Introduction
In recent years, cytokine research has been at the
forefront of cancer research and cytokine approaches are involved
in the treatment of various carcinomas. Cytokine approaches for
cancer therapy have three potential mechanisms of action. They can
i) directly induce cell death programs in tumor cells, ii) increase
the number or activity of immune effector cells, or iii) increase
the recognition of tumor cells by the immune system (1).
Interferons (IFNs) are one of the most important
cytokines. They are naturally secreted glycoproteins produced by
almost every cell type as a mechanism of host defense in response
to microbial attack (2). The IFN
family includes three different groups. IFNα belongs to the type I
IFN group, and was discovered 50 years ago. It was the first
cytokine to be produced by recombinant DNA technology, and it is
used as an important regulator of cell growth and differentiation,
affecting cellular communication and signal transduction pathways
as well as immunological control (3). More recently, IFNα has been applied
in the treatment of multiple carcinomas including leukemia,
hepatocellular carcinoma, bladder cancer, and osteosarcoma
(4–7).
Breast cancer is a leading cause of cancer-related
death in women worldwide. According to GLOBOCAN 2012, there are
approximately 1.67 million newly diagnosed breast cancer patients
every year (8). In China, breast
cancer is the most commonly diagnosed cancer, and the number of
breast cancer patients has been increasing annually (9). Surgery, chemotherapy, radiotherapy,
endocrinotherapy and molecular targeting therapy are the major
treatment modalities for breast cancer. However, challenges remain
in the treatment of breast cancer, and novel therapeutic approaches
are urgently needed.
Although the use of IFN in clinical practice is
widely recommended, its antineoplastic activity and clinical
efficacy for breast cancer is still unclear and controversial
(10,11). In this study, an NEB Ph.D.-12
peptide library was employed to select a short peptide that
specifically binds to the cell membrane of MCF-7 cells, and a
high-affinity IFNα-MCF-7 fusion molecule IMBP was constructed. Our
aim was to investigate whether this reconstructed cytokine had
enhanced cell growth inhibition activity compared to the wild-type
IFNα.
Materials and methods
Cell culture
Human breast cancer cell line MCF-7, lung cancer
cell line A549 and prostate cancer cell line PC-3 were kept in our
lab. Human embryonic kidney 293T cell line was used for lentiviral
packaging. All cell lines were routinely cultivated in Dulbecco's
modified eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) with
10% fetal bovine serum (Invitrogen), 100 U/ml penicillin and 100
μg/ml streptomycin (Invitrogen) at 37°C in a humidified
atmosphere containing 5% CO2.
Phage display library screening of breast
cancer binding peptides
Phage display library (Ph.D.-12 library, #E8110S,
New england Biolabs, Ipswich, MA, USA) was employed to screen a
short 12-peptide that specifically binds to MCF-7 breast cancer
cells (12,13). Briefly, 1×1011 phage in
1 ml DMEM medium was incubated with 1×106 MCF-7 cells at
room temperature (RT) for 1 h to develop phage-cell complexes.
After binding, cells were washed 5 times with 5 ml TBST [TBS (10
mmol/l Tris pH 7.5, 150 mmol/l NaCl) containing 0.1% Tween-20] to
remove the unbound phages. Cells were collected by centrifuging at
3200 × g. The surface-bound phages were eluted in 1 ml elution
buffer [0.2 N Glycine-HCl (pH 2.2), 1 mg/ml BSA] for 20 min and was
neutralized by 150 μl 1 N Tris-HCl, pH 9.0. The eluted
phages were reproduced by infecting E. coli ER2738 and
purified using polyethylene glycol (PEG)-8000/NaCl solution (20%
PEG-8000 and 2.5 N NaCl).
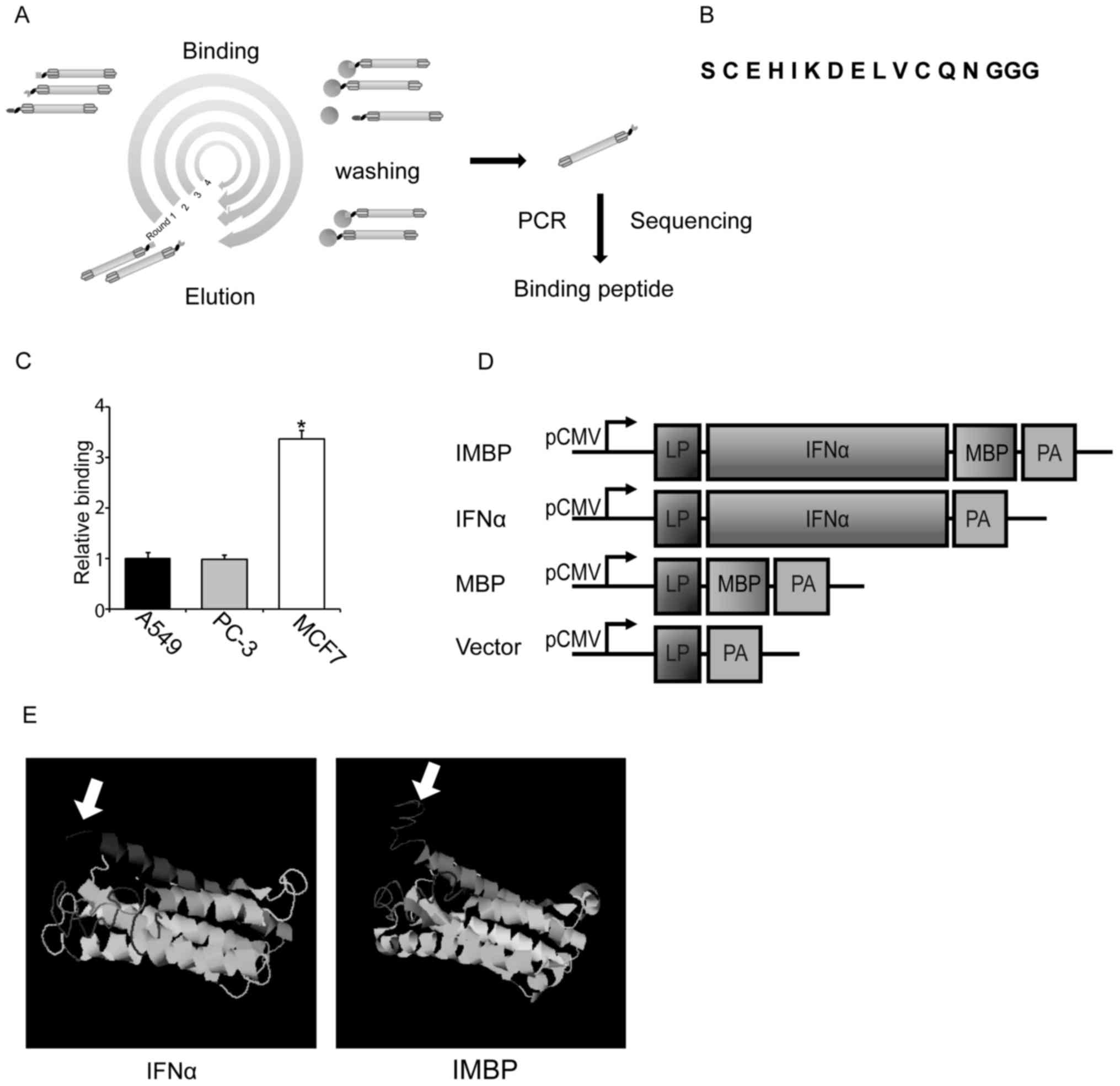
After 4 rounds of surface panning, the DNA sequences
of MCF-7-binding peptide (MBP) were amplified by PCR (Fig. 1A). Primers used for PCR
amplification were as follows: 5′-CTTTAGTGGTACCTTTCTATTCTCGAGTCT-3′ (forward primer
with Xho I) and 5′-CTTTCAACAGTTTCGTCTAGAACCTCCACC-3′ (reverse
primer with XbaI). The PCR production was purified from 3%
agrose gel and cloned into pJet vector using CloneJET PCR Cloning
kit (Thermo Fisher Scientific, Waltham, MA, USA) for sequencing and
constructing the engineered IFNα molecules.
Phage-ELISA assay
Phage-ELISA was carried out to determine the binding
affinity of the isolated MBP. Approximately 2×104 MCF-7
cells were collected into a 96-well V-bottom tissue culture plate
and incubated with blocking buffer (5% BSA/PBS, Sigma, Shanghai,
China) at 37°C for 1 h. For comparison, A549 lung cancer and PC-3
prostate cancer cells were used as negative controls. Cells were
pelleted by centrifuging at 3,200 × g and resuspended in 100
μl PBS buffer (Thermo Fisher Scientific, Beijing, China)
containing 1×109 phage particles. After incubated at RT
for 1 h, cells were pelleted and washed with PBST for 5×5 min. For
quantitation, cells were incubated with 100 μl PBS buffer
containing HRP conjugated anti-M13 monoclonal antibody (1:5,000, GE
Healthcare, Piscataway, NJ, USA) at 37°C for 45 min. Cells were
centrifuged and washed by PBST for 5×5 min. Then 100 μl of
TMB solution (BD Biosciences, San Jose, CA, USA) was added to each
well and the plate was kept in the dark for 15 min until blue color
was developed. Reaction was stopped by adding 50 μl 1 N
sulfuric acid. The absorbance of 450 nm and 630 nm were measured by
microplate reader (Bioteck, Beijing, China). Final optical density
(OD) was designated as
OD450-OD630-ODblank.
Recombinant plasmid construction
After the phage library screening and sequencing,
one of the MBP sequence was selected for the following cell
studies. We ligated MBP to the C-terminus of IFNα to construct an
engineered synthetic IFNα fusion protein. The fusion protein was
generated by ligating the MBP sequence into IFNα vector at the
XhoI/XbaI restriction sites and was designated as
IMBP. The wild-type IFNα expression plasmid was prepared as
previous described (14,15) and kept in our laboratory. The
vector containing the isolated MBP sequence and the empty
lentivirus vector were constructed in parallel and used as control
groups. All plasmid constructs were confirmed by DNA
sequencing.
The putative structure of the IMBP
The putative structure of the synthetic IMBP was
predicted using online I-TASSER server (http://zhanglab.ccmb.med.umich.edu).
Lentivirus production
293T cells were seeded into a 6-well plate at a
proper density 24 h pre-transfection. Lentiviruses were packaged by
the co-transfection of the plasmids constructed above and
pSPAX2/pMD2.G packing vectors (System Biosciences, Mountain View,
CA, USA) using Lipofectamine 3000 reagent (Thermo Fisher
Scientific). The viral supernatants were collected at 24 and 48 h
post-transfection and used for cell transfection as previously
described (16,17).
Infection of recombinant lentivirus
Approximately 2×105 MCF-7 cells were
plated into a 6-well plate 24 h pre-transfection. Cells were
infected with lentiviruses carrying MBP, IFNα, IMBP and empty
vector DNA in the presence of 8 μg/ml polybrene (Sigma).
After two rounds of infection with lentiviruses at 37°C for 24 h,
the tissue culture medium was replaced with fresh complete medium.
Three days after viral infection, the stable MCF-7 cells were
selected by puromycin and used for the following cell
experiments.
Detection of the secreted IMBP fusion
protein
On day 5 post-transduction, 100 μl
supernatant of MCF-7 cells was collected and the secreted IFNα and
IMBP fusion protein were quantitated with IFNα assay kit according
to the manufacture's instruction (Cusabio, Hubei, China).
Western blotting
Protein were extracted with RIPA buffer (KeyGEN
Biotech, Jiangsu, China) supplemented with cocktail protease
inhibitor (Roche, Shanghai, China) and quantified with BCA protein
assay kit (Beyotime Biotechnology, Jiangsu, China). Whole cell
lysates were resolved on 5–10% or 5–12% SDS polyacrylamide gel
electrophoresis (SDS-PAGE). To detect the secreted IFNα and IMBP
proteins in cell supernatants, on day 5 post-transduction, 100
μl MCF-7 cell supernatant was collected and condensed to 20
μl in a vacuum-freeze dryer (Boyikang, Beijing, China).
Solutions were resolved on Mini-PROTEIN TGX gradient gel (Bio-Rad,
Beijing, China). Proteins were transferred to 0.45 μm PVDF
membranes (Roche) and immunoblotted at 4°C overnight or at RT for 1
h with the following antibodies: IFNα (1:1,000, Abcam, Shanghai,
China), STAT1 (phospho Y701, 1:1,000, Abcam), Cleaved CASP-9
(1:1,000, Cell Signaling Technology, Beijing, China), p53 (1:1,500,
Santa Cruz Biotechnology, Santa Cruz, CA, USA), p21 (1:500, Santa
Cruz Biotechnology), CDK2 (1:1500, Cell Signaling Technology),
Cyclin A (1:1,500, Abcam) and β-actin (1:3,000, Santa Cruz
Biotechnology). Membranes were incubated with horseradish
peroxidase-conjugated secondary antibody (1:3,000, ZSGB-BIO,
Beijing, China) at 37°C for 1 h before chemiluminescence reading.
Protein expression levels were determined semi-quantitatively by
densitometric analysis with the Quantity One software (Bio-Rad).
Western blotting was performed in triplicate, and data showed a
representative finding of these triplicate analyses.
Cell binding assay of IMBP
The binding affinity of the engineered hybrid
molecule to the MCF-7 cell membrane was measured by FACS. MCF-7
cells were collected and stained with Trypan blue to make sure that
viable cells were more than ninety percent. Approximately
1×106 normal MCF-7 cells were incubated with equal
amount of the secreted IFNα or IMBP at 37°C for 1 h. After washed
with PBS, cells were incubated with FITC-conjugated IFNα antibody
(PBL, Piscataway, NJ, USA). The FITC-conjugated mouse IgG (Abcam)
was used as the isotype control. Cells were analyzed using BD LSRII
Fortesa flow cytometer (BD Biosciences) and FlowJo software
(FlowJo, OR, Ashland, USA) to calculate the fluorescence intensity
(14,15).
Cell viability assay
On day 7 post-transfection, approximately
5×103 viable stable MCF-7 cells transduced by lentivirus
carrying IMBP, IFNα, MBP and empty vector DNA were seeded into a
96-well plate 24 h before cell viability assay. Then, cell growth
was analyzed by WST-1 Cell Proliferation Reagent (Roche). According
to the manufacturer's instructions, 20 μl WST-1 reagent was
added to 200 μl cell culture medium and incubated at 37°C in
the dark for 2 h. The absorbances of 450 and 630 nm were measured
with microplate reader (Bioteck). Final OD was designated as
OD450-OD630-ODblank.
Cell cycle assay
On day 7 post-transfection, 1×106 stable
MCF-7 cells were collected and washed by cold PBS. Cells were
resuspended in 1 ml fixation solution (300 μl PBS and 700
μl ethanol). After incubation at 4°C for 4 h, cells were
centrifuged and the fixation solution was removed. After washed
twice with PBS, cells were pelleted, stained with 0.5 ml propidium
iodide (PI, Sigma) staining solution [50 μg/ml PI, 20
μg/ml RNase A (Takara, Liaoning, China) and 0.2% Triton
X-100 (Sigma)], and incubated in the dark at 37°C for 10 min. Cell
suspensions were filtered with a 400-mesh sieve. Cell cycle was
analyzed by BD LSRII Fortesa flow cytometer.
Flow cytometry for cell apoptosis
Annexin V-FITC and PI double staining flow cytometry
analyses were used for cell apoptosis assay. Approximately
1×106 stable MCF-7 cells were collected and washed three
times with cold PBS and binding buffer. Then cells were stained
with Annexin V-FITC and PI (Annexin V-FITC Apoptosis Detection kit,
BD Bioscience) for apoptosis detection. Briefly, MCF-7 cells were
first resuspended in 1 ml binding buffer. Then, 5 μl of
Annexin V-FITC was added to the tubes, and cells were incubated for
10 min at RT followed by the addition of 5 μl PI. After 15
min incubation in PI buffer at RT, cells were immediately analyzed
with a flow cytometer (BD Biosciences). The cells in the different
portions represented the different cell states as follows: the
late-apoptotic cells were present in the upper right portion, the
viable cells were present in the lower left portion, and the early
apoptotic were cells present in the lower right portion.
Treatment with chemotherapeutic drug
doxorubicin
The anticancer drug doxorubicin (DOX, Sigma) was
dissolved in water. We harvested 12 ml of cell supernatant
containing the secreted interferons from the stable MCF-7 cells
transduced by lentivirus carrying IMBP, IFNα, MBP and empty vector.
The cell supernatant was centrifuged at 3,500 × g for 5 min to
remove cell debris. Then the supernatant was successively passed
through two centrifugal filter units with MW cut-off 30 and 10 kDa
(Millipore, Temecula, CA, USA) to remove large and small molecules.
The interceptions were dissolved in 6 ml DMEM complete medium and
kept in 4°C. Normal MCF-7 cells were seeded into 6-well plates at a
density of 3×105 cells/well. After 24 h, cell medium was
changed to the interception-DMEM supplemented with 0.1 μg/ml
DOX. MCF-7 cells were treated with this therapeutic medium for 3
days, with the medium changed every day. After that, cells were
harvested and applied for the next step of cell viability assay or
molecule detection.
Real-time PCR
Total cellular RNA was isolated using an Eastep
Super total RNA isolation kit (Promega, Beijing, China).
First-strand cDNA was synthesized from 800 ng of total RNA using
Transcriptor First Strand cDNA Synthesis kit (Roche). Real-time PCR
was performed using an aliquot of first-strand cDNA as a template
in a 20 μl reaction system containing 10 μl 2X SYBR
premixed buffer (Roche), 2 μl forward and reverse primers.
The primers were as follows: IFIT1 (interferon induced protein with
tetratricopeptide repeats) sense 5′-TCTCAGAGGAGCCTGGCTAA-3′, IFIT1
antisense 5′-CCAGACTATCCTTGACCTGATGA-3′; MX1 (MX dynamin-like
GTPase 1) sense: 5′-CTTTCCAGTCCAGCTCGGCA-3′, MX1 antisense:
5′-AGCTGCTGGCCGTACGTCTG-3′); STAT1 sense:
5′-GGCACCAGAACGAATGAGGG-3′, STAT1 antisense:
5′-CCATCGTGCACATGGTGGAG-3′ (18);
IFITM1 (interferon-induced transmembrane protein 1) sense:
5′-ACAGGAAGATGGTTGGCGAC-3′, IFITM1 antisense:
5′-ATGGTAGACTGTCACAGAGC-3′; β-actin sense:
5′-TCACCCACACTGTGCCCATCTACGA-3′, β-actin antisense:
5′-CAGCGGAACCGCTCATTGCCAA TGG-3′ (19). The PCR amplification process was
one cycle at 95°C 10 min, 40 cycles at 95°C for 10 sec and 60°C for
30 sec (ABI StepOnePlus, Beijing, China).
Statistical analysis
All data and results were calculated from at least
three replicate measurements and presented as means ± SD. The
significance was determined by SPSS 20.0 (IBM). Student's t-test
was used to compare statistical differences for variables among
treatment groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Construction of the IFNα/MCF-7-binding
peptide fusion protein
In order to enhance the antitumor activity of IFNα
in breast cancer therapy, we used a NEB Ph.D.-12 peptide library to
select a short peptide with 12 amino acids that specifically binds
to the cell membrane of MCF-7 cells. Our aim is to add a short,
high-affinity peptide to the C-terminus of IFNα and construct an
IFNα-MCF-7 fusion molecule (IMBP), which may facilitate the binding
of IFNα to the MCF-7 cell membranes and enhance the cell growth
inhibition activity of IFNα.
According to the manual of NEB Ph.D.-12 peptide
library, MCF-7 cells were incubated with 1×1011 phage
particles in DMEM tissue culture medium. The unbounded phage
particles were stripped off by stripping buffer. The bound phages
on the cell membrane were eluted and recovered for the second round
of screening. After four rounds of screening, the phages
specifically binding to MCF-7 cells were enriched and identified
(Fig. 1A). After cloning and
sequencing, one sort of phage encodes short peptide with the
sequence of 'C E H I K D E L V C Q N' was selected for further cell
studies (Fig. 1B). Using
phage-ELISA, we showed that this phage displayed short peptide was
able to bind preferentially to MCF-7 cells compared to A549 lung
cancer and PC-3 prostate cancer cells (Fig. 1C).
In order to examine the role of this short peptide
and compare the antitumor activity of wild-type IFNα and IMBP, we
linked the short peptide to the C-terminus of IFNα and the IMBP
fusion molecule was synthesized (Fig.
1D). Then we used software on the website (http://zhanglab.ccmb.med.umich.edu) to predict
the protein structure of the synthetic IMBP. The structure of
wild-type IFNα and the putative structure of IMBP is illustrated in
Fig. 1E. Arrow indicates the short
peptide fused into wild-type IFNα.
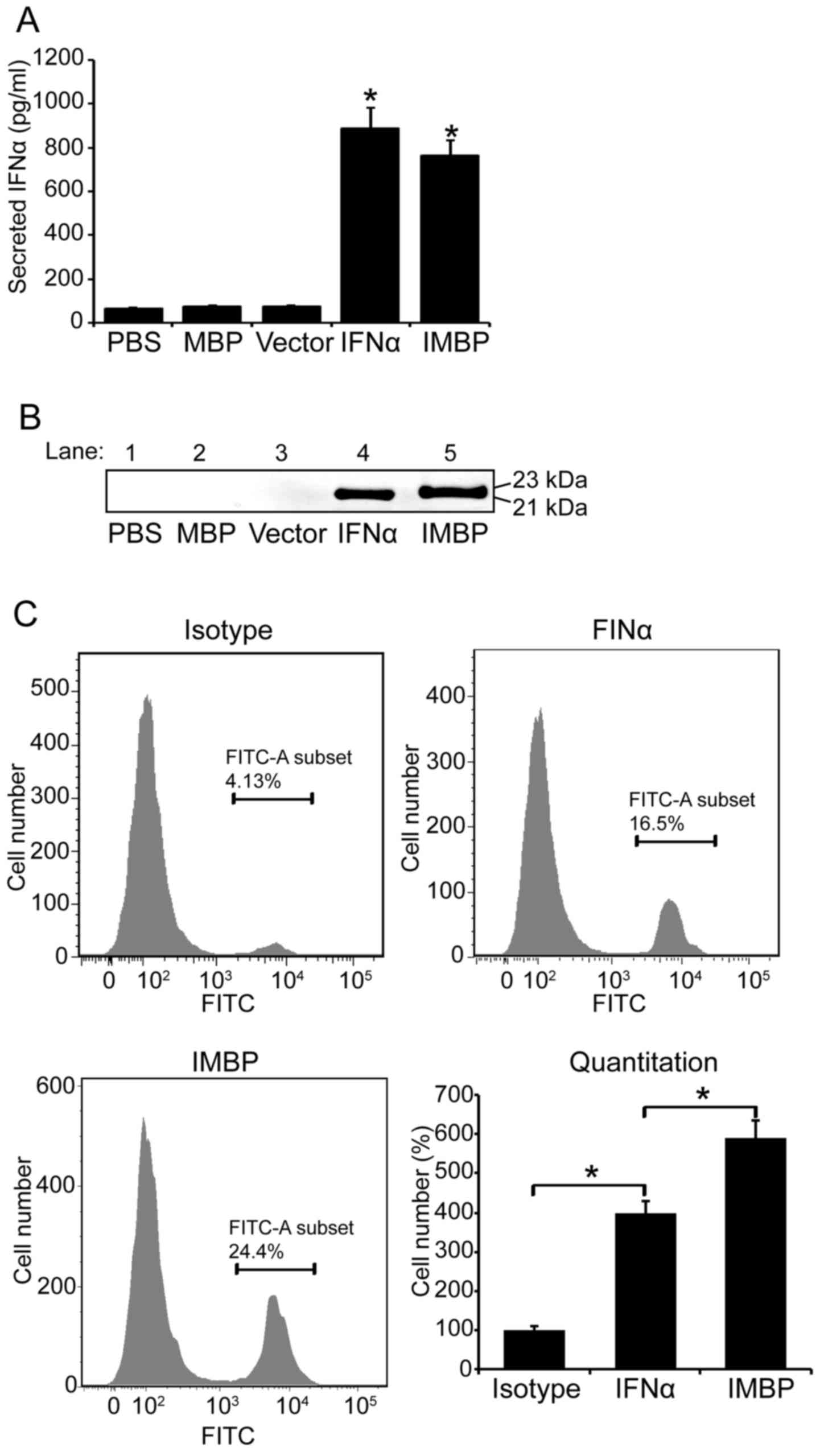
Detection of the secreted IFNα and IMBP
fusion protein
We used 293T cells to produce lentivirus carring the
DNA sequences of IMBP, wild-type IFNα, MBP and empty vector. Then,
MCF-7 cells were transducted by lentiviruses and the stable cells
were selected with puromycin. On day 5 post-transduction, 100
μl supernatant of MCF-7 cells was collected and the secreted
IFNα and IMBP fusion protein were quantitated with IFNα assay kit.
Based on the result of IFNα ELISA, the concentration of secreted
wild-type IFNα in MCF-7 supernatant was 885 pg/ml and that of the
IMPB fusion molecule was 763 pg/ml (Fig. 2A). We further used western blotting
to confirm the secretion of wild-type IFNα and IMPB fusion
molecule. The blots are shown in Fig.
2B.
Since the phage particles bind to MCF-7 cell
membrane specifically, we used FACS to further examine whether the
IMBP fusion protein binds to MCF-7 cell surface easier than the
wild-type IFNα. Therefore, FITC conjugated-IFNα antibody was used
in FACS assay and the binding affinity of the two proteins was
quantitatively evaluated according to the fluorescence intensity in
flow cytometry (Fig. 2C).
Quantitative analysis result showed that the binding affinity of
the IMBP fusion protein was 1.5-fold increase compared to the
wild-type IFNα (P<0.05), and the binding ability of both the
wild-type IFNα and the IMBP fusion protein was significantly higher
than the isotype control (Fig. 2C,
P<0.05).
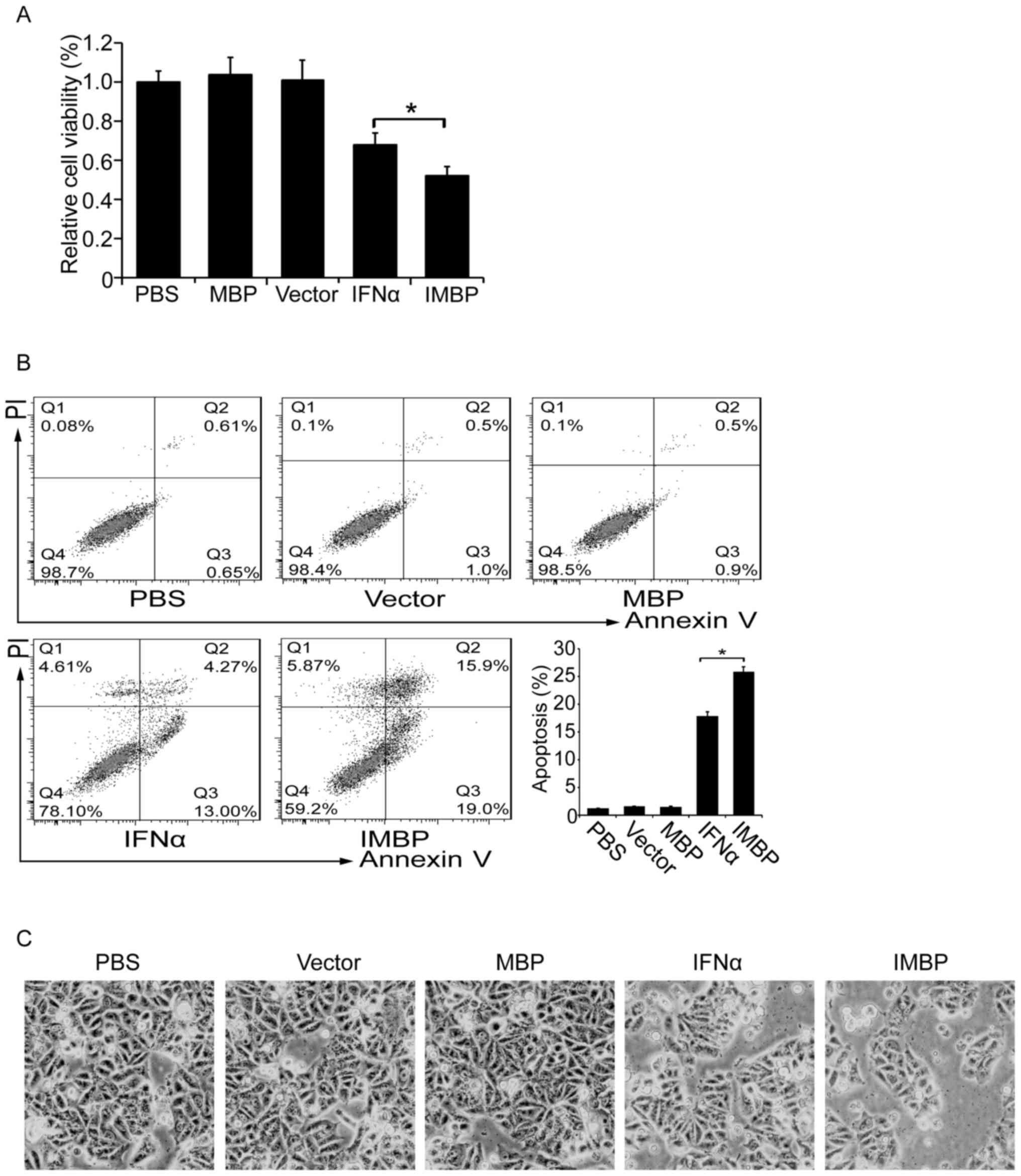
Synthetic IMBP fusion protein inhibits
cell growth of MCF-7 cells
We compared the cell growth inhibition effect
between the IMBP and the wild-type IFNα. MCF-7 breast cancer cells
were transfected with lentiviruses carrying IMBP, IFNα, MBP and
empty vector, respectively. Cell proliferation was determined by
WST-1 assay. We found that although both the secreted IFNα and IMBP
inhibited the growth of MCF-7 cells, IMBP was superior to IFNα
(Fig. 3A, P<0.05).
Then we examined cell apoptosis of the stable
transfected MCF-7 cells with flow cytometry. As was shown in
Fig. 3B, we found that the
transfection of both IFNα and IMBP induced an increased apoptosis
ratio, but the IMBP group showed a significantly higher apoptosis
ratio than the IFNα group (26 vs. 18%, P<0.05). The cell
morphology also showed a similar result (Fig. 3C).
IMBP potentiates the therapeutic efficacy
of doxorubicin-based chemotherapy
Since doxorubicin is one of widely used
anthracyclines to treat breast cancer, we examined the synergistic
effect on cell killing by the combined use of DOX and IMBP fusion
molecule. First, using WST-1 cell proliferation assay, we tested a
serial working concentration of DOX including 0.02, 0.05, 0.1, 0.2,
0.5 and 1.0 μg/ml for the cell growth inhibition of MCF-7
cells. We selected a relative low concentration of 0.1 μg/ml
of DOX with a cell growth inhibition ratio approximately 25% for
the chemotherapy (data not shown). Then, we investigated the cell
killing ability of the co-administration of DOX/IFNα or DOX/IMBP.
The WST-1 assay result showed that cell proliferation in both the
DOX/IFNα and DOX/IMBP treatment groups was apparently inhibited
compared to the PBS control group and the DOX group. Of note, cell
killing ability of the co-administration of DOX and IMBP was
superior to the co-administration of DOX and IFNα (Fig. 4A, 48 vs. 32%, P<0.05).
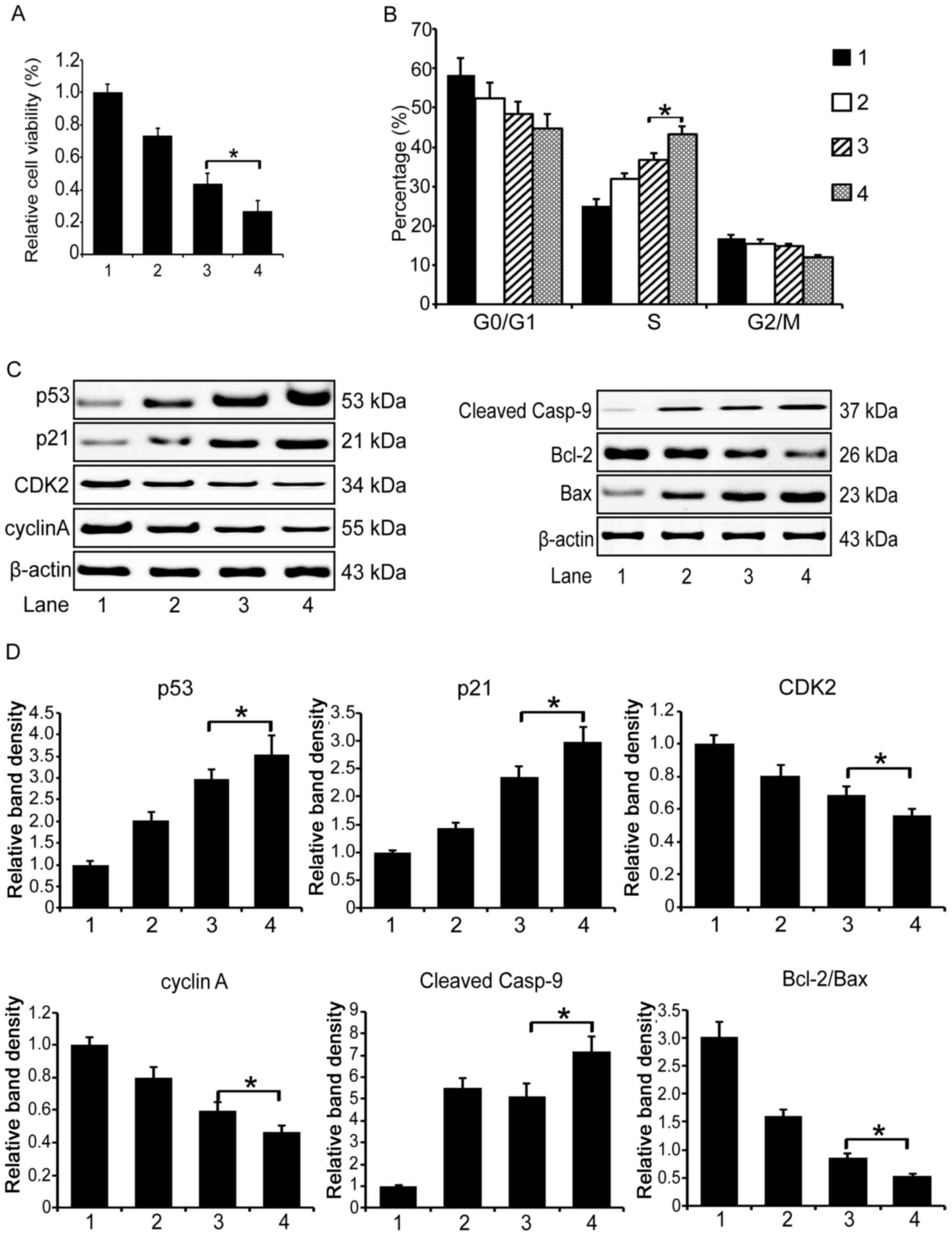
Then we examined the cell cycle distribution in the
treated MCF-7 cells with flow cytometry. We found that the
co-administration apparently induced an S phase cell cycle arrest,
and the DOX/IMBP treatment group showed a more obvious arrest
effect than the DOX/IFNα group (Fig.
4B, 43 vs. 37%, P<0.05).
To unveil the underlying molecular mechanism that
explains the enhanced therapeutic efficacy of the combined use of
DOX and IMBP in chemotherapy, we first examined the expression of
the cell cycle pathway related genes by western blotting. We found
that the expression of p53 and p21 in the DOX/IMBP chemotherapy
group was dramatically up-regulated compared to the DOX/IFNα group;
the expression of CDK2 and cyclin A was dramatically down regulated
(Fig. 4C left panel and D,
P<0.05). We then examined the expression of the cell apoptosis
pathway-related genes and found that the caspase 9 and Bcl-2/Bax
apoptosis pathways were activated (Fig. 4C right panel and D, P<0.05).
IMBP fusion molecule activates the STAT1
pathway
To further delineate the mechanism of the enhanced
activity of the combination chemotherapy, we used real-time PCR to
examine the expression of the interferon pathway genes in the
treated MCF-7 cells. The quantitation result showed that both the
DOX/IMBP and DOX/IFNα co-administration activate several inducible
genes of the STAT1 pathway, including STAT1, IFIT1, IFITM1, and
MX1. However, DOX/IMBP group showed a stronger activation effect
than the DOX/IFNα group (Fig. 5A).
The western blot analysis result also showed an activation of the
phosphorylated STAT1 (Y701, Fig.
5B).
Discussion
Anthracyclines, one type of anticancer therapy, are
commonly used to treat both early and metastatic breast cancer;
however, their toxicity, especially cardiac toxicity, is high
(20,21). Additionally, frequent use of single
therapeutic agent may result in chemoresistance, which is a major
obstacle to the successful treatment of breast cancer. To minimize
the toxicity of DOX and maximize the therapeutic efficiency of DOX,
a combination therapeutic strategy is important.
The functions of IFNs are represented by three major
biological activities, antiviral activity, antitumor activity and
immunoregulatory activity (22).
Although IFNα is widely used in the treatment of various types of
cancer, its antineoplastic potency for breast cancer is low.
Therefore, improvements in this cytokine are valuable and further
investigation is still needed.
In our previous study, using a cDNA in-frame library
screening approach, we identified a short peptide derived from
placental growth factor-2 (PLGF-2). We demonstrated that fusing
this short peptide to IFNα and IFNγ induced greater activity than
the wild-type counterparts in inhibiting of tumor cell growth,
invasion and colony formation (14,15).
In this study, using a Ph.D.-12 peptide library, we selected a
short peptide that specifically binds to the cell membrane of MCF-7
cells, and we synthesized a high-affinity IFNα-MCF-7 fusion
molecule (IMBP). Using lentiviral DNA delivery system, we obtained
a stable MCF-7 breast cancer cell line that secretes IMBP. Compared
with wild-type IFNα, the IMBP fusion molecule demonstrated higher
antitumor activity potency in the inhibition of cell growth,
promotion of cell cycle arrest and induction of cell apoptosis.
Recently, using the same approach, we also identified a short
peptide that specifically binds to Jurkat T lymphocyte leukemia
cells (JBP). We demonstrated that a JBP and IFNα fusion protein
(IFNP) also had significantly better antitumor activity than
wild-type IFNα (unpublished data). Through this series study, we
evaluated the value of engineering an IFNα fusion molecule, which
may serve as a novel antitumor agent in cancer treatment.
IFNs bind to receptors on the cell surface, activate
specific signaling pathways and exert antitumor actions (23). Unfortunately, we still do not know
the function of how this Ph.D.-12 peptide binds to the cell
membrane. According to the result of 'Blastp' on the NCBI website,
this 12-peptide is not homologous to any existing protein
sequences. We speculate that it may bind to some unknown
receptor(s) or biomacromolecule(s) on the surface of the cell
membrane. Using a cell binding assay, we demonstrated that MBP
promoted the adherence affinity of IFNα to its receptor (Fig. 2C), which is probably why we
detected a lower level of free IMBP in the cell supernatant
(Fig. 2A). We speculate that
because more IMBP molecules adhere to the cell surface of MCF-7
cells than IFNα, the IMBP possesses a higher tumor cell killing
ability than wild-type IFNα.
The STAT pathway plays a critical role in the
anti-infection role of IFNs (24–27).
When binding to its receptor on the cell surface, IFNα initiates
the phosphorylation of TYK2 and JAK1 kinases, which is followed by
the activation of STAT family transcription factors (28,29).
Several IFN inducible genes can be co-activated in the STAT
pathway, including IFIT1, IFIT3, MX1, IFITM1, OAS1
(2′–5′-oligoadenylate synthetase 1), PARP9 and PARP12
[poly(ADP-ribose) polymerase family] (30). It was reported that IFIT3 promotes
IFNα adjuvant therapeutic effects by strengthening IFNα effector
signaling in hepatocellular carcinoma patients (31); IFNγ inhibits the growth of human
liver cancer cells through activating IFITM1, which enhances the
transcriptional activity of p53 and stabilizes the p53 protein by
inhibiting p53 phosphorylation on Thr55 (19).
The JAK-STAT signaling pathway also plays an
important role in doxorubicin-based breast cancer chemotherapy
(32). It was demonstrated that
the combined use of doxorubicin and IFNs may have a significant
synergistic effect. Thomas et al showed that doxorubicin
potentiates STAT1 activation in response to IFNγ, and the
combination of doxorubicin and IFNγ enhances apoptosis in breast
cancer cells (33). Hannesdóttir
et al reported that STAT1 is crucial to the susceptibility
of breast cancer cells to chemotherapeutic drugs (e.g.,
doxorubicin) by contributing to the induction of a productive
antitumor immune response, which is based on IFNγ-producing T cells
(34). In this study, we found
that a combination of low dose DOX antitumor drug (0.1
μg/ml) and endogenous secretory IFNα had an advantage over
the single use of either DOX or IFNα in inhibiting breast cancer
cell growth. Of note, we found that low dose DOX combined with
synthetic IMBP fusion protein had a significantly better
therapeutic effect than the DOX/IFNα combination. When we examined
the molecular mechanism of this enhanced antitumor activity, we
observed that the DOX/IMBP combination activated cell cycle arrest
and cell apoptosis pathway genes more efficiently than the DOX/IFNα
combination (Fig. 4), which was
also correlated with the activation of STAT1 pathway target genes
(Fig. 5).
In summary, by using a phage library screening, we
have identified a short peptide that specifically binds to MCF-7
breast cancer cells. Fusion of this short peptide to IFNα
significantly enhanced the antitumor activity. The combined use of
DOX and IMBP fusion protein potentiated the effectiveness of
chemotherapy. Since using lentivirus to deliver molecules into
cells is hard to apply in clinical gene therapy, construction of an
in vitro expression and purification system to produce
purified IMBP recombinant protein is needed in our further study.
We consider this type of purified IMBP molecule may have good
clinical application prospect and the co-administration of DOX and
IMBP provides new insight into breast cancer treatment.
Acknowledgments
The present study was supported by grants from the
National Science Foundation of China (81302380, to D.-H.Y.) and
Grant from the Health and Family Planning Commission in Jilin
Province of China (2016Q035, to L.Z.).
References
|
1
|
Roychowdhury S and Caligiuri MA: Cytokine
therapy for cancer: Antigen presentation. Cancer Treat Res.
123:249–266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kotredes KP and Gamero AM: Interferons as
inducers of apoptosis in malignant cells. J Interferon Cytokine
Res. 33:162–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gutterman JU: Cytokine therapeutics:
Lessons from interferon alpha. Proc Natl Acad Sci USA.
91:1198–1205. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lamm D, Brausi M, O'Donnell MA and Witjes
JA: Interferon alfa in the treatment paradigm for
non-muscle-invasive bladder cancer. Urol Oncol. 32:e21–e30. 2014.
View Article : Google Scholar
|
|
5
|
Whelan J, Patterson D, Perisoglou M,
Bielack S, Marina N, Smeland S and Bernstein M: The role of
interferons in the treatment of osteosarcoma. Pediatr Blood Cancer.
54:350–354. 2010. View Article : Google Scholar
|
|
6
|
Wang L, Jia D, Duan F, Sun Z, Liu X, Zhou
L, Sun L, Ren S, Ruan Y and Gu J: Combined anti-tumor effects of
IFN-α and sorafenib on hepatocellular carcinoma in vitro and in
vivo. Biochem Biophys Res Commun. 422:687–692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simonsson B, Hjorth-Hansen H, Bjerrum OW
and Porkka K: Interferon alpha for treatment of chronic myeloid
leukemia. Curr Drug Targets. 12:420–428. 2011. View Article : Google Scholar
|
|
8
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:e359–e386. 2015.
View Article : Google Scholar
|
|
9
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ramos MC, Mardegan MC, Tirone NR, Michelin
MA and Murta EF: The clinical use of type 1 interferon in
gynecology. Eur J Gynaecol Oncol. 31:145–150. 2010.PubMed/NCBI
|
|
11
|
Wang BX, Rahbar R and Fish EN: Interferon:
Current status and future prospects in cancer therapy. J Interferon
Cytokine Res. 31:545–552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X and Mao C: Using phage as a platform
to select cancer cell-targeting peptides. Methods Mol Biol.
1108:57–68. 2014. View Article : Google Scholar
|
|
13
|
Cao B, Yang M and Mao C: Phage as a
genetically modifiable supramacromolecule in chemistry, materials
and medicine. Acc Chem Res. 49:1111–1120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin H, Chen N, Guo R, Wang H, Li W, Wang
G, Cui J, Jin H and Hu JF: Antitumor potential of a synthetic
interferon-alpha/PLGF-2 positive charge peptide hybrid molecule in
pancreatic cancer cells. Sci Rep. 5:169752015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Chen N, Yin H, Zhang L, Li W, Wang
G, Cui J, Yang B and Hu JF: A placental growth factor-positively
charged peptide potentiates the antitumor activity of
interferon-gamma in human brain glioblastoma U87 cells. Am J Cancer
Res. 6:214–225. 2016.PubMed/NCBI
|
|
16
|
Sun J, Li W, Sun Y, Yu D, Wen X, Wang H,
Cui J, Wang G, Hoffman AR and Hu JF: A novel antisense long
noncoding RNA within the IGF1R gene locus is imprinted in
hematopoietic malignancies. Nucleic Acids Res. 42:9588–9601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Li W, Guo R, Sun J, Cui J, Wang G,
Hoffman AR and Hu JF: An intragenic long noncoding RNA interacts
epigenetically with the RUNX1 promoter and enhancer chromatin DNA
in hematopoietic malignancies. Int J Cancer. 135:2783–2794. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi HJ, Lui A, Ogony J, Jan R, Sims PJ
and Lewis-Wambi J: Targeting interferon response genes sensitizes
aromatase inhibitor resistant breast cancer cells to
estrogen-induced cell death. Breast Cancer Res. 17:62015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang G, Xu Y, Chen X and Hu G: IFITM1
plays an essential role in the antiproliferative action of
interferon-gamma. Oncogene. 26:594–603. 2007. View Article : Google Scholar
|
|
20
|
Khasraw M, Bell R and Dang C: Epirubicin:
Is it like doxorubicin in breast cancer? A clinical review Breast.
21:142–149. 2012.
|
|
21
|
Damiani RM, Moura DJ, Viau CM, Caceres RA,
Henriques JA and Saffi J: Pathways of cardiac toxicity: Comparison
between chemotherapeutic drugs doxorubicin and mitoxantrone. Arch
Toxicol. 90:2063–2076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chelbi-Alix MK and Wietzerbin J:
Interferon, a growing cytokine family: 50 years of interferon
research. Biochimie. 89:713–718. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parker BS, Rautela J and Hertzog PJ:
Antitumour actions of interferons: Implications for cancer therapy.
Nat Rev Cancer. 16:131–144. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szelag M, Piaszyk-Borychowska A,
Plens-Galaska M, Wesoly J and Bluyssen HA: Targeted inhibition of
STATs and IRFs as a potential treatment strategy in cardiovascular
disease. Oncotarget. 7:48788–48812. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Merches K, Khairnar V, Knuschke T,
Shaabani N, Honke N, Duhan V, Recher M, Navarini AA, Hardt C,
Häussinger D, et al: Virus-induced type I interferon deteriorates
control of systemic pseudomonas aeruginosa infection. Cell Physiol
Biochem. 36:2379–2392. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heim MH and Thimme R: Innate and adaptive
immune responses in HCV infections. J Hepatol. 61(Suppl 1):
S14–S25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu Y, Jia H, Chen J, Cui G, Gao H, Wei Y,
Lu C, Wang L, Uede T and Diao H: Decreased osteopontin expression
as a reliable prognostic indicator of improvement in pulmonary
tuberculosis: impact of the level of interferon-gamma-inducible
protein 10. Cell Physiol Biochem. 37:1983–1996. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao LJ, He SF, Liu Y, Zhao P, Bian ZQ and
Qi ZT: Inhibition of STAT pathway impairs anti-hepatitis C virus
effect of interferon alpha. Cell Physiol Biochem. 40:77–90. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takaoka A and Yanai H: Interferon
signalling network in innate defence. Cell Microbiol. 8:907–922.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Legrier ME, Bièche I, Gaston J, Beurdeley
A, Yvonnet V, Déas O, Thuleau A, Château-Joubert S, Servely JL,
Vacher S, et al: Activation of IFN/STAT1 signalling predicts
response to chemotherapy in oestrogen receptor-negative breast
cancer. Br J Cancer. 114:177–187. 2016. View Article : Google Scholar :
|
|
31
|
Yang Y, Zhou Y, Hou J, Bai C, Li Z, Fan J,
Ng IOL, Zhou W, Sun H, Dong Q, et al: Hepatic IFIT3 predicts
interferon-α therapeutic response in patients of hepatocellular
carcinoma. Hepatology. Mar 13–2017.Epub ahead of print. View Article : Google Scholar
|
|
32
|
Lee SC, Xu X, Lim YW, Iau P, Sukri N, Lim
SE, Yap HL, Yeo WL, Tan P, Tan SH, et al: Chemotherapy-induced
tumor gene expression changes in human breast cancers.
Pharmacogenet Genomics. 19:181–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thomas M, Finnegan CE, Rogers KM, Purcell
JW, Trimble A, Johnston PG and Boland MP: STAT1: A modulator of
chemotherapy-induced apoptosis. Cancer Res. 64:8357–8364. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hannesdóttir L, Tymoszuk P, Parajuli N,
Wasmer MH, Philipp S, Daschil N, Datta S, Koller JB, Tripp CH,
Stoitzner P, et al: Lapatinib and doxorubicin enhance the
Stat1-dependent antitumor immune response. Eur J Immunol.
43:2718–2729. 2013. View Article : Google Scholar : PubMed/NCBI
|