Introduction
Lung cancer is globally the leading cause of
cancer-related deaths (1).
Non-small cell lung cancer (NSCLC), which accounts for more than
80% of all cases, is divided in adenocarcinoma (ADC), squamous cell
carcinoma (SCC) and large cell carcinoma (LCLC). The 5-year
survival rate of lung cancer with 15% is still poor (2). A deregulated gene and protein
expression of mitosis associated factors to force chromosomal
segregation during the cell division is frequently observed in
cancer. The analysis of the molecular landscape revealed a high
mutation rate of cell cycle genes in ADC (64%) (3). Furthermore, it was shown that the
proliferation rate of NSCLC is predictive for the survival of the
patients (4). A high expression of
Ki-67 resulted in a poor prognosis in patients with ADC while it
was associated with better prognosis in SCC patients (5). Cell cycle associated proteins
including components that contribute to the execution of mitosis
are indispensable for the proliferation of tumor cells. During
mitosis, the kinetochore properly attaches to the chromosomes in
microtubules to ensure correct segregation (6). The formation of the bipolar spindle
and the segregation process are coordinated by the Aurora A kinase
(AURKA) (7). AURKA promotes the
entry into mitosis under the control of cyclin B/CDK1 and regulates
the progression of mitosis by phosphorylation of multiple targets
(8–10). Several cofactors influence the
activity of the AURKA during this process. The targeting protein
for Xklp2 (TPX2) is one of these AURKA cofactors. During mitosis,
TPX2 activity is controlled by the importin α/β-Ran system
(11). TPX2 causes AURKA to adopt
an active conformation and protects it from dephosphorylation
(12). A direct target that is
phosphorylated by the AURKA is the disks large-associated protein 5
(DLGAP5) (13). DLGAP5 is a
mitotic spindle protein that promotes the formation of tubulin
polymers resulting in tubulin sheets around the end of the
microtubules (14). The
association of DLGAP5 with the mitotic spindle is stabilized
through the phosphorylation by AURKA (15). The kinesin family member 11 (KIF11)
is a motor protein of the kinesin-5-family which cross-links
antiparallel microtubules in the spindle (16). Its movement is regulated by TPX2
binding (17). The cytoskeleton
associated protein 5 (CKAP5) protects the kinetochore microtubules
from depolymerization and plays a role in the assembly of central
microtubules (18,19). A protein complex named EXATH
(comprising AURKA, TPX2 and DLGAP5, KIF11 and CKAP5) which was
described in Xenopus egg extract, was shown to exhibit
MT-stabilizing and organizing activities, and to contribute to the
maturation and formation of the mitotic spindle (20). Current studies primarily targeted
AURKA. In vitro studies with NSCLC cell lines indicated that
the AURKA inhibitor MK-5108 promises to be a potent drug in
combination with docetaxel (21).
However, clinical trials with the AURKA inhibitors VX-680 and
AT9283 had to be terminated because of severe toxicities in
patients with chronic myelogenous leukemia (22). Furthermore, the AURKA inhibitor
alisertib displayed a response rate of only 9% in a phase I/II
study (23). A knockdown of KIF11
with siRNA as well as its inhibition with ispinesib resulted in a
strong decrease of the viability of NSCLC cells (24). Therefore, we investigated in this
study the expression of AURKA, DLGAP5, TPX2, KIF11 and CKAP5 in a
large cohort of adeno- and squamous cell carcinoma patients (n=362)
to evaluate their prognostic significance and the potential as
therapeutic targets for NSCLC.
Materials and methods
Tissue sample collection,
characterization and preparation
Tissue samples were provided by the Lung Biobank
Heidelberg, a member of the accredited Tissue Bank of the National
Center for Tumor Diseases (NCT) Heidelberg, the BioMaterialBank
Heidelberg, and the Biobank platform of the German Center for Lung
Research (DZL). Tumor and matched distant (>5 cm) normal lung
tissue samples from NSCLC patients (n=362) who underwent resection
for primary lung cancer at the Thoraxklinik at University Hospital,
Heidelberg, Germany, were collected. All diagnoses were made
according to the 2004 WHO classification for lung cancer by at
least two experienced pathologists (25). Tumor stage was designated according
to the 7th edition of the UICC tumor, node and metastasis (26). Tissues were snap-frozen within 30
min after resection and stored at −80°C until the time of analysis.
For nucleic acid isolation 10–15 tumor cryosections (10–15
µm each) were prepared for each patient. The first and the
last sections in each series were stained with hematoxylin and
eosin (H&E) and were reviewed by an experienced lung
pathologist to determine the proportions of viable tumor cells,
stromal cells, normal lung cell cells, infiltrating lymphocytes and
necrotic areas. Only samples with a viable tumor content of ≥50%
were used for subsequent analyses.
Total RNA isolation and cDNA
synthesis
Total RNA was isolated from tissue using an RNeasy
Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's
instructions. For RNA isolation from cell lines, the AllPrep
DNA/RNA Mini kit (Qiagen) was used. Total RNA was transcribed to
sscDNA with a Transcriptor First Strand cDNA Synthesis kit (Roche
Diagnostics GmbH, Mannheim, Germany). More detailed information is
described elsewhere (27).
Quantitative real-time PCR (qPCR)
Real-time quantitative PCR (qPCR) was performed in
accordance with the MIQE-guidelines (28) using a LightCycler® 480
real-time PCR instrument in a 384-well plate format (Roche
Diagnostics). Gene-specific primers and probes (Universal
ProbeLibrary; Roche Diagnostics) were used in combination with the
ABsolute™ qPCR Mix (Fisher Scientific GmbH, Schwerte, Germany). CT
values were calculated with the LightCycler® 480
software version 1.5 using the 2nd derivative maximum method (Roche
Diagnostics). Detailed information is described elsewhere (27).
Statistical analyses
Data of qPCR analyses were statistically analyzed
using REMARK criteria (29) with
the SPSS 22.0 for Windows (IBM, Ehningen, Germany). The primary
endpoint of the study was overall survival. Overall survival time
was calculated from the date of surgery until the last date of
contact or death. Univariate analysis of survival data was
performed according to Kaplan-Meier (30) and using the Cox proportional
hazards model. The log-rank test was used to test the significance
between the groups. A P-value of <0.05 was considered
significant. Multivariate survival analysis was performed using the
Cox proportional hazards model. The non-parametric Mann-Whitney U
test (31) as well as the
Kruskal-Wallis test (32) were
used to investigate significant differences between the patient
groups. The Spearman's rank correlation coefficient test was
performed for correlation analyses (33). Visualization of the qPCR data was
performed using the GraphPad Prism 5.
Cell culture
H838 and H1975 cell lines were purchased from the
American Type Culture Collection (ATCC; Wesel, Germany) and
cultivated in DMEM/Ham's F-12 (Life Technologies, Darmstadt,
Germany) with 10% fetal calf serum (FCS; GE Healthcare Life
Sciences, Freiburg, Germany) for not more than 20 passages. H838
cell line was cultivated in RPMI-1640 (Life Technologies) with 10%
FCS.
Immunofluorescence analyses (IF) and
immunohistochemistry (IHC)
For IF, 1.5×105 H1975 or 2×105
H838 were seeded onto a 12-well plate. The next day, cells were
fixed with ice cold methanol for 10 min at −20°C and incubated with
the indicated antibodies overnight at 4°C. Anti-AURKA (#4718; Cell
Signaling Technology, Danvers, MA, USA), anti-TPX2 (NB500-179;
Acris Antibodies GmbH, Herford, Germany), anti-DLGAP5 (sc-68540;
Santa Cruz Biotechnology, Heidelberg, Germany), anti-KIF11
(sc-365681; Santa Cruz Biotechnology) and anti-CKAP5 (sc-240235;
Santa Cruz Biotechnology) antibodies were used to stain the spindle
associated proteins. Cells were incubated with secondary antibodies
(Alexa Fluor dyes; Dianova GmbH, Hamburg, Germany) for 40 min at
37°C. Hoechst 33342 (Sigma-Aldrich, Munich, Germany) was used for
nuclear staining. Cells were covered with coverslips using Fluoprep
(75521; Biomérieux Deutschland GmbH, Nuertingen, Germany).
Pictures were taken with an Olympus IX71 inverted
microscope. Staining was observed with Olympus ColorView II digital
camera and with the Olympus cellSens software (Olympus). Tiffs were
assembled into figures using Photoshop CS6 (Adobe Photoshop).
Results
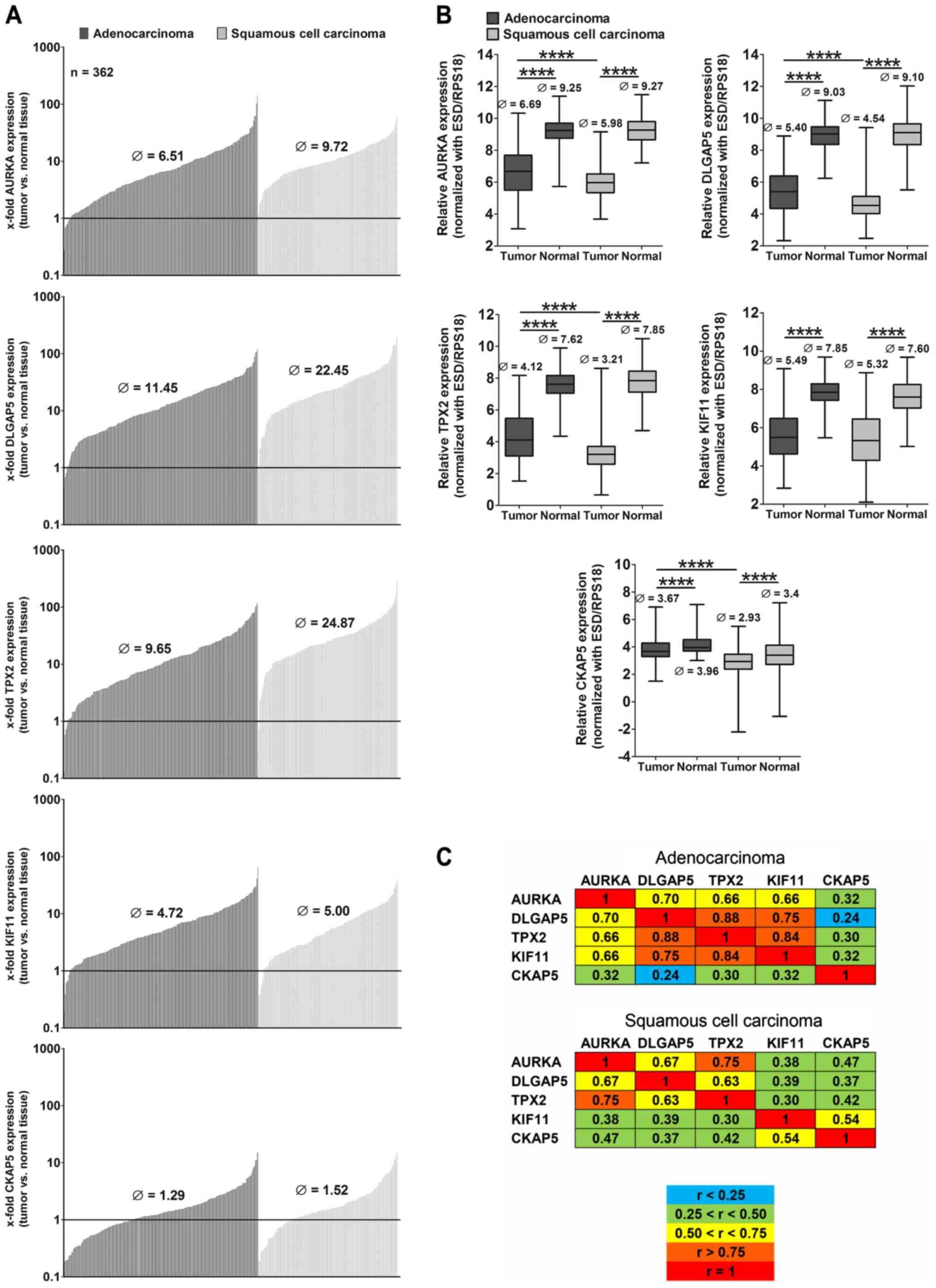
AURKA, DLGAP5, TPX2, KIF11 and CKAP5 are
highly overexpressed in NSCLC
To investigate the role of the above five
mitosis-associated genes in NSCLC, we analyzed the gene expression
in tumor and corresponding normal lung tissue (n=362) of patients
who underwent surgery (Table I) by
qPCR. AURKA, DLGAP5, TPX2, KIF11 were highly overexpressed in
>96% of all tumors compared to normal lung tissue, CKAP5 was
overexpressed in ~70% of the patients, but to a lower level than
the other four genes (Fig. 1A). In
general, the overexpression of the genes was much higher in
squamous cell carcinomas (SCC) compared to adenocarcinomas (ADC)
and differed significantly from gene expression in normal tissue
(Fig. 1B). In SCC we observed a
higher expression in advanced stages compared to stage I with the
exception of KIF11 which was lower expressed in stage III compared
to stage I (data not shown). No significant differences were
observed in the gene expression of tumors between males and females
(data not shown). Since the five proteins were described to
interact and to form a complex during cell mitosis (20), we were interested whether we could
find any correlation between the expression of the genes. The
correlation of the expression of the genes differed with respect to
the different histologies (Fig.
1C). In ADC, AURKA, DLGAP5, TPX2 and KIF11 all showed
correlation values r>0.66. Only CKAP5 expression did not exhibit
significant associations with the other genes. In SCC, CKAP5
slightly correlated with KIF11 (r=0.54) but only AURKA and TPX2
displayed a high correlation in the tumor tissue (r=0.75).
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Parameters | Gene expression
analyses
|
|---|
| n (%) |
|---|
| Median age
(years) | 65 (38–88) |
| Gender | 362 |
| Male | 250 (69) |
| Female | 112 (31) |
| Histology |
|
Adenocarcinoma | 211 (58) |
| Squamous cell
carcinoma | 151 (42) |
| Therapy |
| OP | 212 (59) |
| OP/RT | 13 (4) |
| OP/CT | 100 (28) |
| OP/RT/CT | 37 (10) |
| P-stage |
| IA | 37 (10) |
| IB | 90 (25) |
| IIA | 70 (19) |
| IIB | 51 (14) |
| IIIA | 105 (29) |
| IIIB | 9 (2) |
| ECOG |
| 0 | 320 (88) |
| 1 | 32 (9) |
| 2 | 4 (1) |
| n.d. | 8 (2) |
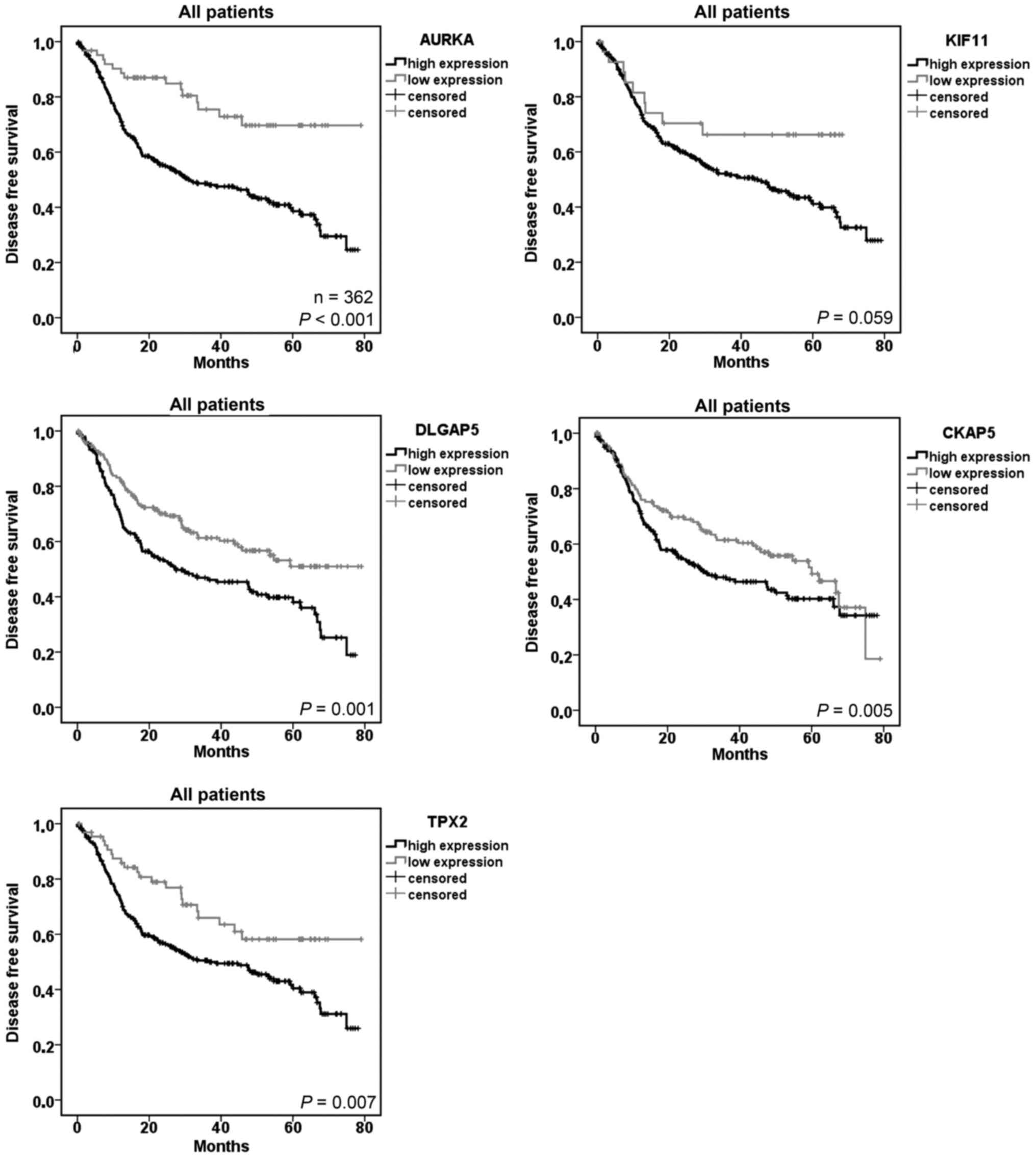
AURKA, DLGAP5, TPX2, KIF11 and CKAP5 are
factors for poor disease-free survival in NSCLC patients
In survival analyses, we studied the association
between the increased gene expression and the survival of the
patients. We used the optimal cut-off value calculated by an
R-based tool (34) to separate the
groups with high and low gene expression in the tumor tissue. In
univariate analyses we found significantly increased hazard ratios
and poor disease-free survival in patients with a high gene
expression of AURKA (HR=2.813, P≤0.001), DLGAP5 (HR=1.669,
P=0.001), TPX2 (HR=1.826, P=0.007) and CKAP5 (P=0.05) (Table II and Fig. 2). In multivariate analyses using
clinical parameters and the AURKA expression, the histology, the
pathological stage (III vs. I) as well as the age were prognostic
factors for disease-free survival. Considering the subhistologies,
a high gene expression of the five mitosis-associated genes
resulted in a poor prognosis of patients with ADC, but not with
SCC. KIF11 barely failed to be a prognostic marker in univariate
analyses.
 | Table IIStatistical analyses (Cox-regression)
of relative tumor expression. |
Table II
Statistical analyses (Cox-regression)
of relative tumor expression.
| Variables | Hazard
ratio
(95% CI) | P-value |
|---|
| Univariate analyses
of disease-free survival |
| AURKA (high vs.
low) | 2.813
(1.656–4.779) |
<0.001 |
| DLGAP5 | 1.669
(1.223–2.276) | 0.001 |
| TPX2 | 1.826
(1.167–2.857) | 0.007 |
| KIF11 | 1.891
(0.966–3.705) | 0.063 |
| CKAP5 | 1.364
(0.998–1.865) | 0.05 |
| Multivariate
analyses of disease-free survival |
| AURKA (high vs.
low) | 2.486
(1.298–4.761) | 0.006 |
| DLGAP5 | 1.338
(0.928–1.928) | 0.119 |
| TPX2 | 0.86
(0.475–1.555) | 0.617 |
| CKAP5 | 1.141
(0.815–1.598) | 0.441 |
| Histology (SCC vs.
ADC) | 0.642
(0.455–0.906) | 0.012 |
| Gender (male vs.
female) | 1.216
(0.858–1.723) | 0.272 |
| pStage (II vs.
I) | 1.468
(0.968–2.226) | 0.071 |
| pStage (III vs.
I) | 2.562
(1.715–3.829) |
<0.001 |
| Age | 1.018
(1.001–1.036) | 0.034 |
| Univariate analyses
of disease-free survival in ADC |
| AURKA (high vs.
low) | 3.498
(1.985–6.164) |
<0.001 |
| DLGAP5 | 2.011
(1.356–2.984) |
<0.001 |
| TPX2 | 2.226
(1.374–3.605) | 0.001 |
| KIF11 | 2.855
(1.246–6.541) | 0.013 |
| CKAP5 | 1.533
(1.031–2.279) | 0.033 |
| Univariate analyses
of disease-free survival in SCC |
| AURKA (high vs.
low) | 0.753
(0.104–5.435) | 0.777 |
| DLGAP5 | 1.293
(0.591–2.827) | 0.519 |
| TPX2 | 1.368
(0.68–2.750) | 0.377 |
| KIF11 | 1.460
(0.904–2.357) | 0.12 |
| CKAP5 | 0.735
(0.403–1.338) | 0.312 |
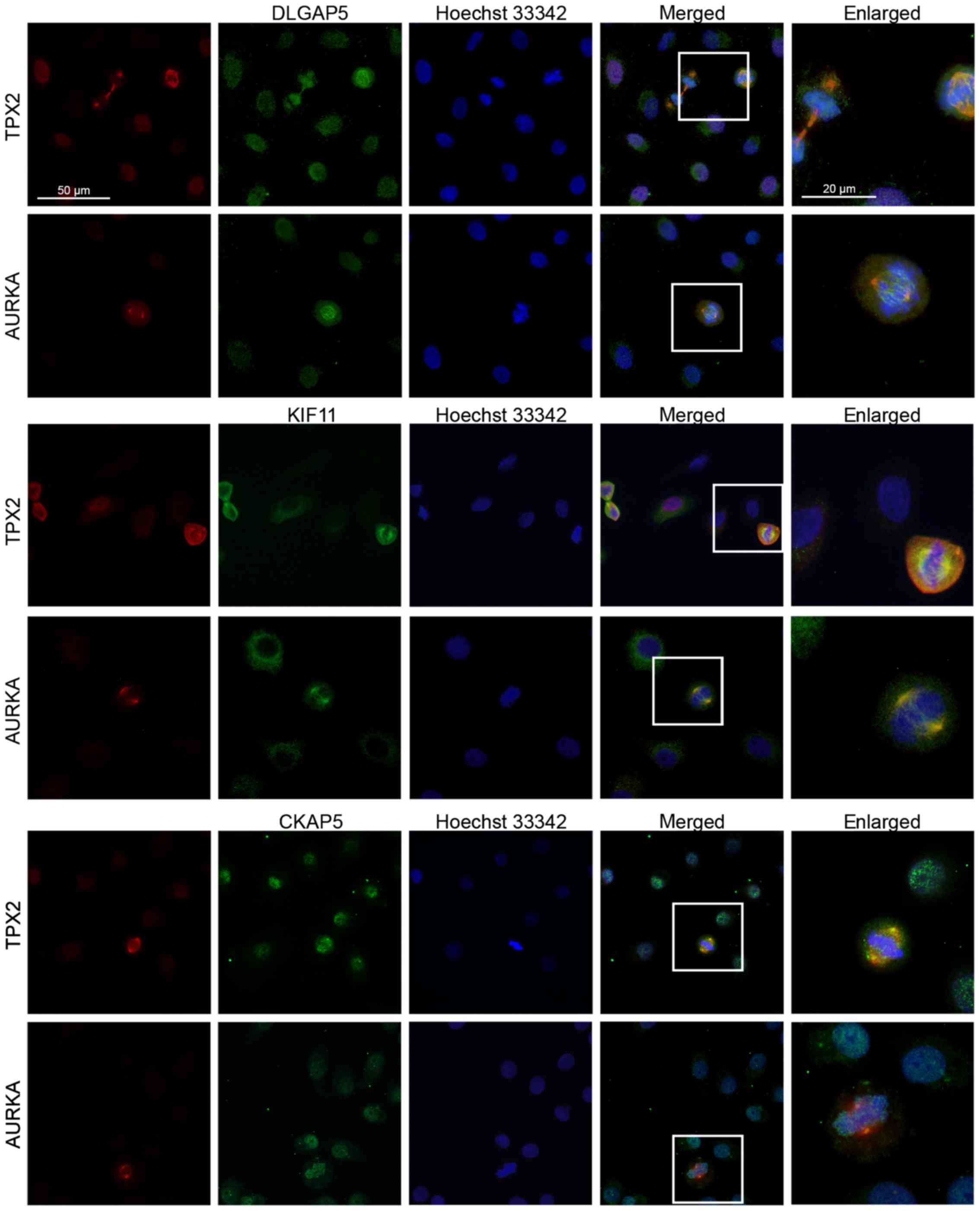
DLGAP5, KIF11 and CKAP5 co-localize with
TPX2 and AURKA at the spindle apparatus during cell division
To clarify whether the five proteins might also form
a complex in lung cancer cells, we performed immunofluorescence
analyses and co-stainings of DLGAP5, KIF11 and CKAP5 with AURKA and
TPX2 in the the lung cancer cell line H838 (Fig. 3). In non-dividing cells, AURKA,
TPX2, DLGAP5 and CKAP5 were localized in the cytoplasm and the
nucleus (Fig. 3, first two
columns). In contrast, KIF11 was mainly expressed in the cytoplasm
of the cells. In cells undergoing mitosis, DLGAP5, KIF11 and CKAP5
co-localized with AURKA and TPX2 at the spindle apparatus and the
microtubules (last two columns). While the AURKA was mainly
localized at the centrosomes (Fig.
3, rows 2, 4 and 6), DLGAP5, KIF11 and TPX2 covered the entire
microtubules. CKAP5 exhibited a more distributed pattern and
co-localized with AURKA and TPX2 only at the centrosomes (rows 5
and 6). Similar results were obtained for the cell line H1975 (data
not shown).
Discussion
In the present study, we demonstrated that the five
cell cycle associated genes AURKA, DLGAP5, TPX2, KIF11 and CKAP5
were overexpressed in NSCLC tumor tissue compared to corresponding
adjacent non-neoplastic lung tissue. Especially AURKA, TPX2 and
DLGAP5 were found to be highly upregulated and correlated with each
other. It was shown that during the cell cycle TPX2 and DLGAP5 are
phosphorylated by AURKA (15,35).
Therefore, a coexpression of AURKA with TPX2 and DLGAP5 seems
reasonable. While KIF11 correlated with these three genes in ADC,
this was not the case in SCC. CKAP5 did not correlate with the
expression of any of the other genes. The five investigated
proteins were shown to form a complex in Xenopus egg extract
(20). Here, the survival analyses
of the qPCR expression data indicated that for NSCLC this complex
might only be prognostically relevant for patients with ADC.
Furthermore, AURKA was the only gene which was prognostic for the
disease-free survival in multivariate analysis. This is in
agreement with the cellular role of AURKA since it was demonstrated
that AURKA phosphorylates important cell cycle proteins including
DLGAP5 (13,22). With immunofluorescence analyses, we
demonstrated that the proteins DLGAP5 and KIF11 co-localized with
AURKA and TPX2 in lung cancer cells during cell mitosis. The
expression pattern of CKAP5 correlated only partly with the
expression pattern of AURKA and TPX2. Neither in ADC nor SCC a
correlation between CKAP5 and the other two genes was observed.
With regard to the potential of the investigated
genes as therapeutic targets for NSCLC, screening studies
frequently discovered some of these genes to be essential for tumor
survival in lung cancer (24,36,37).
A cell cycle progression score (CCP) including DLGAP5 and KIF11 was
shown to be prognostic for different cancer entities (38,39).
Concerning lung cancer, this CCP was prognostic mainly for patients
with ADC (38,40). An extension of the score using TPX2
and AURKA might improve the prognostic value of the CCP score.
In conclusion, in the present study, we demonstrated
for the first time the co-expression of DLGAP5, KIF11 and CKAP5 in
NSCLC patients together with TPX2 and AURKA. All five genes were
prognostic markers for the survival of the patients. For TPX2,
DLGAP5 and CKAP5 so far no in vitro or in vivo
studies concerning drug application have been described. Since our
data indicated that there is a strong correlation between AURKA,
TPX2 and DLGAP5 expression in NSCLC patients, these three genes
might be particularly suitable targets for future therapeutic
studies.
Acknowledgments
We would like to thank Chang Xu, Martin
Fallenbuechel, Jessica Eschenbach, Carmen Hoppstock, Christa Stolp
and Andrea Bopp for their expert technical assistance.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Youlden DR, Cramb SM and Baade PD: The
International Epidemiology of Lung Cancer: geographical
distribution and secular trends. J Thorac Oncol. 3:819–831. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ding L, Getz G, Wheeler DA, Mardis ER,
McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan
MB, et al: Somatic mutations affect key pathways in lung
adenocarcinoma. Nature. 455:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sofocleous CT, Garg SK, Cohen P, Petre EN,
Gonen M, Erinjeri JP, Downey RJ, Travis WD and Solomon SB: Ki-67 is
an independent predictive biomarker of cancer specific and local
recurrence-free survival after lung tumor ablation. Ann Surg Oncol.
20(Suppl 3): S676–S683. 2013. View Article : Google Scholar
|
|
5
|
Warth A, Cortis J, Soltermann A, Meister
M, Budczies J, Stenzinger A, Goeppert B, Thomas M, Herth FJ,
Schirmacher P, et al: Tumour cell proliferation (Ki-67) in
non-small cell lung cancer: A critical reappraisal of its
prognostic role. Br J Cancer. 111:1222–1229. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pinsky BA and Biggins S: The spindle
checkpoint: Tension versus attachment. Trends Cell Biol.
15:486–493. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hannak E, Kirkham M, Hyman AA and Oegema
K: Aurora-A kinase is required for centrosome maturation in
Caenorhabditis elegans. J Cell Biol. 155:1109–1116. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Satinover DL, Brautigan DL and Stukenberg
PT: Aurora-A kinase and inhibitor-2 regulate the cyclin threshold
for mitotic entry in Xenopus early embryonic cell cycles. Cell
Cycle. 5:2268–2274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Macůrek L, Lindqvist A, Lim D, Lampson MA,
Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB and Medema
RH: Polo-like kinase-1 is activated by aurora A to promote
checkpoint recovery. Nature. 455:119–123. 2008. View Article : Google Scholar
|
|
10
|
Mori D, Yano Y, Toyo-oka K, Yoshida N,
Yamada M, Muramatsu M, Zhang D, Saya H, Toyoshima YY, Kinoshita K,
et al: NDEL1 phosphorylation by Aurora-A kinase is essential for
centrosomal maturation, separation, and TACC3 recruitment. Mol Cell
Biol. 27:352–367. 2007. View Article : Google Scholar :
|
|
11
|
Di Fiore B, Ciciarello M and Lavia P:
Mitotic functions of the Ran GTPase network: The importance of
being in the right place at the right time. Cell Cycle. 3:305–313.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bayliss R, Sardon T, Vernos I and Conti E:
Structural basis of Aurora-A activation by TPX2 at the mitotic
spindle. Mol Cell. 12:851–862. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wong J, Lerrigo R, Jang CY and Fang G:
Aurora A regulates the activity of HURP by controlling the
accessibility of its microtubule-binding domain. Mol Biol Cell.
19:2083–2091. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Santarella RA, Koffa MD, Tittmann P, Gross
H and Hoenger A: HURP wraps microtubule ends with an additional
tubulin sheet that has a novel conformation of tubulin. J Mol Biol.
365:1587–1595. 2007. View Article : Google Scholar
|
|
15
|
Yu CT, Hsu JM, Lee YC, Tsou AP, Chou CK
and Huang CY: Phosphorylation and stabilization of HURP by
Aurora-A: Implication of HURP as a transforming target of Aurora-A.
Mol Cell Biol. 25:5789–5800. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Blangy A, Lane HA, d'Hérin P, Harper M,
Kress M and Nigg EA: Phosphorylation by p34cdc2 regulates spindle
association of human Eg5, a kinesin-related motor essential for
bipolar spindle formation in vivo. Cell. 83:1159–1169. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma N, Tulu US, Ferenz NP, Fagerstrom C,
Wilde A and Wadsworth P: Poleward transport of TPX2 in the
mammalian mitotic spindle requires dynein, Eg5, and microtubule
flux. Mol Biol Cell. 21:979–988. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cassimeris L and Morabito J: TOGp, the
human homolog of XMAP215/Dis1, is required for centrosome
integrity, spindle pole organization, and bipolar spindle assembly.
Mol Biol Cell. 15:1580–1590. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barr AR and Gergely F: MCAK-independent
functions of ch-Tog/XMAP215 in microtubule plus-end dynamics. Mol
Cell Biol. 28:7199–7211. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Koffa MD, Casanova CM, Santarella R,
Köcher T, Wilm M and Mattaj IW: HURP is part of a Ran-dependent
complex involved in spindle formation. Curr Biol. 16:743–754. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chinn DC, Holland WS and Mack PC:
Anticancer activity of the Aurora A kinase inhibitor MK-5108 in
non-small-cell lung cancer (NSCLC) in vitro as monotherapy and in
combination with chemotherapies. J Cancer Res Clin Oncol.
140:1137–1149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goldenson B and Crispino JD: The aurora
kinases in cell cycle and leukemia. Oncogene. 34:537–545. 2015.
View Article : Google Scholar
|
|
23
|
Melichar B, Adenis A, Lockhart AC,
Bennouna J, Dees EC, Kayaleh O, Obermannova R, DeMichele A,
Zatloukal P, Zhang B, et al: Safety and activity of alisertib, an
investigational aurora kinase A inhibitor, in patients with breast
cancer, small-cell lung cancer, non-small-cell lung cancer, head
and neck squamous-cell carcinoma, and gastro-oesophageal
adenocarcinoma: A five-arm phase 2 study. Lancet Oncol. 16:395–405.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martens-de Kemp SR1, Nagel R, Stigter-van
Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ and
Brakenhoff RH: Functional genetic screens identify genes essential
for tumor cell survival in head and neck and lung cancer. Clin
Cancer Res. 19:1994–2003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beasley MB, Brambilla E and Travis WD: The
2004 World Health Organization classification of lung tumors. Semin
Roentgenol. 40:90–97. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wittekind C: 2010 TNM system: On the 7th
edition of TNM classification of malignant tumors. Pathologe.
31:331–332. 2010.In German. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schneider MA, Granzow M, Warth A, Schnabel
PA, Thomas M, Herth FJ, Dienemann H, Muley T and Meister M:
Glycodelin: A new biomarker with immunomodulatory functions in
non-small cell lung cancer. Clin Cancer Res. 21:3529–3540. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McShane LM, Altman DG, Sauerbrei W, Taube
SE, Gion M and Clark GM: Statistics Subcommittee of the NCI-EORTC
Working Group on Cancer Diagnostics REporting recommendations for
tumour MARKer prognostic studies (REMARK). Eur J Cancer.
41:1690–1696. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dinse GE and Lagakos SW: Nonparametric
estimation of lifetime and disease onset distributions from
incomplete observations. Biometrics. 38:921–932. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mann HB and Whitney DR: On a test whether
one of two random variables is stochastically larger than the
other. Ann Math Stat. 18:50–60. 1947. View Article : Google Scholar
|
|
32
|
Kruskal WH and Wallis WA: Use of ranks in
one-criterion variance analysis. J Am Stat Assoc. 47:583–621. 1952.
View Article : Google Scholar
|
|
33
|
Fieller EC, Hartley HO and Pearson ES:
Tests for rank correlation coefficients I. Biometrika. 44:470–481.
1957. View Article : Google Scholar
|
|
34
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff Finder: A
comprehensive and straightforward Web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fu J, Bian M, Xin G, Deng Z, Luo J, Guo X,
Chen H, Wang Y, Jiang Q and Zhang C: TPX2 phosphorylation maintains
metaphase spindle length by regulating microtubule flux. J Cell
Biol. 210:373–383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Tang H, Sun Z, Bungum AO, Edell ES,
Lingle WL, Stoddard SM, Zhang M, Jen J, Yang P, et al:
Network-based approach identified cell cycle genes as predictor of
overall survival in lung adenocarcinoma patients. Lung Cancer.
80:91–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen L, Zhuo D, Chen J and Yuan H:
Screening feature genes of lung carcinoma with DNA microarray
analysis. Int J Clin Exp Med. 8:12161–12171. 2015.PubMed/NCBI
|
|
38
|
Eguchi T, Kadota K, Chaft J, Evans B, Kidd
J, Tan KS, Dycoco J, Kolquist K, Davis T, Hamilton SA, et al: Cell
cycle progression score is a marker for five-year lung
cancer-specific mortality risk in patients with resected stage I
lung adenocarcinoma. Oncotarget. 7:35241–35256. 2016.PubMed/NCBI
|
|
39
|
Cuzick J, Swanson GP, Fisher G, Brothman
AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E,
Foster CS, et al Transatlantic Prostate Group: Prognostic value of
an RNA expression signature derived from cell cycle proliferation
genes in patients with prostate cancer: A retrospective study.
Lancet Oncol. 12:245–255. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wistuba II, Behrens C, Lombardi F, Wagner
S, Fujimoto J, Raso MG, Spaggiari L, Galetta D, Riley R, Hughes E,
et al: Validation of a proliferation-based expression signature as
prognostic marker in early stage lung adenocarcinoma. Clin Cancer
Res. 19:6261–6271. 2013. View Article : Google Scholar : PubMed/NCBI
|