Introduction
Esophageal cancer is a serious malignancy and the
eighth most common cancer worldwide, with an estimated 456,000 new
cases, and the sixth most common cause of death from cancer with an
estimated 400,000 deaths in 2012 (1). The global incidence of esophageal
squamous cell carcinoma (ESCC) was 5.2 per 100,000 in the same
year. Approximately 80% of cases of global ESCC occurred in the
Central and East Asian region (2).
The overall 5-year survival rate for ESCC ranges from 15 to 25%
(3). Biomarker discovery for the
malignancy could potentially lead to earlier diagnosis as well as
allowing the monitoring of cancer recurrence (4).
Tumor necrosis factor α induced protein 3 (TNFAIP3
or A20) was first identified as a gene that is activated in
response to TNFα in human umbilical vein endothelial cells
(5). TNFAIP3 protein is composed
of seven Cys2/Cys2 zinc-fingers (ZnFs) (6) that are induced by TNF-mediated NF-κB
activation (7), and has a
dual-function as a ubiquitin-editing enzyme to regulate NF-κB
through several molecules involved in the NF-κB pathway (8,9).
TNFAIP3 was originally characterized as a protein
that protects cells from the cytotoxic effect of TNF (10), and regulates TNF receptor signals
by interactions with TNF receptor-associated factor-2 (11), interleukin 1, A20 binding inhibitor
of NF-κB activation (ABIN) (12),
inhibitor of NF-κB kinase γ (13),
and stress-activated protein kinase (14). TNFAIP3 was also demonstrated to
interact with TXBP151 protein to mediate the anti-apoptotic process
through cleavage of caspase-3, -6 and -7 (15), suggesting that overexpression of
TNFAIP3 is correlated with inflammatory and malignant diseases
(16).
Recently, we reported the correlation of TNFAIP8
overexpression and cancer progression and poor prognosis in ESCC
clinical samples. TNFAIP8 was also demonstrated as an effective
therapeutic target for ESCC. TNFAIP8 is an apoptosis regulator and
contains a death-effector domain that is also induced by NF-κB
activation (17). Although TNFAIP3
and −8 have a different mechanism, both were reported to play a
role in multiple myeloma (18). As
with TNFAIP8, TNFAIP3 may also be a promising therapeutic target
for malignant diseases.
Overexpression of TNFAIP3 was also found in breast
tumor (19), pancreatic cancer
(20), hepatocellular carcinoma
(21) and bladder cancer (22). Also, single nucleotide
polymorphisms in TNFAIP3 (TNFAIP3-SNPs) were reported to be
associated with advanced disease stage and survival in
surgically-treated esophageal adenocarcinoma and squamous cell
carcinoma (23). However, the
clinical study and function of TNFAIP3 in malignant disease is very
limited, especially in ESCC. In this study, we evaluated the
correlation between TNFAIP3 expression and cancer progression in
ESCC clinical samples. We also investigated the TNFAIP3 function in
ESCC cells in vitro.
Materials and methods
Patients and specimens
Surgical specimens for immunohistochemical (IHC)
study were obtained from 149 ESCC patients (134 males and 15
females) who underwent potentially curative surgery; no evidence of
residual tumors and the resected margins were free of tumors by
microscopic examination (R0) at the Department of General Surgical
Science, Gunma University, between 2000 and 2010, after obtaining
written informed consent. The patients were aged from 41 to 83
years (mean, 64.1 years). Tumor stage and disease grade of clinical
samples were classified according to the 6th edition of the TNM
classification of the International union against Cancer (UICC).
The tumor differentiation evaluation was based on the histological
criteria outlined by the Japanese Society for Esophageal
Disease.
Surgical specimens for RNA samples were obtained
from 83 ESCC patients (72 males and 11 females) that were a subset
of IHC samples. The patients were aged from 42 to 83 years (mean,
65.2 years). Normal tissues were obtained far from the margin of
the cancer in surgical specimens. All specimens for RNA extraction
were immediately frozen in liquid nitrogen and stored at −80°C
until RNA extraction.
None of the patients had received radiotherapy or
chemotherapy prior to surgery, nor did any have hematogenous
metastases at the time of surgery. Patients who had undergone
non-curative surgery and/or who had received inadequate follow-up
were excluded from the study.
Immunohistochemistry
Resected specimens were fixed with 10% formaldehyde,
embedded in paraffin blocks, cut into 4 µm thick sections,
and mounted onto platinum pro-micro slide glass (Matsunami Glass
Ind., Ltd., Osaka, Japan). We examined sections containing portions
of both tumor and its normal esophageal epithelium as inner
control. Each section was deparaffinized, rehydrated, and incubated
with fresh 0.3% hydrogen peroxide in methanol for 30 min at room
temperature to block endogenous peroxidase activity. After being
rehydrated through a graded series of ethanol concentrations, the
sections were autoclaved in 10 mM citrate buffer (pH 6.0) at 120°C
for 2 min and then cooled to 30°C. After rinsing the sections in
0.1 M phosphate-buffered saline (PBS; pH 7.4), non-specific binding
sites were blocked by incubation with 10% normal goat serum for 30
min. The sections were then incubated at 4°C overnight with rabbit
anti-TNFAIP3 polyclonal antibody (1:150; Abcam, Cambridge, UK) in
PBS containing 1% bovine serum albumin. Negative controls were
obtained by absence of the specific primary antibody. The sections
were then washed in PBS and incubated with biotinylated anti-rabbit
IgG for 30 min at room temperature. IHC was performed using a
Histofine streptavidin-biotin peroxidase (SAB-PO) complex solution
kit (Nichirei Co., Tokyo, Japan). The sections were then lightly
counterstained with Mayer's hematoxylin and mounted.
The intensity and area of TNFAIP3 staining in tumor
tissues was scored between 0–3, as follows; 0, no staining, 0%; 1,
weak, <30%; 2, moderate, 30–60%; and 3, strong intensity,
>60%. Low TNFAIP3 expression was scored 0–1, while high TNFAIP3
expression was 2–3. TNFAIP3 staining evaluation was performed by
three experienced researchers well trained in pathology, in a
blinded manner; i.e., they did not have any knowledge of the
clinical or pathological backgrounds of the patients, or the
assessment of each other.
RNA extraction and quantitative real-time
reverse transcriptase PCR
Total RNA was extracted from the tissue and cells
using the RNeasy plus Mini kit (Qiagen, Hilden, Germany). The
quantity of isolated RNA was measured using a ND-1000
spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
cDNA for TNFAIP3 mRNA quantitative real-time reverse
transcriptase PCR (RT-PCR) was synthesized from 1 µg total
RNA with the Omniscript Reverse Transcriptase kit (Qiagen) in a
reaction volume of 20 µl (60 min at 37°C and 5 min at 93°C
before being put on ice). The TNFAIP3-specific
oligonucleotide primers were designed as follows: TNFAIP3
forward, 5′-TGCACACTGTGTTTCATCGAC-3′; reverse,
5′-ACGCTGTGGGACTGACTTTC-3′. GAPDH (258 bp) forward,
5′-AAGGTGAAGGTCGGAGTCAAC-3′; reverse, 5′-CTTGATTTTGGAGGGATCTCG-3′.
PCR amplification to quantify the levels of TNFAIP3 and
GAPDH mRNA in the clinical samples was performed using a
Light cycler 480 Real-Time PCR system and the LightCycler 480 SYBR
Green I Master kit (Roche Applied Science, Mannheim, Germany). The
amplification conditions consisted of initial denaturation at 95°C
for 10 min followed by 40 cycles of denaturation at 95°C for 10
sec, annealing at 60°C for 10 sec, and elongation at 65°C for 10
sec. All of the samples, expression of TNFAIP3 mRNA was calculated
by dividing the quantity of TNFAIP3 mRNA with the quantity of GAPDH
mRNA. To investigate the correlation of TNFAIP3 mRNA expression
levels with clinicopathological features, we divided the tumor
TNFAIP3 mRNA expression value with the corresponding non-cancerous
ones in each samples. The results of samples with TNFAIP3 mRNA
expression value higher than the median value of all samples was
considered high expression, while that lower than the median value
was low expression.
Cell lines
Four esophageal cancer cell lines were established
from squamous cell carcinoma KYSE-70, TE-1, TE-8, TE-15 cell lines,
and the non-cancerous immortalized esophageal cell line Het-1A, was
used. Het-1A, TE-1, TE-8, TE-15 and KYSE-70 cells were provided
from the American Type Culture Collection (Manassas, VA, USA), JCRB
Cell Bank, and RIKEN BioResource Center (Ibaraki, Japan) through
the National Bio-Resource Project of the MEXT, Japan. TE-1, and
TE-15 cells are well-differentiated ESCC primary lesion cells, TE-8
cells are moderately differentiated ESCC primary lesion cells,
while KYSE-70 cells are from a poorly-differentiated ESCC sample.
All cell lines were cultured in RPMI-1640 (Wako, Osaka, Japan)
containing 10% fetal bovine serum and antibiotics (100 U/ml
penicillin and 100 µg/ml streptomycin). The cells were
cultured in a humidified 5% CO2 incubator at 37°C.
Western blot analyses
Het-1A, KYSE-70, TE-1, TE-8 and TE-15 cells
(1×106 cell/ml) were washed twice in ice-cold medium and
cell lysates were prepared after solubilizing cells with PRO-PREP
Protein Extraction Solution (iNtRON Biotechnology, Gyeonggi-do,
Korea). Protein concentrations of the lysates were determined with
a Pierce BCA protein assay kit (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) using bovine serum albumin as a standard. Forty
microgram of each extract was subjected to electrophoresis on a
NuPAGE Novex 4–20% Bis-Tris gel (Thermo Fisher Scientific) and the
proteins were electrotransferred to a Hybond ECL (7×8 cm) membrane
(GE Life Science Healthcare, Little Chalfont, UK). The membranes
were then incubated overnight at 4°C with rabbit polyclonal
antibody against TNFAIP3 (1:1,000; Abcam) and mouse monoclonal
antibody against β-actin (1:2,000; Sigma-Aldrich, St. Louis, MO,
USA). Bands on the membrane were detected using an Image Quant
LAS4000 (GE Life Science) with the aid of an enhanced
chemiluminescence detection system.
Small interfering RNA
TNFAIP3 small interfering RNA (siRNA)
(hTNFAIP3_#1: sense 5′-CAAAGUUGGAUGAAGCUAAtt and antisense
5′-UUAGCUUCAUCCAACUUUGtt, hTNFAIP3_#2: sense
5′-GCACCAUGUUUGAAGGAUAtt and antisense 5′-UAUCCUUCAAACAUGGUGCtt and
hTNFAIP3_#3: sense 5′-GAGCAGGAGAGGAAAGAUAtt and antisense
5′-UAUCUUUCCUCUCCUGCUCtt) siRNAs were purchased from GeneDesign
(Osaka, Japan). TE-15 cells were seeded in 6-well flat-bottomed
microtiter plates at a density of 1.5×105 cells per well
in a volume of 2 ml and incubated in a humidified atmosphere (37°C
and 5% CO2). After incubation, TE-15 cells were treated
with siRNAs according to the manufacturer's instructions to final
concentration 30 nM per well, by adding Opti-MEM I Reduced-Serum
Medium liquid (Thermo Fisher Scientific) mixed with Lipofectamine
RNAi MAX (Thermo Fisher Scientific). The experiments were then
performed after 48 h incubation.
Cell proliferation assay
Cell proliferation analysis was performed using
TE-15 cells transfected with TNFAIP3 siRNA; 100 µl of
a cell suspension (1×104 cells) was seeded into each
well of a 96-well plate (Falcon, Franklin Lakes, NJ, USA) and
incubated at 37°C overnight. Cell viability was determined using a
Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) according to the
manufacturer's instructions. Proliferation index were assessed as
absorbance at 450 nm (OD450) the reference wavelength at
620 nm was read and the results were derived from three sets of
triplicate experiments.
Wound healing assay
To determine the effect of TNFAIP3 on esophageal
cell migration, a wound-healing assay was performed in TE-15 cells
transfected with TNFAIP3 siRNAs. The cells were seeded in
6-well plates and incubated until 80% confluence. A wound was made
through the monolayer using a sterile 200 µl pipette tip and
cell debris removed by washing the cells with PBS three times.
Wounds were observed under a microscope and measured over a time
course to calculate the migration rate according to the following
formula: percentage wound healing for 24 and 48 h = [(wound length
at 0 h) − (wound length at 24 or 48 h)]/(wound length at 0 h) ×
100. The experiments were performed three times with triplicate
samples.
Cell invasion assay
Cell invasion was examined using the BD BioCoat
Matrigel invasion chamber (8.0 µm, BD Bioscience, San Jose,
CA, USA) according to the manufacturer's instructions. Five hundred
microliter containing 2.5×104 TE-15 cells transfected
with TNFAIP3 siRNA were added to each invasion chamber.
After incubation for 12 h, the cells were stained with a Diff-Quick
kit (Sysmex Corp., Kobe, Japan), and then observed and counted
under the microscope. The parental groups were used for
normalization. All samples were tested twice in triplicate.
Statistical analysis
The χ2 test and t-test were used to
assess the statistical significance of the correlations between
TNFAIP3 expression and clinicopathological parameters. Kaplan-Meier
curves were generated for disease-specific survival. In addition,
univariate and multivariate survival analyses were performed using
Cox's proportional hazards regression model. Statistical analyses
were performed using JMP5.0 software (SAS Institute Inc., NC, USA).
Differences were considered statistically significant when the
P-value was <0.05.
Results
TNFAIP3 protein and mRNA expression
levels in ESCC tissue samples
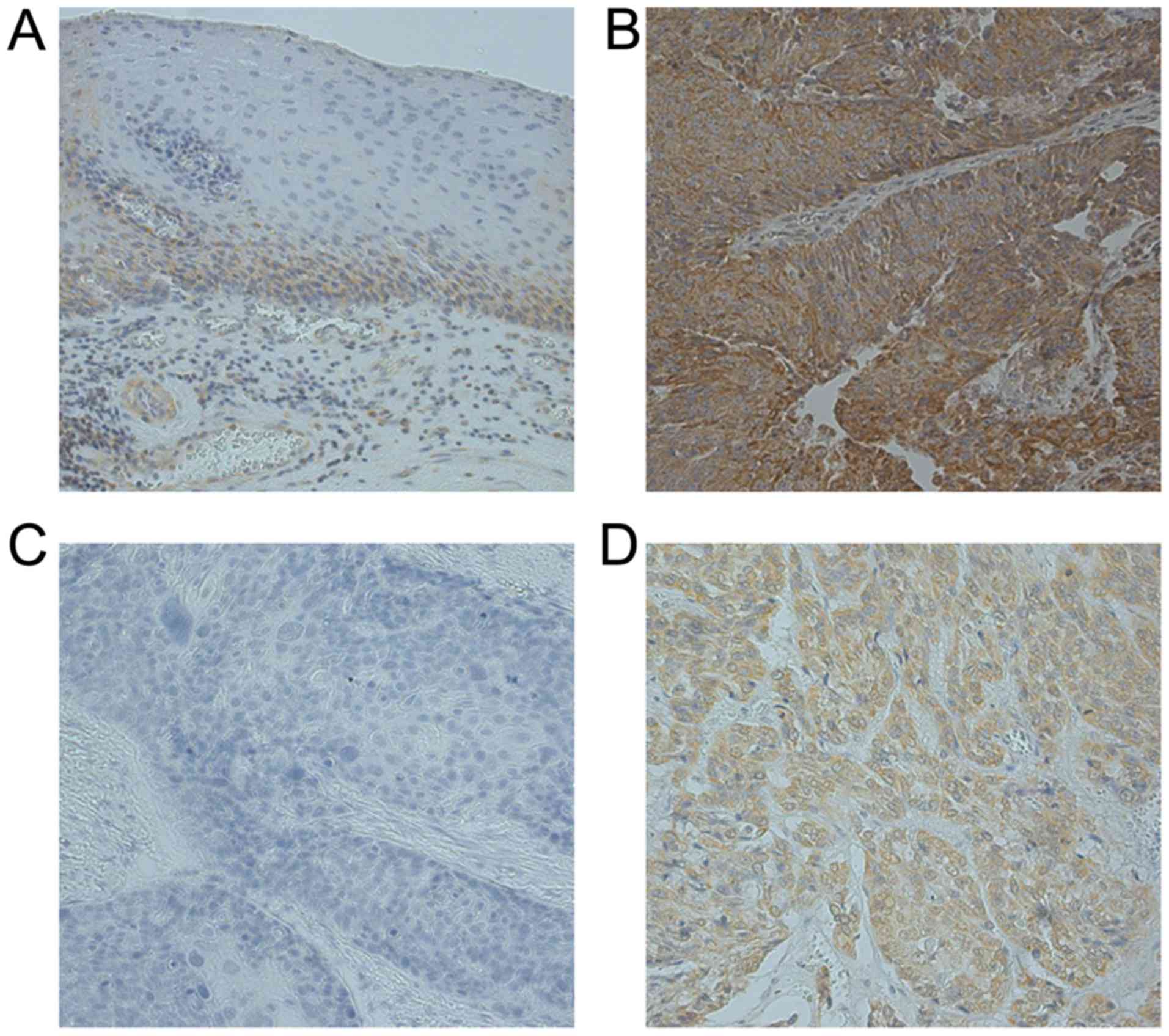
In normal esophageal epithelium, expression of
TNFAIP3 was low in the basal layer of the epithelium (Fig. 1A). TNFAIP3 expression was localized
in the cytoplasmic components of tumor cells (Fig. 1B and D). The expression of TNFAIP3
was investigated by IHC in 149 ESCC specimens, and we found that
TNFAIP3 protein expression was high in 71 specimens (47.65%).
TNFAIP3 high expression is correlated
with differentiation of ESCC pathological features in protein
level, but not in mRNA level
The correlations between TNFAIP3 expression and the
clinicopathological characteristics of ESCC patients (age, gender,
differentiation, TNM stage, tumor depth, lymph node metastasis,
distant metastasis, lymphatic invasion, and venous invasion) that
were investigated by IHC are shown in Table I. A significant correlation between
TNFAIP3 expression and tumor differentiation status (P=0.041) was
identified, whereas there were no significant correlation between
TNFAIP3 expression and age (P=0.137), or gender (P=0.935), TNM
stage (P=0.603), tumor depth (P=0.381), lymph node metastasis
(P=0.534), distant metastasis (P=0.299), lymphatic invasion
(P=0.391), or venous invasion (P=0.075). However, no significant
correlations were found between TNFAIP3 mRNA expression and
the clinicopathological characteristics of ESCC patients or patient
survival rates (data not shown).
 | Table ICorrelations between TNFAIP3
expression and clinicopathological features. |
Table I
Correlations between TNFAIP3
expression and clinicopathological features.
|
Characteristics | TNFAIP3 low
n=78 | TNFAIP3 high
n=71 | P-value |
|---|
| Age (years) (mean ±
SD) | 63.14±0.90 | 65.09±0.94 | 0.137 |
| Gender | | | |
| Male | 70 | 64 | 0.935 |
| Female | 8 | 7 | |
|
Differentiation | | | |
| Well | 8 | 17 | 0.041a |
| Moderate | 41 | 37 | |
| Poor | 29 | 17 | |
| TNM stage | | | |
| I | 18 | 60 | 0.603 |
| II, III, IV | 60 | 52 | |
| Tumor depth | | | |
| T1 | 36 | 24 | 0.381 |
| T2 | 9 | 8 | |
| T3 | 31 | 35 | |
| T4 | 2 | 4 | |
| Lymph node
metastasis | | | |
| N0 | 28 | 29 | 0.534 |
| N1 | 50 | 42 | |
| Distant metastasis
(LYM) | | | |
| M0 | 62 | 61 | 0.299 |
| M1 | 16 | 10 | |
| Lymphatic
invasion | | | |
| Negative | 14 | 10 | 0.391 |
| Positive | 64 | 61 | |
| Venous
invasion | | | |
| Negative | 23 | 13 | |
| Positive | 55 | 58 | 0.075 |
TNFAIP3 high expression is correlated
with poor survival as independent factor
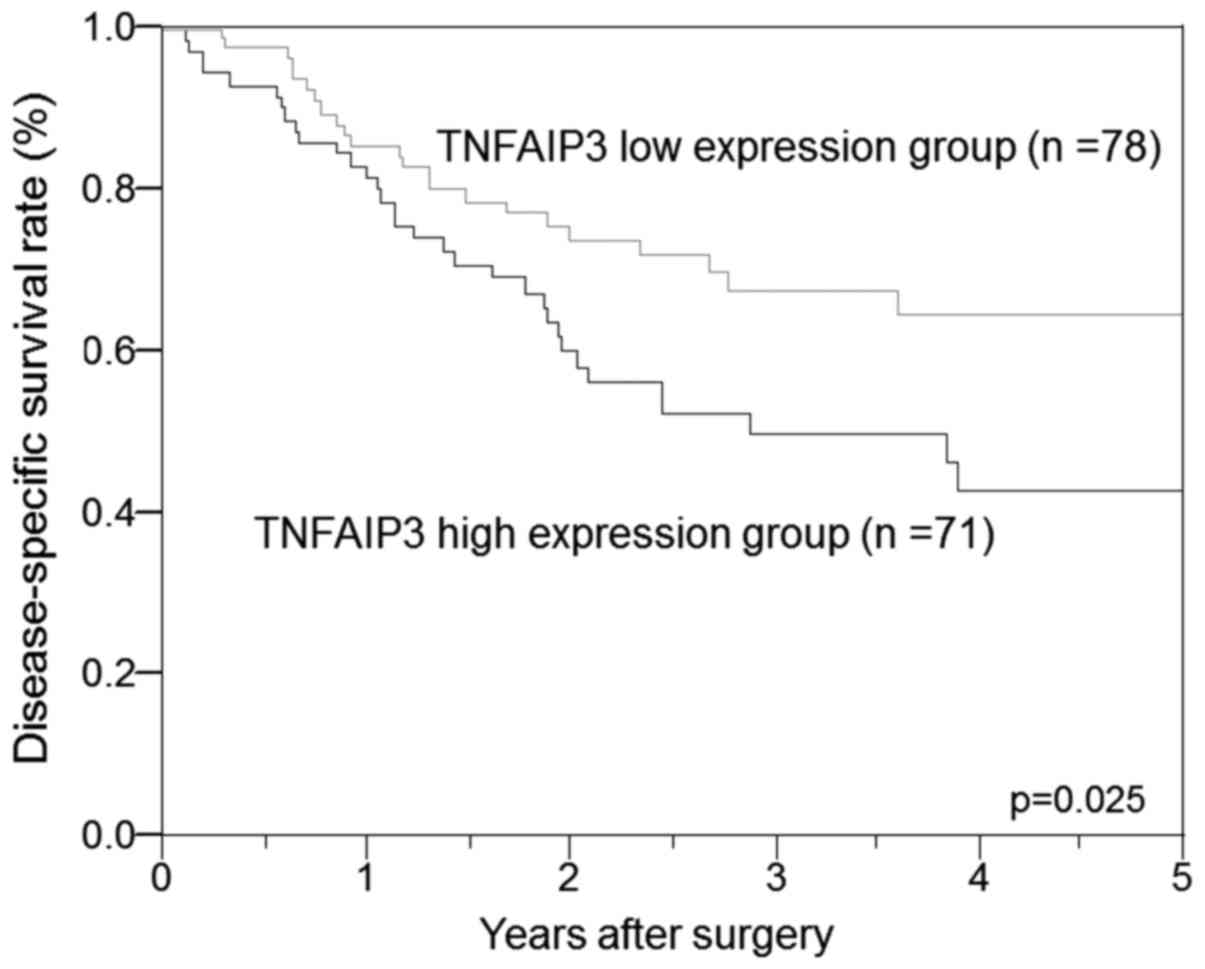
The disease-specific survival rate of the ESCC
patients with high TNFAIP3 expression was significantly poorer than
that of the patients with low TNFAIP3 expression (P=0.025; Fig. 2). In univariate analysis, high
TNFAIP3 expression was found to be a significant prognostic factor
for poor survival, in addition to tumor depth, the presence of
lymph node metastasis, distant metastasis (cervical lymph node
metastasis, not distant organ metastasis), lymphatic invasion, and
venous invasion. Moreover, multivariate analysis of the six factors
revealed that the presence of distant metastasis was significant
(P=0.008) and showed that positive TNFAIP3 expression was an
independent prognostic factor (P=0.021; Table II).
 | Table IIUnivariate and multivariate analyses
of survival in 149 ESCC patients. |
Table II
Univariate and multivariate analyses
of survival in 149 ESCC patients.
| Characteristic | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| TNFAIP3
expression | | | | |
| Low vs high | 1.34
(1.03–1.76) | 0.025a | 1.37
(1.04–1.82) | 0.021a |
| Tumor depth | | | | |
| T1 vs T2–4 | 0.65
(0.48–0.87) | 0.003a | 0.97
(0.69–1.33) | 0.879 |
| Lymph node
metastasis | | | | |
| N0 vs N1 | 0.52
(0.35–0.73) | 0.0001a | 0.68
(0.43–1.00) | 0.056 |
| Distant metastasis
(LYM) | | | | |
| M0 vs M1 | 0.56
(0.42–0.74) | 0.0001a | 0.66
(0.49–0.89) | 0.008a |
| Lymphatic
invasion | | | | |
| Negative vs
positive | 2.34
(1.30–5.79) | 0.001a | 1.05
(0.45–2.99) | 0.908 |
| Venous
invasion | | | | |
| Negative vs
positive | 2.42
(1.46–4.90) | 0.0001a | 1.63
(0.86–3.76) | 0.139 |
TNFAIP3 mRNA expression in ESCC tissues
was significantly higher than in non-cancerous tissue
Whereas, TNFAIP3 mRNA expression was
determined in clinical samples from 83 patients using real-time
RT-PCR, and 53 patients (63.85%) showed higher TNFAIP3 mRNA
expression levels in their ESCC tissue specimens (Fig. 3). The mean TNFAIP3 mRNA
level in the ESCC tissue was significantly higher than that in the
corresponding non-cancerous tissue (P=0.027).
Expression of TNFAIP3 protein in ESCC
cells and noncancerous immortalized esophageal cells and knockdown
of TNFAIP3 in TE-15 cells
We examined TNFAIP3 protein expression in four ESCC
cell lines (KYSE70, TE-1, TE-8 and TE-15) and the non-cancerous
immortalized esophageal cell line Het-1A. TNFAIP3 protein was
expressed at low levels in Het-1A and TE-8 cells, whereas it was
moderately expressed in KYSE70 and TE-1 cells and showed extremely
high expression in TE-15 cells (Fig.
4A). Based on these results, we used TE-15 cells to analyze
cell behavior following knockdown of TNFAIP3.
To determine the contribution of high expression
levels of TNFAIP3 in ESCC cells, we used siRNA to knock down
TNFAIP3 expression in TE-15 cancer cells. The suppression of
TNFAIP3 by siRNAs was checked using western blotting
(Fig. 4B). Since TNFAIP3
siRNA3 did not show any significant reduction, we used
TNFAIP3 siRNA1 and siRNA2 for the subsequent analysis.
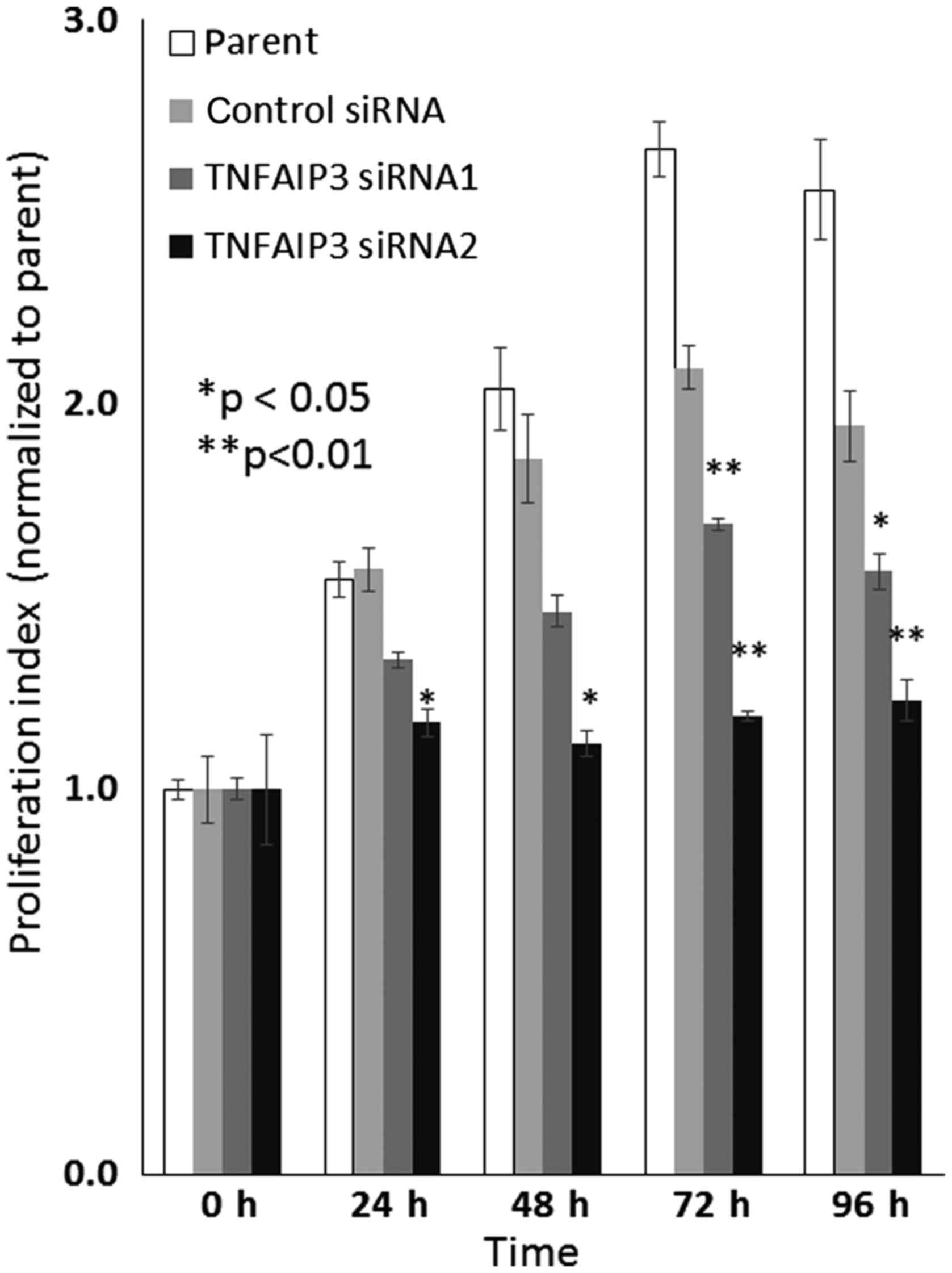
Depletion of TNFAIP3 expression inhibits
cell proliferation, migration, and invasion
TE-15 cells treated with TNFAIP3 siRNA1 and
siRNA2 showed a significant reduction in their cell proliferation
rate compared with cells transfected with the negative control
siRNA and the parental group (Fig.
5).
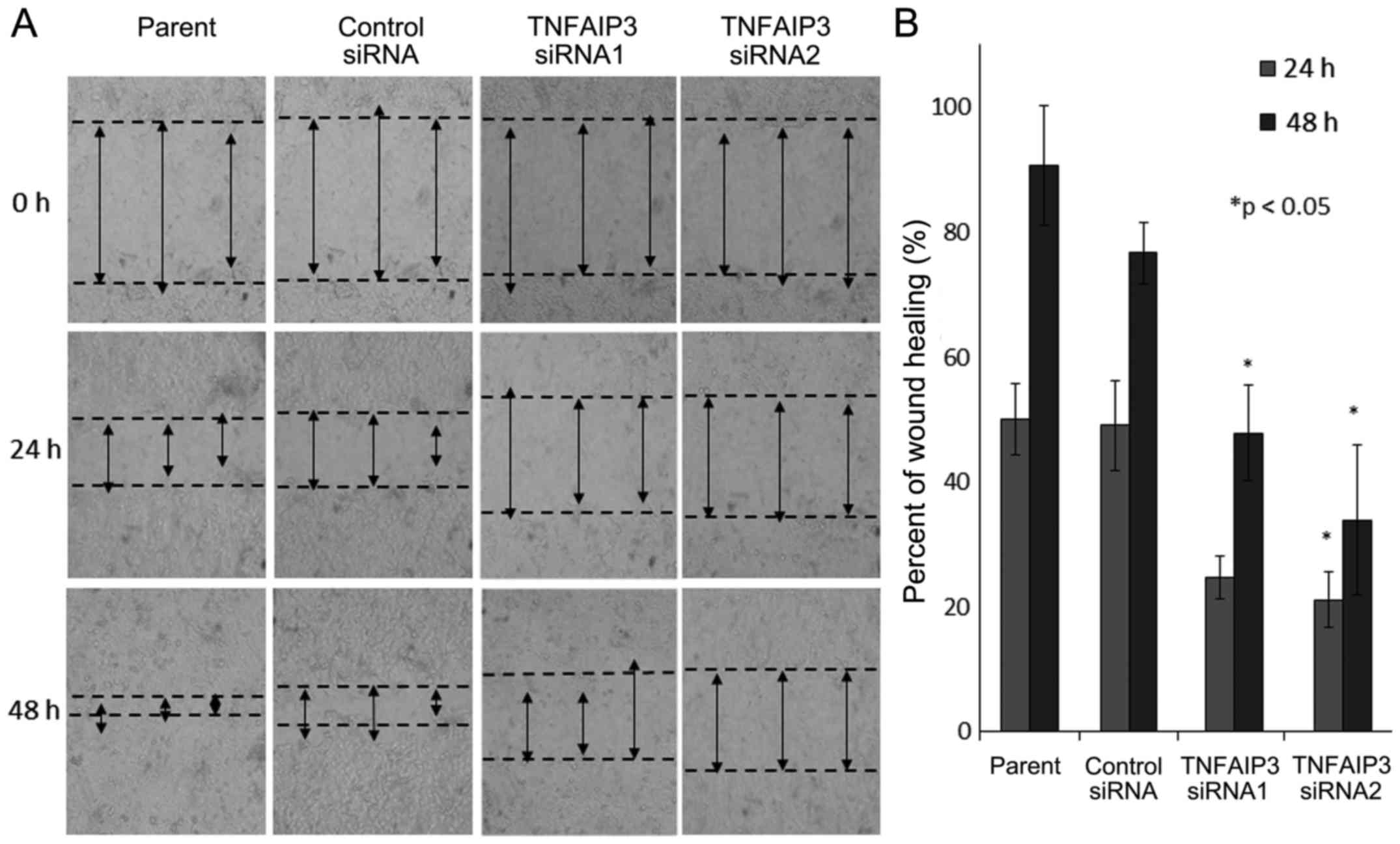
A significant reduction in cell migration ability
was demonstrated in TE-15 cells that were treated with
TNFAIP3 siRNA1 and siRNA2 compared with cells transfected
with the negative control siRNA and the parental group (Fig. 6).
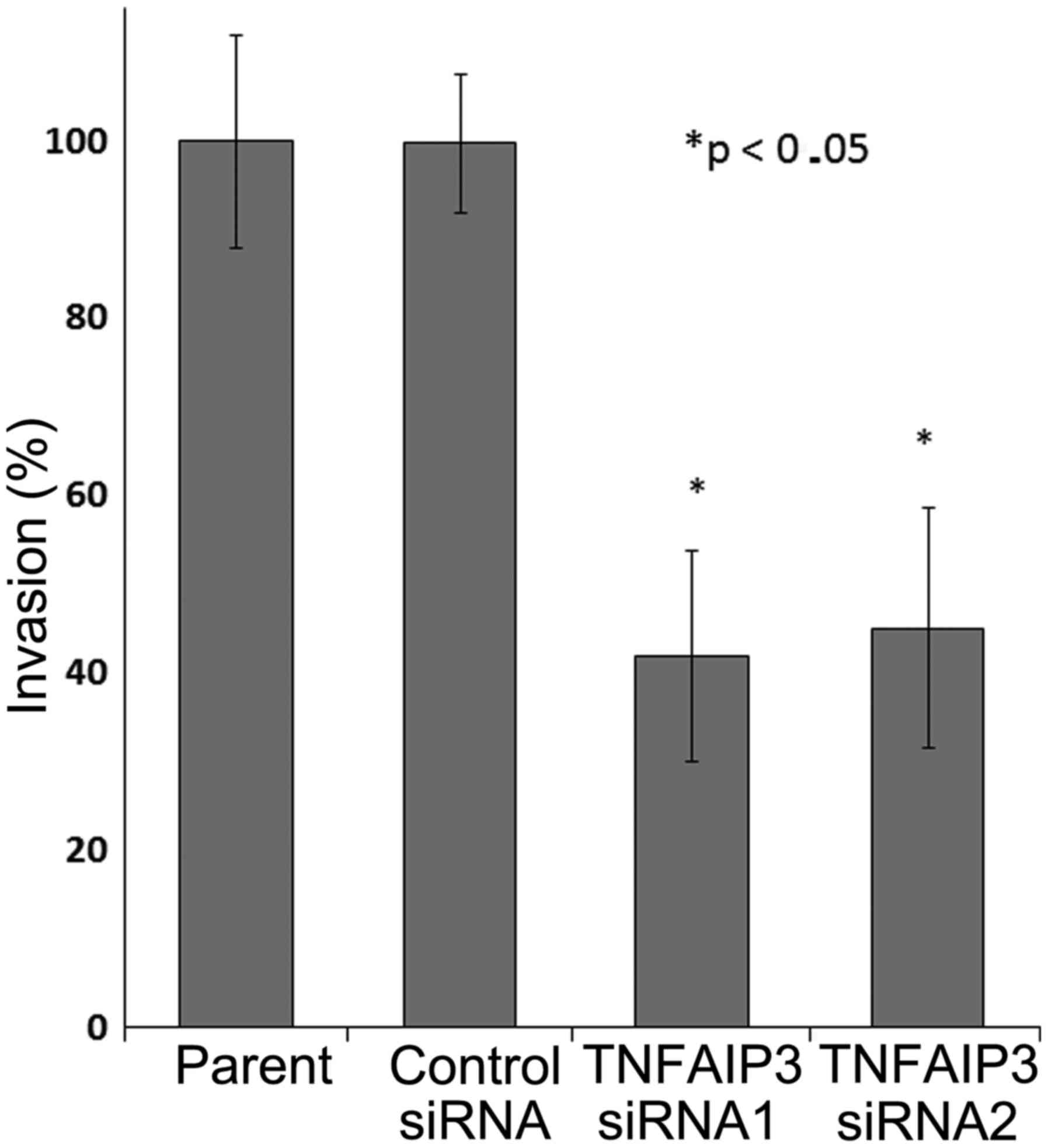
The weakening of the cell ability to invade was
shown in TE-15 cells treated with TNFAIP3 siRNA1 and siRNA2
compared with cells transfected with the negative control siRNA and
the parental group (Fig. 7). Based
on the in vitro experiment results above, the depletion of
TNFAIP3 protein expression seems to decrease cancer cell
proliferation, migration and invasion ability.
Discussion
In the present study, we found that high expression
of TNFAIP3 in ESCC clinical samples was significantly correlated
with tumor differentiation that is associated with tumor growth and
spread. The disease-specific survival rates of patient with high
TNFAIP3 expression were significantly poorer than those of patients
with low TNFAIP3. Moreover, multivariate analysis showed that
positive TNFAIP3 expression was an independent prognostic factor
for poor survival. According to clinical results, we investigated
whether the TNFAIP3 overexpression accelerate tumor growth and
spread enhancing the ability of tumor cells to migrate and invade.
In vitro analysis showed that proliferation, migration and
invasion were significantly reduced following TNFAIP3 siRNA
knockdown compared with the control groups. Our results indicated
that TNFAIP3 plays a role in enhancing the ability of cells to
expand, that might be the reason why of the tumor appears to grow
slowly in the primary tumor, but actually tumor cells migrate to
other organs.
TNFAIP3 is an anti-inflammatory signaling molecule
that was reported to protect cells from cytotoxic effects (10). Since TNFAIP3 was found to regulate
NF-κB signaling through ubiquitination to form germline
polymorphisms and somatic mutations, TNFAIP3 has been linked to
multiple diseases. TNFAIP3-SNPs that might reduce TNFAIP3 function
and expression were linked with multiple inflammatory diseases such
as systemic lupus erythematosus, rheumatoid arthritis, psoriasis,
and type 1 diabetes (16). TNFAIP3
with missense mutations was reported in breast cancer cells at
14-3-3 binding sites by changing the crucial serine residue to
arginine (24). TNFAIP3 mutations
cause overexpression and confer an anti-apoptotic function on
TNFAIP3 to mediate RIP1 ubiquitination and hence inhibit caspase-8
cleavage in hepatocellular carcinoma cells (21). In endothelial cells, TNFAIP3
overexpression was showed to restrict TNF- and FAS-induced
apoptosis by preventing subsequent activation of the apical
caspases-8 and -2 to the downstream effectors (25). High TNFAIP3 expression caused by
TNFAIP3 mutations were also reported in glioma stem cells (26). However, no study has been reported
on high expression of TNFAIP3 associated with mutations in
ESCC.
A previous study reported that germline TNFAIP3-SNPs
in esophageal cancer had a relationship with AJCC disease stages,
lymph node metastasis, and recurrence (23); however, in the present study, the
overexpression of TNFAIP3 protein was correlated with
differentiation. TNFAIP3 expression was high in well-differentiated
ESCC and low in poorly and moderately differentiated cancer in the
present study. This finding was reinforced by the findings in
vitro that showed high TNFAIP3 expression was found in TE-15
cells, which are well-differentiated ESCC cells. Although these
studies are different, the prediction obtained from previous
studies demonstrated that TNFAIP3 regulates NF-κB signaling
(8,16), with downstream effects on TGF-β
signaling (27) and Wnt/β-catenin
signaling (28), to influence
tumor differentiation. However, for details of the mechanism
further investigation is required.
In head and neck squamous cell carcinoma, TNFAIP3
was expressed in poorly differentiated and undifferentiated, but
not in well-differentiated cancers in mRNA level (29). However, there was no significance
difference between TNFAIP3 mRNA levels and
clinicopathological factors in this study, including tumor
differentiation. Nonetheless, TNFAIP3 protein expression has a
relationship with clinicopathological factors such as tumor
differentiation. A previous study reported that a truncation mutant
form of TNFAIP3 containing four ZnFs is responsible for the
ubiquitination that can regulate TNFAIP3 protein expression levels
(6). TNFAIP3 consists of two
ubiquitin-editing domains, an N-terminal deubiquitinating domain of
the ovarian tumor (OTU) family and a C-terminal ubiquitin ligase
domain. In a previous study, the N terminal domain was shown to
deubiquitinate K63-polyubiquitinated RIP1. The C-terminal
ZnF-containing domain functions as a ubiquitin ligase that
catalyzes K48-linked RIP1, targeting RIP1 for proteasomal
degradation. Wild-type TNFAIP3 disassembles K63-linked ubiquitin
chains on RIP1 and increases K48-linked RIP1 ubiquitination.
Furthermore, TNFAIP3 with a ZnF4 mutation at the OTU-domain
inhibits the disassembling of K63-linked ubiquitin chains on RIP1
and diminishes the ability of TNFAIP3 to ubiquitinate RIP1 with
K48-linked ubiquitin chains (8,9). The
dual-ubiquitination of mutant TNFAIP3 is predicted to cause
differences in TNFAIP3 mRNA and protein expression levels in our
study (16).
TNFAIP3-SNP was also found to be an independent
prognostic factor, but AC and CC genotypes had a relationship with
disease-free and overall survival in esophageal cancer (23). Even though it is not on the same
condition that deserves to be compared, the present study revealed
that TNFAIP3 was found to be an independent prognostic factor, and
ESCC patients with high expression of TNFAIP3 had significantly
poorer disease-specific survival than those patients with low
expression ones. Although TNFAIP3 was not significantly correlated
with metastases (lymph node and distant) or invasion (lymphatic and
venous) in ESCC clinicopathological features, but in multivariate
analysis TNFAIP3 showed significance (P=0.021) after distant
metastases (P=0.008). Since activation of JAK/STAT pathway is
predicted to play roles in malignancies, including tumor
proliferation, invasion and metastases, activation of STAT3 was
also considered a responsible mechanism for TNFAIP3 becoming an
independent prognostic factor in the present study (30). According to a previous study, SNPs
of TNFAIP3 in the C-allele and alteration of the binding of
transcription factors reduce TNFAIP3 expression (16). It means this study results would be
reasonable if that happens is the opposite of the previous results
studied in TNFAIP3-SNP.
From our clinicopathological result in patients with
ESCC, we expected that TNFAIP3 plays a role in cell growth and
spread in ESCC. In breast cancer, overexpression of TNFAIP3 induced
cell proliferation (19),
consistent with a study of hepatocellular carcinoma cells that were
reported with increased proliferation of TNFAIP3-overexpressing
cells. So it is reasonable when in the present study, the depletion
of TNFAIP3 expression in TE-15 cells resulted in a significant
reduction in cell proliferation. These results suggest that TNFAIP3
plays a role in the regulation of cell proliferation. Based on a
previous study, the mechanism of ABIN is by mimicking NF-κB and
combined with TNFAIP3 inhibiting bond formation of TNFAIP3 and
NF-κB that cause apoptosis occurrence (12). Different from ABIN mechanism,
TNFAIP3 depletion lead to inhibit cell proliferation through
attenuation of TNFAIP3 that increased SOCS3 expression, which
inhibited STAT3 phosphorylation and suppressed cell proliferation.
Because, overexpression of TNFAIP3 regulated
lipopolysaccharide/TNF-induced IL-6 inhibition and induced STAT3
phosphorylation. Thus, high and sustained STAT3 phosphorylation
induced cell proliferation (30).
Moreover, when combined with cisplatin, IC50 of
cisplatin in TE-15 cells is more cytotoxic compared with
combination of cisplatin and TNFAIP3 siRNA treated TE-15 cells
(data not shown).
Even though there are no previous reports describing
the relationship between TNFAIP3 and the invasive or migratory
ability of cancer cells, one study demonstrated microbial invasion
of the intestinal inner mucus layer in TNFAIP3 transgenic mice
(31). Another report showed that
TNFAIP3 influenced the ability of intestinal cell migration in
TNFAIP3 knockout mice (32). The
present study demonstrated that the invasive and migratory ability
of TE-15 cells in the TNFAIP3 siRNA groups were
significantly reduced. Suppression of TNFAIP3 protein inhibits
activation of NF-κB, which inhibits the activation of IFNα/STAT1
signaling (32) then resulting in
a reduction of cell migration ability.
Clinical report of the TNFAIP3 overexpression is
limited. The finding of TNFAIP3 affects in migration and invasion
of esophageal cancer cells is a novelty of this study.
Investigation to identify a suitable mechanism for cancer therapy
is now in progress in order to apply this biomarker into a novel
therapy against malignant disease. Furthermore, study that
describes TNFAIP3 expression in patients with therapy adjuvant is
required in future research.
Our data implied that the expression of TNFAIP3 in
ESCC cancer tissues was significantly higher compared with the
normal esophageal epithelial tissues, and was significantly
correlated with tumor differentiation and found to be an
independent prognostic marker for disease survival in ESCC. Our
TNFAIP3 knockdown data demonstrated that TNFAIP3 expression may
relate, not only to proliferation, but also to migration and
invasion in ESCC cells. Taken together, these results suggest that
TNFAIP3 could be used as a promising marker for diagnosis and as an
effective therapeutic target for ESCC in the future.
Acknowledgments
This study was supported by Grants-in-Aid for
Scientific Research from the Japan Society for the Promotion of
Science (JSPS no. 22591450).
Glossary
Abbreviations
Abbreviations:
|
ESCC
|
esophageal squamous cell carcinoma
|
|
TNFAIP3
|
tumor necrosis factor α induced
protein 3
|
|
ZnF
|
zinc-finger
|
|
ABIN
|
A20 binding inhibitor of NF-κB
activation
|
|
TNFAIP3-SNPs
|
TNFAIP3 in single nucleotide
polymorphisms
|
|
IHC
|
immunohistochemical
|
|
UICC
|
International Union against Cancer
|
|
RT-PCR
|
reverse transcriptase PCR
|
|
siRNA
|
small-interfering RNA
|
|
OTU
|
ovarian tumor
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Arnold M, Soerjomataram I, Ferlay J and
Forman D: Global incidence of oesophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dixit VM, Green S, Sarma V, Holzman LB,
Wolf FW, O'Rourke K, Ward PA, Prochownik EV and Marks RM: Tumor
necrosis factor-α induction of novel gene products in human
endothelial cells including a macrophage-specific chemotaxin. J
Biol Chem. 265:2973–2978. 1990.PubMed/NCBI
|
|
6
|
Opipari AW Jr, Boguski MS and Dixit VM:
The A20 cDNA induced by tumor necrosis factor α encodes a novel
type of zinc finger protein. J Biol Chem. 265:14705–14708.
1990.PubMed/NCBI
|
|
7
|
Krikos A, Laherty CD and Dixit VM:
Transcriptional activation of the tumor necrosis factor α-inducible
zinc finger protein, A20, is mediated by kappa B elements. J Biol
Chem. 267:17971–17976. 1992.PubMed/NCBI
|
|
8
|
Wertz IE, O'Rourke KM, Zhou H, Eby M,
Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al:
De-ubiquitination and ubiquitin ligase domains of A20 downregulate
NF-kappaB signalling. Nature. 430:694–699. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bosanac I, Wertz IE, Pan B, Yu C, Kusam S,
Lam C, Phu L, Phung Q, Maurer B, Arnott D, et al: Ubiquitin binding
to A20 ZnF4 is required for modulation of NF-κB signaling. Mol
Cell. 40:548–557. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Opipari AW Jr, Hu HM, Yabkowitz R and
Dixit VM: The A20 zinc finger protein protects cells from tumor
necrosis factor cytotoxicity. J Biol Chem. 267:12424–12427.
1992.PubMed/NCBI
|
|
11
|
Song HY, Rothe M and Goeddel DV: The tumor
necrosis factor-inducible zinc finger protein A20 interacts with
TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci
USA. 93:6721–6725. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Heyninck K, De Valck D, Vanden Berghe W,
Van Criekinge W, Contreras R, Fiers W, Haegeman G and Beyaert R:
The zinc finger protein A20 inhibits TNF-induced
NF-kappaB-dependent gene expression by interfering with an RIP- or
TRAF2-mediated transactivation signal and directly binds to a novel
NF-kappaB-inhibiting protein ABIN. J Cell Biol. 145:1471–1482.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang SQ, Kovalenko A, Cantarella G and
Wallach D: Recruitment of the IKK signalosome to the p55 TNF
receptor: RIP and A20 bind to NEMO (IKKgamma) upon receptor
stimulation. Immunity. 12:301–311. 2000. View Article : Google Scholar
|
|
14
|
Lee EG, Boone DL, Chai S, Libby SL, Chien
M, Lodolce JP and Ma A: Failure to regulate TNF-induced NF-kappaB
and cell death responses in A20-deficient mice. Science.
289:2350–2354. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
De Valck D, Jin DY, Heyninck K, Van de
Craen M, Contreras R, Fiers W, Jeang KT and Beyaert R: The zinc
finger protein A20 interacts with a novel anti-apoptotic protein
which is cleaved by specific caspases. Oncogene. 18:4182–4190.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma A and Malynn BA: A20: Linking a complex
regulator of ubiquitylation to immunity and human disease. Nat Rev
Immunol. 12:774–785. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hadisaputri YE, Miyazaki T, Suzuki S,
Yokobori T, Kobayashi T, Tanaka N, Inose T, Sohda M and Kuwano H:
TNFAIP8 overexpression: Clinical relevance to esophageal squamous
cell carcinoma. Ann Surg Oncol. 19(Suppl 3): S589–S596. 2012.
View Article : Google Scholar
|
|
18
|
Wang MC, Liu SX and Liu PB: Gene
expression profile of multiple myeloma cell line treated by
realgar. J Exp Clin Cancer Res. 25:243–249. 2006.PubMed/NCBI
|
|
19
|
Vendrell JA, Ghayad S, Ben-Larbi S,
Dumontet C, Mechti N and Cohen PA: A20/TNFAIP3, a new
estrogen-regulated gene that confers tamoxifen resistance in breast
cancer cells. Oncogene. 26:4656–4667. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Yuan L, Liu Z, Yin J, Jiang X and
Lu J: Expression of A20 is reduced in pancreatic cancer tissues. J
Mol Histol. 43:319–325. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong B, Lv G, Wang Q, Wei F, Bellail AC,
Hao C and Wang G: Targeting A20 enhances TRAIL-induced apoptosis in
hepatocellular carcinoma cells. Biochem Biophys Res Commun.
418:433–438. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang M and Li S: Bladder polypoid
cystitis-derived A20 associates with tumorigenesis. Cell Biochem
Biophys. 67:669–673. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ghadban T, Schmidt-Yang M, Uzunoglu FG,
Perez DR, Tsui TY, El gammal AT, Erbes PJ, Zilbermints V, Wellner
U, Pantel K, et al: An A/C germline single-nucleotide polymorphism
in the TNFAIP3 gene is associated with advanced disease stage and
survival in only surgically treated esophageal cancer. J Hum Genet.
59:661–666. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lademann U, Kallunki T and Jäättelä M: A20
zinc finger protein inhibits TNF-induced apoptosis and stress
response early in the signaling cascades and independently of
binding to TRAF2 or 14-3-3 proteins. Cell Death Differ. 8:265–272.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Daniel S, Arvelo MB, Patel VI, Longo CR,
Shrikhande G, Shukri T, Mahiou J, Sun DW, Mottley C, Grey ST, et
al: A20 protects endothelial cells from TNF-, Fas-, and NK-mediated
cell death by inhibiting caspase 8 activation. Blood.
104:2376–2384. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hjelmeland AB, Wu Q, Wickman S, Eyler C,
Heddleston J, Shi Q, Lathia JD, Macswords J, Lee J, McLendon RE, et
al: Targeting A20 decreases glioma stem cell survival and tumor
growth. PLoS Biol. 8:e10003192010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun X, Chen E, Dong R, Chen W and Hu Y:
Nuclear factor (NF)-κB p65 regulates differentiation of human and
mouse lung fibroblasts mediated by TgF-β. Life Sci. 122:8–14. 2015.
View Article : Google Scholar
|
|
28
|
Murtas D, Maric D, De Giorgi V, Reinboth
J, Worschech A, Fetsch P, Filie A, Ascierto ML, Bedognetti D, Liu
Q, et al: IRF-1 responsiveness to IFN-γ predicts different cancer
immune phenotypes. Br J Cancer. 109:76–82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Codd JD, Salisbury JR, Packham G and
Nicholson LJ: A20 RNA expression is associated with
undifferentiated nasopharyngeal carcinoma and poorly differentiated
head and neck squamous cell carcinoma. J Pathol. 187:549–555. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
da Silva CG, Studer P, Skroch M, Mahiou J,
Minussi DC, Peterson CR, Wilson SW, Patel VI, Ma A, Csizmadia E, et
al: A20 promotes liver regeneration by decreasing SOCS3 expression
to enhance IL-6/STAT3 proliferative signals. Hepatology.
57:2014–2025. 2013. View Article : Google Scholar :
|
|
31
|
Murphy SF, Rhee L, Grimm WA, Weber CR,
Messer JS, Lodolce JP, Chang JE, Bartulis SJ, Nero T, Kukla RA, et
al: Intestinal epithelial expression of TNFAIP3 results in
microbial invasion of the inner mucus layer and induces colitis in
IL-10-deficient mice. Am J Physiol Gastrointest Liver Physiol.
307:G871–G882. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Moll HP, Lee A, Minussi DC, da Silva CG,
Csizmadia E, Bhasin M and Ferran C: A20 regulates atherogenic
interferon (IFN)-γ signaling in vascular cells by modulating basal
IFNβ levels. J Biol Chem. 289:30912–30924. 2014. View Article : Google Scholar : PubMed/NCBI
|