol Cancer. 12:1552013. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Xia Z, Liu F, Zhang J and Liu L: Decreased
expression of MiRNA-204-5p contributes to glioma progression and
promotes glioma cell growth, migration and invasion. PLoS One.
10:e01323992015. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Gandellini P, Folini M, Longoni N, Pennati
M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta
P, et al: miR-205 exerts tumor-suppressive functions in human
prostate through down-regulation of protein kinase Cepsilon. Cancer
Res. 69:2287–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Chen QY, Jiao DM, Yan L, Wu YQ, Hu HZ,
Song J, Yan J, Wu LJ, Xu LQ and Shi JG: Comprehensive gene and
microRNA expression profiling reveals miR-206 inhibits MET in lung
cancer metastasis. Mol Biosyst. 11:2290–2302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang
DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, et al: Identification
of microRNA-214 as a negative regulator of colorectal cancer liver
metastasis by way of regulation of fibroblast growth factor
receptor 1 expression. Hepatology. 60:598–609. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S,
Guo X, Wang B, Gang Y, Zhang Y, et al: MiR-218 inhibits invasion
and metastasis of gastric cancer by targeting the Robo1 receptor.
PLoS Genet. 6:e10008792010. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Wei JJ, Wu X, Peng Y, Shi G, Basturk O,
Yang X, Daniels G, Osman I, Ouyang J, Hernando E, et al: Regulation
of HMGA1 expression by microRNA-296 affects prostate cancer growth
and invasion. Clin Cancer Res. 17:1297–1305. 2011. View Article : Google Scholar :
|
|
211
|
Wang L, Yao J, Shi X, Hu L, Li Z, Song T
and Huang C: MicroRNA-302b suppresses cell proliferation by
targeting EGFR in human hepatocellular carcinoma SMMC-7721 cells.
BMC Cancer. 13:4482013. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Hurst DR, Edmonds MD and Welch DR:
Metastamir: The field of metastasis-regulatory microRNA is
spreading. Cancer Res. 69:7495–7498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Li KK, Pang JC, Lau KM, Zhou L, Mao Y,
Wang Y, Poon WS and Ng HK: MiR-383 is downregulated in
medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol.
23:413–425. 2013. View Article : Google Scholar
|
|
215
|
Bou Kheir T, Futoma-Kazmierczak E,
Jacobsen A, Krogh A, Bardram L, Hother C, Grønbæk K, Federspiel B,
Lund AH and Friis-Hansen L: miR-449 inhibits cell proliferation and
is down-regulated in gastric cancer. Mol Cancer. 10:292011.
View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Luo W, Huang B, Li Z, Li H, Sun L, Zhang
Q, Qiu X and Wang E: MicroRNA-449a is downregulated in non-small
cell lung cancer and inhibits migration and invasion by targeting
c-Met. PLoS One. 8:e647592013. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Okamoto K, Ishiguro T, Midorikawa Y, Ohata
H, Izumiya M, Tsuchiya N, Sato A, Sakai H and Nakagama H: miR-493
induction during carcinogenesis blocks metastatic settlement of
colon cancer cells in liver. EMBO J. 31:1752–1763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Gu Y, Cheng Y, Song Y, Zhang Z, Deng M,
Wang C, Zheng G and He Z: MicroRNA-493 suppresses tumor growth,
invasion and metastasis of lung cancer by regulating E2F1. PLoS
One. 9:e1026022014. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Sakai H1, Sato A, Aihara Y, Ikarashi Y,
Midorikawa Y, Kracht M, Nakagama H and Okamoto K: MKK7 mediates
miR-493-dependent suppression of liver metastasis of colon cancer
cells. Cancer Sci. 105:425–430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Kikkawa N, Kinoshita T, Nohata N, Hanazawa
T, Yamamoto N, Fukumoto I, Chiyomaru T, Enokida H, Nakagawa M,
Okamoto Y, et al: microRNA-504 inhibits cancer cell proliferation
via targeting CDK6 in hypopharyngeal squamous cell carcinoma. Int J
Oncol. 44:2085–2092. 2014.PubMed/NCBI
|
|
221
|
Keklikoglou I, Koerner C, Schmidt C, Zhang
JD, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T,
Schneeweiss A, et al: MicroRNA-520/373 family functions as a tumor
suppressor in estrogen receptor negative breast cancer by targeting
NF-κB and TGF-β signaling pathways. Oncogene. 31:4150–4163. 2012.
View Article : Google Scholar
|
|
222
|
Song B, Ji W, Guo S, Liu A, Jing W, Shao
C, Li G and Jin G: miR-545 inhibited pancreatic ductal
adenocarcinoma growth by targeting RIG-I. FEBS Lett. 588:4375–4381.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Bowen D, Zhe W, Xin Z, Shipeng F, Guoxin
W, Jianxing H and Zhang B: MicroRNA-545 suppresses cell
proliferation by targeting cyclin D1 and CDK4 in lung cancer cells.
PLoS. 9:880222014. View Article : Google Scholar
|
|
224
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et
al: A MicroRNA signature associated with prognosis and progression
in chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar
|
|
232
|
Garzon R, Volinia S, Liu CG,
Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K,
Alder H, Nakamura T, et al: MicroRNA signatures associated with
cytogenetics and prognosis in acute myeloid leukemia. Blood.
111:3183–3189. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
234
|
White NM, Fatoohi E, Metias M, Jung K,
Stephan C and Yousef GM: Metastamirs: A stepping stone towards
improved cancer management. Nat Rev Clin Oncol. 8:75–84. 2011.
View Article : Google Scholar
|
|
235
|
Zhou L, Liu F, Wang X and Ouyang G: The
roles of microRNAs in the regulation of tumor metastasis. Cell
Biosci. 5:322015. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Wang XH, Cai P, Wang MH and Wang Z:
microRNA 25 promotes osteosarcoma cell proliferation by targeting
the cell cycle inhibitor p27. Mol Med Rep. 10:855–859.
2014.PubMed/NCBI
|
|
237
|
Siu MK, Tsai YC, Chang YS, Yin JJ, Suau F,
Chen WY and Liu YN: Transforming growth factor-β promotes prostate
bone metastasis through induction of microRNA-96 and activation of
the mTOR pathway. Oncogene. 34:4767–4776. 2015. View Article : Google Scholar
|
|
238
|
Xia X, Li Y, Wang W, Tang F, Tan J, Sun L,
Li Q, Sun L, Tang B and He S: MicroRNA-1908 functions as a
glioblastoma oncogene by suppressing PTEN tumor suppressor pathway.
Mol Cancer. 14:1542015. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Sachdeva M, Mito JK, Lee CL, Zhang M, Li
Z, Dodd RD, Cason D, Luo L, Ma Y, Van Mater D, et al: MicroRNA-182
drives metastasis of primary sarcomas by targeting multiple genes.
J Clin Invest. 124:4305–4319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H
and Liu Z: MicroRNA-10b promotes migration and invasion through
KLF4 in human esophageal cancer cell lines. J Biol Chem.
285:7986–7994. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Wang YY, Ye ZY, Zhao ZS, Li L, Wang YX,
Tao HQ, Wang HJ and He XJ: Clinicopathologic significance of
miR-10b expression in gastric carcinoma. Hum Pathol. 44:1278–1285.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Liu W, Zabirnyk O, Wang H, Shiao YH,
Nickerson ML, Khalil S, Anderson LM, Perantoni AO and Phang JM:
miR-23b targets proline oxidase, a novel tumor suppressor protein
in renal cancer. Oncogene. 29:4914–4924. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Fletcher CE, Dart DA, Sita-Lumsden A,
Cheng H, Rennie PS and Bevan CL: Androgen-regulated processing of
the oncomir miR-27a, which targets Prohibitin in prostate cancer.
Hum Mol Genet. 21:3112–3127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Ng WL, Yan D, Zhang X, Mo YY and Wang Y:
Over-expression of miR-100 is responsible for the low-expression of
ATM in the human glioma cell line: M059J. DNA Repair (Amst).
9:1170–1175. 2010. View Article : Google Scholar
|
|
246
|
Zheng YS, Zhang H, Zhang XJ, Feng DD, Luo
XQ, Zeng CW, Lin KY, Zhou H, Qu LH, Zhang P, et al: MiR-100
regulates cell differentiation and survival by targeting RBSP3, a
phosphatase-like tumor suppressor in acute myeloid leukemia.
Oncogene. 31:80–92. 2012. View Article : Google Scholar :
|
|
247
|
Knackmuss U, Lindner SE, Aneichyk T,
Kotkamp B, Knust Z, Villunger A and Herzog S: MAP3K11 is a tumor
suppressor targeted by the oncomiR miR-125b in early B cells. Cell
Death Differ. 23:242–252. 2016. View Article : Google Scholar
|
|
248
|
Park JK, Henry JC, Jiang J, Esau C, Gusev
Y, Lerner MR, Postier RG, Brackett DJ and Schmittgen TD: miR-132
and miR-212 are increased in pancreatic cancer and target the
retinoblastoma tumor suppressor. Biochem Biophys Res Commun.
406:518–523. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Kong W, He L, Coppola M, Guo J, Esposito
NN, Coppola D and Cheng JQ: MicroRNA-155 regulates cell survival,
growth, and chemosensitivity by targeting FOXO3a in breast cancer.
J Biol Chem. 285:17869–17879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu
H, Liu MF and Wang ED: MicroRNA-155 functions as an OncomiR in
breast cancer by targeting the suppressor of cytokine signaling 1
gene. Cancer Res. 70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Czyzyk-Krzeska MF and Zhang X: MiR-155 at
the heart of oncogenic pathways. Oncogene. 33:677–678. 2014.
View Article : Google Scholar :
|
|
252
|
Wang J and Wu J: Role of miR-155 in breast
cancer. Front Biosci (Landmark Ed). 17:2350–2355. 2012. View Article : Google Scholar
|
|
253
|
Ling N, Gu J, Lei Z, Li M, Zhao J, Zhang
HT and Li X: microRNA-155 regulates cell proliferation and invasion
by targeting FOXO3a in glioma. Oncol Rep. 30:2111–2118.
2013.PubMed/NCBI
|
|
254
|
Musilova K and Mraz M: MicroRNAs in B-cell
lymphomas: How a complex biology gets more complex. Leukemia.
29:1004–1017. 2015. View Article : Google Scholar
|
|
255
|
Fontana L, Fiori ME, Albini S, Cifaldi L,
Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V,
Giacomini P, et al: AntagomiR-17-5p abolishes the growth of
therapy-resistant neuroblastoma through p21 and BIM. PLoS One.
3:e22362008. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A,
Bogunovic D, et al: Aberrant miR-182 expression promotes melanoma
metastasis by repressing FOXO3 and microphthalmia-associated
transcription factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han
L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, et al: MiR-221 and
miR-222 target PUMA to induce cell survival in glioblastoma. Mol
Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
259
|
Garofalo M, Quintavalle C, Romano G, Croce
CM and Condorelli G: miR221/222 in cancer: Their role in tumor
progression and response to therapy. Curr Mol Med. 12:27–33. 2012.
View Article : Google Scholar
|
|
260
|
Quintavalle C, Garofalo M, Zanca C, Romano
G, Iaboni M, del Basso De Caro M, Martinez-Montero JC, Incoronato
M, Nuovo G, Croce CM, et al: miR-221/222 overexpession in human
glioblastoma increases invasiveness by targeting the protein
phosphate PTPμ. Oncogene. 31:858–868. 2012. View Article : Google Scholar
|
|
261
|
Chen WX, Hu Q, Qiu MT, Zhong SL, Xu JJ,
Tang JH and Zhao JH: miR-221/222: Promising biomarkers for breast
cancer. Tumour Biol. 34:1361–1370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Matsuzaki J and Suzuki H: Role of
MicroRNAs-221/222 in digestive systems. J Clin Med. 4:1566–1577.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Würdinger T, Tannous BA, Saydam O, Skog J,
Grau S, Soutschek J, Weissleder R, Breakefield XO and Krichevsky
AM: miR-296 regulates growth factor receptor overexpression in
angiogenic endothelial cells. Cancer Cell. 14:382–393. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
et al: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Adv Exp Med Biol.
604:17–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
266
|
Xu Y, Jin J, Liu Y, Huang Z, Deng Y, You
T, Zhou T, Si J and Zhuo W: Snail-regulated MiR-375 inhibits
migration and invasion of gastric cancer cells by targeting JAK2.
PLoS One. 9:e995162014. View Article : Google Scholar : PubMed/NCBI
|
|
267
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA.
104:20350–20355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Tu K, Liu Z, Yao B, Han S and Yang W:
MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT
signaling in hepatocellular carcinoma. Int J Oncol. 48:965–974.
2016.
|
|
269
|
Shao J, Cao J, Liu Y, Mei H, Zhang Y and
Xu W: MicroRNA-519a promotes proliferation and inhibits apoptosis
of hepatocellular carcinoma cells by targeting FOXF2. FEBS Open
Bio. 5:893–899. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
270
|
Ward A, Shukla K, Balwierz A, Soons Z,
König R, Sahin O and Wiemann S: MicroRNA-519a is a novel oncomir
conferring tamoxifen resistance by targeting a network of
tumour-suppressor genes in ER+ breast cancer. J Pathol.
233:368–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
271
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar
|
|
272
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
273
|
Chen D, Sun Y, Wei Y, Zhang P, Rezaeian
AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, et al:
LIFR is a breast cancer metastasis suppressor upstream of the
Hippo-YAP pathway and a prognostic marker. Nat Med. 18:1511–1517.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
274
|
White RA, Neiman JM, Reddi A, Han G,
Birlea S, Mitra D, Dionne L, Fernandez P, Murao K, Bian L, et al:
Epithelial stem cell mutations that promote squamous cell carcinoma
metastasis. J Clin Invest. 123:4390–4404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
275
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
276
|
Lin J, Teo S, Lam DH, Jeyaseelan K and
Wang S: MicroRNA-10b pleiotropically regulates invasion,
angiogenicity and apoptosis of tumor cells resembling mesenchymal
subtype of glioblastoma multiforme. Cell Death Dis. 3:e3982012.
View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Zhang WL, Zhang JH, Wu XZ, Yan T and Lv W:
miR-15b promotes epithelial-mesenchymal transition by inhibiting
SMURF2 in pancreatic cancer. Int J Oncol. 47:1043–1053.
2015.PubMed/NCBI
|
|
278
|
Wu Q, Yang Z, An Y, Hu H, Yin J, Zhang P,
Nie Y, Wu K, Shi Y and Fan D: MiR-19a/b modulate the metastasis of
gastric cancer cells by targeting the tumour suppressor MXD1. Cell
Death Dis. 5:e11442014. View Article : Google Scholar : PubMed/NCBI
|
|
279
|
Chang Y, Liu C, Yang J, Liu G, Feng F,
Tang J, Hu L, Li L, Jiang F, Chen C, et al: MiR-20a triggers
metastasis of gallbladder carcinoma. J Hepatol. 59:518–527. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Zhao S, Yao D, Chen J, Ding N and Ren F:
MiR-20a promotes cervical cancer proliferation and metastasis in
vitro and in vivo. PLoS One. 10:e01209052015. View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Dean ZS, Riahi R and Wong PK:
Spatiotemporal dynamics of microRNA during epithelial collective
cell migration. Biomaterials. 37:156–163. 2015. View Article : Google Scholar :
|
|
282
|
Peacock O, Lee AC, Cameron F, Tarbox R,
Vafadar-Isfahani N, Tufarelli C and Lund JN: Inflammation and
MiR-21 pathways functionally interact to downregulate PDCD4 in
colorectal cancer. PLoS One. 9:e1102672014. View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Xu J, Zhang W, Lv Q and Zhu D:
Overexpression of miR-21 promotes the proliferation and migration
of cervical cancer cells via the inhibition of PTEN. Oncol Rep.
33:3108–3116. 2015.PubMed/NCBI
|
|
284
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
285
|
Melnik BC: MiR-21: An environmental driver
of malignant melanoma? J Transl Med. 13:2022015. View Article : Google Scholar : PubMed/NCBI
|
|
286
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L,
Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al:
Cancer-secreted miR-105 destroys vascular endothelial barriers to
promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
287
|
Fong MY, Zhou W, Liu L, Alontaga AY,
Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, et al:
Breast-cancer-secreted miR-122 reprograms glucose metabolism in
premetastatic niche to promote metastasis. Nat Cell Biol.
17:183–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
288
|
Lin CW, Chang YL, Chang YC, Lin JC, Chen
CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, et al: MicroRNA-135b
promotes lung cancer metastasis by regulating multiple targets in
the Hippo pathway and LZTS1. Nat Commun. 4:18772013. View Article : Google Scholar : PubMed/NCBI
|
|
289
|
Taylor MA, Sossey-Alaoui K, Thompson CL,
Danielpour D and Schiemann WP: TGF-β upregulates miR-181a
expression to promote breast cancer metastasis. J Clin Invest.
123:150–163. 2013. View Article : Google Scholar
|
|
290
|
Qiu Y, Luo X, Kan T, Zhang Y, Yu W, Wei Y,
Shen N, Yi B and Jiang X: TGF-β upregulates miR-182 expression to
promote gallbladder cancer metastasis by targeting CADM1. Mol
Biosyst. 10:679–685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
291
|
Ren LH, Chen WX, Li S, He XY, Zhang ZM, Li
M, Cao RS, Hao B, Zhang HJ, Qiu HQ, et al: MicroRNA-183 promotes
proliferation and invasion in oesophageal squamous cell carcinoma
by targeting programmed cell death 4. Br J Cancer. 111:2003–2013.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
292
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
293
|
Korpal M, Ell BJ, Buffa FM, Ibrahim T,
Blanco MA, Celià-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua
Y, et al: Direct targeting of Sec23a by miR-200s influences cancer
cell secretome and promotes metastatic colonization. Nat Med.
17:1101–1108. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
294
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
295
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
296
|
Penna E, Orso F, Cimino D, Tenaglia E,
Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De
Pittà C, et al: microRNA-214 contributes to melanoma tumour
progression through suppression of TFAP2C. EMBO J. 30:1990–2007.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
297
|
Penna E, Orso F, Cimino D, Vercellino I,
Grassi E, Quaglino E, Turco E and Taverna D: miR-214 coordinates
melanoma progression by upregulating ALCAM through TFAP2 and
miR-148b downmodulation. Cancer Res. 73:4098–4111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
298
|
Long H, Wang Z, Chen J, Xiang T, Li Q,
Diao X and Zhu B: microRNA-214 promotes epithelial-mesenchymal
transition and metastasis in lung adenocarcinoma by targeting the
suppressor-of-fused protein (Sufu). Oncotarget. 6:38705–38718.
2015.PubMed/NCBI
|
|
299
|
Liu X, Chen Q, Yan J, Wang Y, Zhu C, Chen
C, Zhao X, Xu M, Sun Q, Deng R, et al: MiRNA-296-3p-ICAM-1 axis
promotes metastasis of prostate cancer by possible enhancing
survival of natural killer cell-resistant circulating tumour cells.
Cell Death Dis. 4:e9282013. View Article : Google Scholar : PubMed/NCBI
|
|
300
|
Vaira V, Faversani A, Martin NM, Garlick
DS, Ferrero S, Nosotti M, Kissil JL, Bosari S and Altieri DC:
Regulation of lung cancer metastasis by Klf4-Numb-like signaling.
Cancer Res. 73:2695–2705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
301
|
Ni F, Zhao H, Cui H, Wu Z, Chen L, Hu Z,
Guo C, Liu Y, Chen Z, Wang X, et al: MicroRNA-362-5p promotes tumor
growth and metastasis by targeting CYLD in hepatocellular
carcinoma. Cancer Lett. 356:809–818. 2015. View Article : Google Scholar
|
|
302
|
Chen D, Dang BL, Huang JZ, Chen M, Wu D,
Xu ML, Li R and Yan GR: MiR-373 drives the
epithelial-to-mesenchymal transition and metastasis via the
miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer.
Oncotarget. 6:32701–32712. 2015.PubMed/NCBI
|
|
303
|
Lu S, Zhu Q, Zhang Y, Song W, Wilson MJ
and Liu P: Dual-functions of miR-373 and miR-520c by differently
regulating the activities of MMP2 and MMP9. J Cell Physiol.
230:1862–1870. 2015. View Article : Google Scholar
|
|
304
|
Glover AR, Zhao JT, Gill AJ, Weiss J,
Mugridge N, Kim E, Feeney AL, Ip JC, Reid G, Clarke S, et al:
MicroRNA-7 as a tumor suppressor and novel therapeutic for
adrenocortical carcinoma. Oncotarget. 6:36675–36688.
2015.PubMed/NCBI
|
|
305
|
Babae N, Bourajjaj M, Liu Y, Van Beijnum
JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH,
Van Haastert RJ, et al: Systemic miRNA-7 delivery inhibits tumor
angiogenesis and growth in murine xenograft glioblastoma.
Oncotarget. 5:6687–6700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
306
|
Wang W, Dai LX, Zhang S, Yang Y, Yan N,
Fan P, Dai L, Tian HW, Cheng L, Zhang XM, et al: Regulation of
epidermal growth factor receptor signaling by plasmid-based
microRNA-7 inhibits human malignant gliomas growth and metastasis
in vivo. Neoplasma. 60:274–283. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
307
|
Cortez MA, Valdecanas D, Zhang X, Zhan Y,
Bhardwaj V, Calin GA, Komaki R, Giri DK, Quini CC, Wolfe T, et al:
Therapeutic delivery of miR-200c enhances radiosensitivity in lung
cancer. Mol Ther. 22:1494–1503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
308
|
Wu X, Liu T, Fang O, Dong W, Zhang F,
Leach L, Hu X and Luo Z: MicroRNA-708-5p acts as a therapeutic
agent against metastatic lung cancer. Oncotarget. 7:2417–2432.
2016.
|
|
309
|
Ge YF, Sun J, Jin CJ, Cao BQ, Jiang ZF and
Shao JF: AntagomiR-27a targets FOXO3a in glioblastoma and
suppresses U87 cell growth in vitro and in vivo. Asian Pac J Cancer
Prev. 14:963–968. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
310
|
Shu M, Zheng X, Wu S, Lu H, Leng T, Zhu W,
Zhou Y, Ou Y, Lin X, Lin Y, et al: Targeting oncogenic miR-335
inhibits growth and invasion of malignant astrocytoma cells. Mol
Cancer. 10:592011. View Article : Google Scholar : PubMed/NCBI
|
|
311
|
Rather MI, Nagashri MN, Swamy SS, Gopinath
KS and Kumar A: Oncogenic microRNA-155 down-regulates tumor
suppressor CDC73 and promotes oral squamous cell carcinoma cell
proliferation: Implications for cancer therapeutics. J Biol Chem.
288:608–618. 2013. View Article : Google Scholar :
|
|
312
|
Haug BH, Henriksen JR, Buechner J, Geerts
D, Tømte E, Kogner P, Martinsson T, Flægstad T, Sveinbjørnsson B
and Einvik C: MYCN-regulated miRNA-92 inhibits secretion of the
tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma.
Carcinogenesis. 32:1005–1012. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
313
|
Tang H, Liu X, Wang Z, She X, Zeng X, Deng
M, Liao Q, Guo X, Wang R, Li X, et al: Interaction of hsa-miR-381
and glioma suppressor LRRC4 is involved in glioma growth. Brain
Res. 1390:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
314
|
Ma L, Reinhardt F, Pan E, Soutschek J,
Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW and Weinberg RA:
Therapeutic silencing of miR-10b inhibits metastasis in a mouse
mammary tumor model. Nat Biotechnol. 28:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
315
|
Mercatelli N, Coppola V, Bonci D, Miele F,
Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria
R, et al: The inhibition of the highly expressed miR-221 and
miR-222 impairs the growth of prostate carcinoma xenografts in
mice. PLoS One. 3:e40292008. View Article : Google Scholar : PubMed/NCBI
|
|
316
|
Zhao Y, Zhao L, Ischenko I, Bao Q, Schwarz
B, Nieß H, Wang Y, Renner A, Mysliwietz J, Jauch KW, et al:
Antisense inhibition of microRNA-21 and microRNA-221 in
tumor-initiating stem-like cells modulates tumorigenesis,
metastasis, and chemotherapy resistance in pancreatic cancer.
Target Oncol. 10:535–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
317
|
Wagenaar TR, Zabludoff S, Ahn SM, Allerson
C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, et
al: Anti-miR-21 suppresses hepatocellular carcinoma growth via
broad transcriptional network deregulation. Mol Cancer Res.
13:1009–1021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
318
|
Fabani MM, Abreu-Goodger C, Williams D,
Lyons PA, Torres AG, Smith KG, Enright AJ, Gait MJ and Vigorito E:
Efficient inhibition of miR-155 function in vivo by peptide nucleic
acids. Nucleic Acids Res. 38:4466–4475. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
319
|
Brognara E, Fabbri E, Aimi F, Manicardi A,
Bianchi N, Finotti A, Breveglieri G, Borgatti M, Corradini R,
Marchelli R, et al: Peptide nucleic acids targeting miR-221
modulate p27Kip1 expression in breast cancer MDA-MB-231
cells. Int J Oncol. 41:2119–2127. 2012.PubMed/NCBI
|
|
320
|
Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ,
Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, et al: Knockdown of miR-21 in
human breast cancer cell lines inhibits proliferation, in vitro
migration and in vivo tumor growth. Breast Cancer Res. 13:R22011.
View Article : Google Scholar : PubMed/NCBI
|
|
321
|
Zhang J and Ma L: MicroRNA control of
epithelial-mesenchymal transition and metastasis. Cancer Metastasis
Rev. 31:653–662. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
322
|
Zaravinos A, Radojicic J, Lambrou GI,
Volanis D, Delakas D, Stathopoulos EN and Spandidos DA: Expression
of miRNAs involved in angiogenesis, tumor cell proliferation, tumor
suppressor inhibition, epithelial-mesenchymal transition and
activation of metastasis in bladder cancer. J Urol. 188:615–623.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
323
|
Kiesslich T, Pichler M and Neureiter D:
Epigenetic control of epithelial-mesenchymal-transition in human
cancer. Mol Clin Oncol. 1:3–11. 2013.PubMed/NCBI
|
|
324
|
Lei C, Wang Y, Huang Y, Yu H, Huang Y, Wu
L and Huang L: Up-regulated miR155 reverses the
epithelial-mesenchymal transition induced by EGF and increases
chemo-sensitivity to cisplatin in human Caski cervical cancer
cells. PLoS One. 7:e523102012. View Article : Google Scholar
|
|
325
|
Koutsaki M, Spandidos DA and Zaravinos A:
Epithelial-mesenchymal transition-associated miRNAs in ovarian
carcinoma, with highlight on the miR-200 family: Prognostic value
and prospective role in ovarian cancer therapeutics. Cancer Lett.
351:173–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
326
|
Gao H, Teng C, Huang W, Peng J and Wang C:
SOX2 promotes the epithelial to mesenchymal transition of
esophageal squamous cells by modulating Slug expression through the
activation of STAT3/HIF-α signaling.
|
|
327
|
Lambertini E, Lolli A, Vezzali F,
Penolazzi L, Gambari R and Piva R: Correlation between Slug
transcription factor and miR-221 in MDA-MB-231 breast cancer cells.
BMC Cancer. 12:4452012. View Article : Google Scholar : PubMed/NCBI
|
|
328
|
Qiu G, Lin Y, Zhang H and Wu D: miR-139-5p
inhibits epithelial-mesenchymal transition, migration and invasion
of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2.
Biochem Biophys Res Commun. 463:315–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
329
|
Bezzerri V, Borgatti M, Finotti A,
Tamanini A, Gambari R and Cabrini G: Mapping the transcriptional
machinery of the IL-8 gene in human bronchial epithelial cells. J
Immunol. 187:6069–6081. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
330
|
Raychaudhuri B and Vogelbaum MA: IL-8 is a
mediator of NF-κB induced invasion by gliomas. J Neurooncol.
101:227–235. 2011. View Article : Google Scholar
|
|
331
|
Xie TX, Xia Z, Zhang N, Gong W and Huang
S: Constitutive NF-κB activity regulates the expression of VEGF and
IL-8 and tumor angiogenesis of human glioblastoma. Oncol Rep.
23:725–732. 2010.PubMed/NCBI
|
|
332
|
Sun S, Wang Q, Giang A, Cheng C, Soo C,
Wang CY, Liau LM and Chiu R: Knockdown of CypA inhibits
interleukin-8 (IL-8) and IL-8-mediated proliferation and tumor
growth of glioblastoma cells through down-regulated NF-κB. J
Neurooncol. 101:1–14. 2011. View Article : Google Scholar
|
|
333
|
Gabellini C, Castellini L, Trisciuoglio D,
Kracht M, Zupi G and Del Bufalo D: Involvement of nuclear
factor-kappa B in bcl-xL-induced interleukin 8 expression in
glioblastoma. J Neurochem. 107:871–882. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
334
|
Yang TQ, Lu XJ, Wu TF, Ding DD, Zhao ZH,
Chen GL, Xie XS, Li B, Wei YX, Guo LC, et al: MicroRNA-16 inhibits
glioma cell growth and invasion through suppression of BCL2 and the
nuclear factor-κB1/MMP9 signaling pathway. Cancer Sci. 105:265–271.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
335
|
Fang L, Deng Z, Shatseva T, Yang J, Peng
C, Du WW, Yee AJ, Ang LC, He C, Shan SW, et al: MicroRNA miR-93
promotes tumor growth and angiogenesis by targeting integrin-β8.
Oncogene. 30:806–821. 2011. View Article : Google Scholar
|
|
336
|
Magge SN, Malik SZ, Royo NC, Chen HI, Yu
L, Snyder EY, O'Rourke DM and Watson DJ: Role of monocyte
chemoattractant protein-1 (MCP-1/CCL2) in migration of neural
progenitor cells toward glial tumors. J Neurosci Res. 87:1547–1555.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
337
|
Nazarenko I, Hede SM, He X, Hedrén A,
Thompson J, Lindström MS and Nistér M: PDGF and PDGF receptors in
glioma. Ups J Med Sci. 117:99–112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
338
|
Cai JJ, Qi ZX, Chen LC, Yao Y, Gong Y and
Mao Y: miR-124 suppresses the migration and invasion of glioma
cells in vitro via Capn4. Oncol Rep. 35:284–290. 2015.PubMed/NCBI
|
|
339
|
Cheng Y, Li Y, Nian Y, Liu D, Dai F and
Zhang J: STAT3 is involved in miR-124-mediated suppressive effects
on esophageal cancer cells. BMC Cancer. 15:3062015. View Article : Google Scholar : PubMed/NCBI
|
|
340
|
Dong LL, Chen LM, Wang WM and Zhang LM:
Decreased expression of microRNA-124 is an independent unfavorable
prognostic factor for patients with breast cancer. Diagn Pathol.
10:452015. View Article : Google Scholar : PubMed/NCBI
|
|
341
|
Long QZ, Du YF, Liu XG, Li X and He DL:
miR-124 represses FZD5 to attenuate P-glycoprotein-mediated
chemo-resistance in renal cell carcinoma. Tumour Biol.
36:7017–7026. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
342
|
Lu SH, Jiang XJ, Xiao GL, Liu DY and Yuan
XR: miR-124a restoration inhibits glioma cell proliferation and
invasion by suppressing IQGAP1 and β-catenin. Oncol Rep.
32:2104–2110. 2014.PubMed/NCBI
|
|
343
|
Chen SM, Chou WC, Hu LY, Hsiung CN, Chu
HW, Huang YL, Hsu HM, Yu JC and Shen CY: The Effect of MicroRNA-124
overexpression on anti-tumor drug sensitivity. PLoS One.
10:e01284722015. View Article : Google Scholar : PubMed/NCBI
|
|
344
|
Fabbri E, Brognara E, Montagner G,
Ghimenton C, Eccher A, Cantù C, Khalil S, Bezzerri V, Provezza L,
Bianchi N, et al: Regulation of IL-8 gene expression in gliomas by
microRNA miR-93. BMC Cancer. 15:6612015. View Article : Google Scholar : PubMed/NCBI
|
|
345
|
Fabbri E, Montagner G, Bianchi N, Finotti
A, Borgatti M, Lampronti I, Cabrini G and Gambari R: MicroRNA
miR-93-5p regulates expression of IL-8 and VEGF in neuroblastoma
SK-N-AS cells. Oncol Rep. 35:2866–2872. 2016.PubMed/NCBI
|
|
346
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J
Biol Chem. 282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
347
|
Lu X, Zhao P, Zhang C, Fu Z, Chen Y, Lu A,
Liu N, You Y, Pu P and Kang C: Analysis of miR-221 and p27
expression in human gliomas. Mol Med Rep. 2:651–656.
2009.PubMed/NCBI
|
|
348
|
Gillies JK and Lorimer IA: Regulation of
p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle.
6:2005–2009. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
349
|
Wang Y, Wang X, Zhang J, Sun G, Luo H,
Kang C, Pu P, Jiang T, Liu N and You Y: MicroRNAs involved in the
EGFR/PTEN/AKT pathway in gliomas. J Neurooncol. 106:217–224. 2012.
View Article : Google Scholar
|
|
350
|
Ueda R, Kohanbash G, Sasaki K, Fujita M,
Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze
MT, et al: Dicer-regulated microRNAs 222 and 339 promote resistance
of cancer cells to cytotoxic T-lymphocytes by down-regulation of
ICAM-1. Proc Natl Acad Sci USA. 106:10746–10751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
351
|
Zhang J, Han L, Ge Y, Zhou X, Zhang A,
Zhang C, Zhong Y, You Y, Pu P and Kang C: miR-221/222 promote
malignant progression of glioma through activation of the Akt
pathway. Int J Oncol. 36:913–920. 2010.PubMed/NCBI
|
|
352
|
Zhang C, Jiang T, Wang J, Cheng J, Pu P
and Kang C: MiR-221/222 promote the growth of malignant glioma
cells by regulating its target genes, molecular targets of CNS
tumors. Dr Garami Miklos : ISBN: 978-953-307-736-9InTech; pp.
461–482. 2011
|
|
353
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer
Res. 3:465–477. 2013.PubMed/NCBI
|
|
354
|
Zhang C, Kang C, You Y, Pu P, Yang W, Zhao
P, Wang G, Zhang A, Jia Z, Han L, et al: Co-suppression of
miR-221/222 cluster suppresses human glioma cell growth by
targeting p27Kip1 in vitro and in vivo. Int J Oncol.
34:1653–1660. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
355
|
Zhang R, Pang B, Xin T, Guo H, Xing Y, Xu
S, Feng B, Liu B and Pang Q: Plasma miR-221/222 family as novel
descriptive and prognostic biomarkers for glioma. Mol Neurobiol.
53:1452–1460. 2016. View Article : Google Scholar
|
|
356
|
Yang Y, Li F, Saha MN, Abdi J, Qiu L and
Chang H: miR-137/197 induce apoptosis and suppress tumorigenicity
by targeting MCL-1 in multiple myeloma. Clin Cancer Res.
21:2399–2411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
357
|
Lee SH, Jung YD, Choi YS and Lee YM:
Targeting of RUNX3 by miR-130a and miR-495 cooperatively increases
cell proliferation and tumor angiogenesis in gastric cancer cells.
Oncotarget. 6:33269–33278. 2015.PubMed/NCBI
|
|
358
|
Brognara E, Fabbri E, Montagner G,
Gasparello J, Manicardi A, Corradini R, Bianchi N, Finotti A,
Breveglieri G, Borgatti M, et al: High levels of apoptosis are
induced in human glioma cell lines by co-administration of peptide
nucleic acids targeting miR-221 and miR-222. Int J Oncol.
48:1029–1038. 2016.
|
|
359
|
Giunti L, da Ros M, Vinci S, Gelmini S,
Iorio AL, Buccoliero AM, Cardellicchio S, Castiglione F, Genitori
L, de Martino M, et al: Anti-miR21 oligonucleotide enhances
chemosensitivity of T98G cell line to doxorubicin by inducing
apoptosis. Am J Cancer Res. 5:231–242. 2014.
|
|
360
|
Gao C, Peng FH and Peng LK: MiR-200c
sensitizes clear-cell renal cell carcinoma cells to sorafenib and
imatinib by targeting heme oxygenase-1. Neoplasma. 61:680–689.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
361
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
362
|
Suto T, Yokobori T, Yajima R, Morita H,
Fujii T, Yamaguchi S, Altan B, Tsutsumi S, Asao T and Kuwano H:
MicroRNA-7 expression in colorectal cancer is associated with poor
prognosis and regulates cetuximab sensitivity via EGFR regulation.
Carcinogenesis. 36:338–345. 2015. View Article : Google Scholar
|
|
363
|
Liu R, Liu X, Zheng Y, Gu J, Xiong S,
Jiang P, Jiang X, Huang E, Yang Y, Ge D, et al: MicroRNA-7
sensitizes non-small cell lung cancer cells to paclitaxel. Oncol
Lett. 8:2193–2200. 2014.PubMed/NCBI
|
|
364
|
Gomes SE, Simões AE, Pereira DM, Castro
RE, Rodrigues CM and Borralho PM: miR-143 or miR-145 overexpression
increases cetuximab-mediated antibody-dependent cellular
cytotoxicity in human colon cancer cells. Oncotarget. Jan
25–2016.(Epub ahead of print).
|
|
365
|
Costa PM, Cardoso AL, Nóbrega C, Pereira
de Almeida LF, Bruce JN, Canoll P and Pedroso de Lima MC:
MicroRNA-21 silencing enhances the cytotoxic effect of the
antiangiogenic drug sunitinib in glioblastoma. Hum Mol Genet.
22:904–918. 2013. View Article : Google Scholar :
|
|
366
|
Qian X, Ren Y, Shi Z, Long L, Pu P, Sheng
J, Yuan X and Kang C: Sequence-dependent synergistic inhibition of
human glioma cell lines by combined temozolomide and miR-21
inhibitor gene therapy. Mol Pharm. 9:2636–2645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
367
|
Zhang S, Han L, Wei J, Shi Z, Pu P, Zhang
J, Yuan X and Kang C: Combination treatment with doxorubicin and
microRNA-21 inhibitor synergistically augments anticancer activity
through upregulation of tumor suppressing genes. Int J Oncol.
46:1589–1600. 2015.PubMed/NCBI
|
|
368
|
Zhang Q, Ran R, Zhang L, Liu Y, Mei L,
Zhang Z, Gao H and He Q: Simultaneous delivery of therapeutic
antagomirs with paclitaxel for the management of metastatic tumors
by a pH-responsive anti-microbial peptide-mediated liposomal
delivery system. J Control Release. 197:208–218. 2015. View Article : Google Scholar
|
|
369
|
Fan L, Yang Q, Tan J, Qiao Y, Wang Q, He
J, Wu H and Zhang Y: Dual loading miR-218 mimics and Temozolomide
using AuCOOH@FA-CS drug delivery system: Promising
targeted anti-tumor drug delivery system with sequential release
functions. J Exp Clin Cancer Res. 34:1062015. View Article : Google Scholar
|
|
370
|
Xue W, Dahlman JE, Tammela T, Khan OF,
Sood S, Dave A, Cai W, Chirino LM, Yang GR, Bronson R, et al: Small
RNA combination therapy for lung cancer. Proc Natl Acad Sci USA.
111:E3553–E3561. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
371
|
Nishimura M, Jung EJ, Shah MY, Lu C,
Spizzo R, Shimizu M, Han HD, Ivan C, Rossi S, Zhang X, et al:
Therapeutic synergy between microRNA and siRNA in ovarian cancer
treatment. Cancer Discov. 3:1302–1315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
372
|
Hu X, Li W, Liu G, Wu H, Gao Y, Chen S, He
D and Zhang Y: The effect of Bcl-2 siRNA combined with miR-15a
oligonucleotides on the growth of Raji cells. Med Oncol.
30:4302013. View Article : Google Scholar : PubMed/NCBI
|