Introduction
The incidence and mortality associated with
hepatocellular carcinoma (HCC) has increased in China, with
>400,000 patients succumbing to the disease (1). HCC is becoming a major public health
concern in China. Despite the use of modern therapies to improve
the outcomes of patients with HCC, distant metastasis and a high
rate of recurrence restrict the 5-year survival rate (2). Therefore, the identification of novel
anti-metastasis targets is urgently required to improve the
prognosis of patients with HCC patients.
MicroRNAs (miRNAs or miRs) are a class of
endogenous, highly conserved, short non-coding RNAs (3). They can bind to the 3′-untranslated
region (3′-UTR) of mRNAs to decrease target gene expression
(4). Various cellular processes
have been reported to be regulated by different miRNAs in HCC.
miR-218 has been shown to decrease tumor growth (5), miR-26a to inhibit metastasis
(6) and miR-214 to prevent
angiogenesis (7) in HCC. Moreover,
certain miRNAs, including miR-155 (8), miR-137 (9) and miR-135a (10) have been shown to be associated with
the recurrence of and poor survival in HCC. Hence, miRNAs have the
potential to act as biomarkers of and therapeutic targets in
HCC.
Mounting evidence indicates that miR-1271, a newly
identified miRNA, is downregulated in endometrial cancer, prostate
cancer and myeloma (11–13). The re-induction of miR-1271 has
been shown to inhibit proliferation in ovarian cancer (14) and invasion in pancreatic cancer
(15). However, Wang et al
(16) reported a promoting effect
of miR-1271 in the regulation of non-small cell lung cancer (NSCLC)
proliferation and invasion by targeting HOXA5. Thus, these data
seem to suggest that miR-1271 is a 'friend or foe' depending on the
cancer type.
The pathophysiological program that transforms
epithelial cell types into cells with a mesenchymal phenotype is
identified as epithelial-mesenchymal transition (EMT) (17). EMT frequently occurs in cancer
cells, particularly in metastatic cells. EMT contributes to the
progression of cancers from the initial to the advanced grade, and
is considered a critical mechanism for the migration, invasion and
distinct metastasis of cancer cells (18).
In the present study, we examined the functions of
miR-1271 in metastasis and EMT in HCC. We found that the
downregulation of miR-1271 was a common occurrence in HCC tissues.
The downregulation of miR-1271 was significantly associated with
venous infiltration, an advanced TNM stage and a short survival
time of patients with HCC. Furthermore, miR-1271 was demonstrated
to significantly inhibit tumor migration, invasion and EMT, as well
as the formation of lung metastatic clusters of HCC by targeting
protein tyrosine phosphatase type IVA member 1 (PTP4A1).
Materials and methods
Patients and cell lines
HCC tissues and matched non-tumor tissues were
collected from 65 patients who underwent radical resection for HCC
at the Department of Hepatobiliary Surgery, First Affiliated
Hospital of Xi'an Jiaotong University (Xi'an, China) between
January 2011 and January 2013. None of the patient received any
anticancer therapy prior to surgery. All patients underwent a
complete 3-year follow-up, unless they succumbed to the disease
before the end of the 3 years. Written informed consents were
obtained from all patients enrolled in this study. The use of
clinical samples was approved by the Ethics Committee of First
Affiliated Hospital of Xi'an Jiaotong University. In addition, 4
HCC cell lines (MHCC97H, SMMC7721, Huh-7 and Hep3B), and the human
immortalized normal hepatocyte cell line (LO2) were obtained from
the Shanghai Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences (Shanghai, China) and maintained in our
laboratory. The cells were cultured in Dulbecco's modified Eagle's
liquid medium (DMEM; Gibco, Carlsbad, CA, USA) containing 10% fetal
bovine serum (FBS; Gibco) with 1% v/c penicillin and streptomycin
(Sigma-Aldrich, St. Louis, MO, USA) in a humidified air containing
5% CO2 at 37°C.
Cancer Genome Atlas dataset
Pre-analyzed The Cancer Genome Atlas (TCGA) mature
microRNA RNA-Seq data of 271 HCC samples and 39 normal liver
samples were collected from the UCSC Xena (http://xena.ucsc.edu).
Lentiviruses infection
miR-1271 vectors (miR-1271), negative control
vectors (miR-ctrl), miR-1271 inhibitory vectors (anti-miR-1271) and
negative control inhibitory vectors (anti-miR-ctrl) were purchased
from GeneCopoeia (Guangzhou, China) and cloned into the pEZX-MR03
and pEZX-AM03 lentiviral expression vectors, respectively. MHCC97-H
and Hep3B cells were infected with recombinant lentiviruses at an
MOI of 10.
Plasmids and siRNA transfection
PTP4A1 plasmids was obtained from GeneCopoeia and
cloned into the pcDNA3.1 expression vectors (Invitrogen, Carlsbad,
CA, USA). PTP4A1 specific siRNA and negative control siRNA were
purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The
PTP4A1 expression vector or PTP4A1 siRNA were transfected into the
HCC cells using Lipofectamine 3000 (Invitrogen) according to the
manufacturer's instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cultured cells was
isolated using TRIzol reagent (Invitrogen). The Bulge-Loop miR-1271
qRT-PCR Primer Set (Guangzhou RiboBio) was used to determine
miR-1271 expression. PTP4A1 mRNA levels normalized to GAPDH were
measured by RT-qPCR using the SuperScrip III Reverse Transcriptase
kit (Invitrogen) and the iTaq Universal SYBR-Green Supermix kit
(Bio-Rad Laboratories, Hercules, CA, USA). The primer sequences
were as follows: PTP4A1 forward, 5′-ACCAATGCGACCTTAAC AAA-3′ and
reverse, 5′-ATCTGGTTGGATGGTGGTG-3′; and GAPDH forward,
5′-CCAGGGCTGCTTTTAACTCT-3′ and reverse, 5′-GGACTCCACGACGTACTCA-3′).
The thermocycling conditions are: Holding stage 50°C for 2 min,
95°C for 10 min; PCR stage (40 times) 95°C for 15 sec, 60°C for 1
min.
Western blot analysis
Western blot analysis was used to detect the
expression of PTP4A1, E-cadherin, N-cadherin, vimentin, c-Src, the
phosphorylation of c-Src (Y416), Snail, matrix metalloproteinase
(MMP)-9 and GAPDH in cultured cell lysates. The cells were lysed by
RIPA buffer (Heart Biological Technology Co., Ltd., Xian, China)
and quantified using the BCA protein assay kit II (#5000002;
Bio-Rad Laboratories). The protein sample (40 µg) was
separated on a 10% SDS-PAGE gel and transferred onto nitrocellulose
membranes (Invitrogen) using the Bio-Rad Tank Blotting system
(Bio-Rad Laboratories). The membranes were respectively incubated
with primary antibodies at a 1:1,000 dilution overnight at 4°C. The
purchased primary antibodies were listed as follows: PTP4A1
(11508-1-AP) and N-cadherin (22018-1-AP) from Proteintech Group,
Inc. (Rosemont, IL, USA); N-cadherin (#3195), vimentin (#3932),
c-Src (#2109) and phosphorylated c-Src (p-c-Src;Y416, #6943) from
Cell Signaling Technology (Danvers, MA, USA); Snail (ab53519)
antibody from Abcam (Cambridge, MA, USA); MMP-9 (sc-21733) and
GAPDH (sc-47724) antibodies were from Santa Cruz Biotechnology
(Dallas, TX, USA). Horseradish peroxidase-conjugated secondary
antibodies at a 1:2,000 dilution were used to incubate the
membranes for 1 h at room temperature after washing them by TBST 3
times for 10 min. The targeting proteins on the membrane were
visualized with ECL reagents (Millipore, Plano, TX, USA).
Immunohistochemistry
Briefly, paraformaldehyde-fixed paraffin tumor
tissue sections were performed for immunohistochemical staining.
PTP4A1 primary antibodies were diluted at 1:100 with
phosphate-buffered saline (PBS) to label the antigens at 4°C
overnight. Biotinylated goat anti-rabbit secondary antibodies
(ZSGB-Bio, Beijing, China) were used to label the combined primary
antibodies. Complexes were detected by HRP-streptavidin conjugates
(ZSGB-Bio) and visualized by DAB (ZSGB-Bio). The final scores were
calculated by the product of staining intensity and positive
staining cell percentage as previously described (19).
In vitro migration and invasion
assays
For in vitro experiments, wound healing
assay, Transwell migration assay and Transwell invasion assay were
used to determine cell migration and invasion.
For wound healing assay, a wound line across the
middle of 6-well plates with confluent cells was created using a
200 µl sterile tip. The cells were cultured in reduced serum
DMEM medium in a humidified 5% CO2 incubator at 37°C for
48 h, and then images were taken with a phase-contrast
microscope.
Transwell inserts (Nalge Nunc International,
Penfield, New York, NY, USA) were coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) at 1 mg/ml on the upper layer
for invasion assay. Uncoated inserts were used for migration assay.
Briefly, 5×104 cells were seeded into the upper chamber
with reduced serum DMEM medium. Subsequenlty, 750 µl DMEM
medium containing 10% FBS was added to the lower chamber. The cells
were incubated in a humidified 5% CO2 incubator at 37°C
for 24 h. The chambers were fixed in 4% paraformaldehyde for 5 min
and then stained with 0.3% crystal violet dye for 10 min. The cells
on the upper layer were removed with a cotton swab. Migrating or
invading cells were counted under a light microscope.
In vivo metastasis analysis
For the in vivo metastasis model, 16 BALB/cA,
4–6-week-old, 16–18 g, female nude mice were housed in sterilized
cages with appropriate environment (25°C, 45% humidity) and fed a
regular chow diet with water ad libitum. All animal
protocols were approved by the Biomedical Ethics Committee of Xi'an
Jiaotong University Health Science Center. The mice were randomly
divided into 4 groups as follows: i) those injected with MHCC97-H
cells transfected with miR-1271 overexpression vector; ii) those
injected with MHCC97H cells transfected with miR-ctrl vector; iii)
those injected with Hep3B cells transfected with anti-miR-1271
vector; and iv) those injected with Hep3B cells transfected with
anti-miR-ctrl vector. The cells were suspended in cold 1X PBS
buffer at a concentration of 1×105/ml. Each mouse was
injected with a 100 µl cell suspension (~1×104
cells) into the tail vein very slowly to establish the lung
metastasis model. After 28 days, the weight of all mice increased
up to 20–23 g. The mice were sacrificed by CO2
euthanasia (the flow rate of CO2 was 20%
displacement/min). The lungs were then removed and formalin-fixed,
paraffin-embedded sections were created for H&E staining (Heart
Biological Technology Co., Ltd.).
Dual-Luciferase reporter assay
Two online MicroRNA targets prediction tools,
TargetScan (http://www.targetscan.org/) and MiRanda (http://www.microrna.org/), were used to located the
binding sites between miR-1271 and PTP4A1 3′-UTR region. The 3′-UTR
sequence of PTP4A1 predicted to interact with miR-1271 and the
designed mutant 3′-UTR sequence were synthesized and inserted into
the pEZX-MT06 vector (GeneCopoeia). These two recombinant
constructs were identified as wild-type 3′-UTR vector and mutant
type 3′-UTR vector, respectively and transfected into the MHCC-97H
cells. miR-1271 mimics (Guangzhou Ribobio) were transfected into
these cells at the same time. After 48 h, the cells were harvested
and luciferase activity was measured using the
Luc-Pair™Duo-Luciferase Assay kit 2.0 (GeneCopoeia). Firefly
luciferase activity was normalized to Renilla luciferase
activity.
Statistical analyses
Continuous variables are presented as the means ±
standard deviation (SD). Statistical analysis was performed using
the SPSS version 21.0 software (SPSS Inc., Chicago, IL, USA) or
GraphPad PRISM 5 software (GraphPad Software, La Jolla, CA, USA).
Correlations between miR-1271 and clinicopathological data were
analyzed by the Pearson's Chi-square test. The differences between
groups were analyzed using a Student's t-test. One-way ANOVA was
used to analyze the data from more than two groups. Bonferroni
method was used as a post hoc test. Survival analysis was performed
using a Kaplan-Meier curve and log-rank test. A value of P<0.05
was considered to indicate a statistically significant
difference.
Results
Downregulation of miR-1271 is associated
with metastasis in human HCC
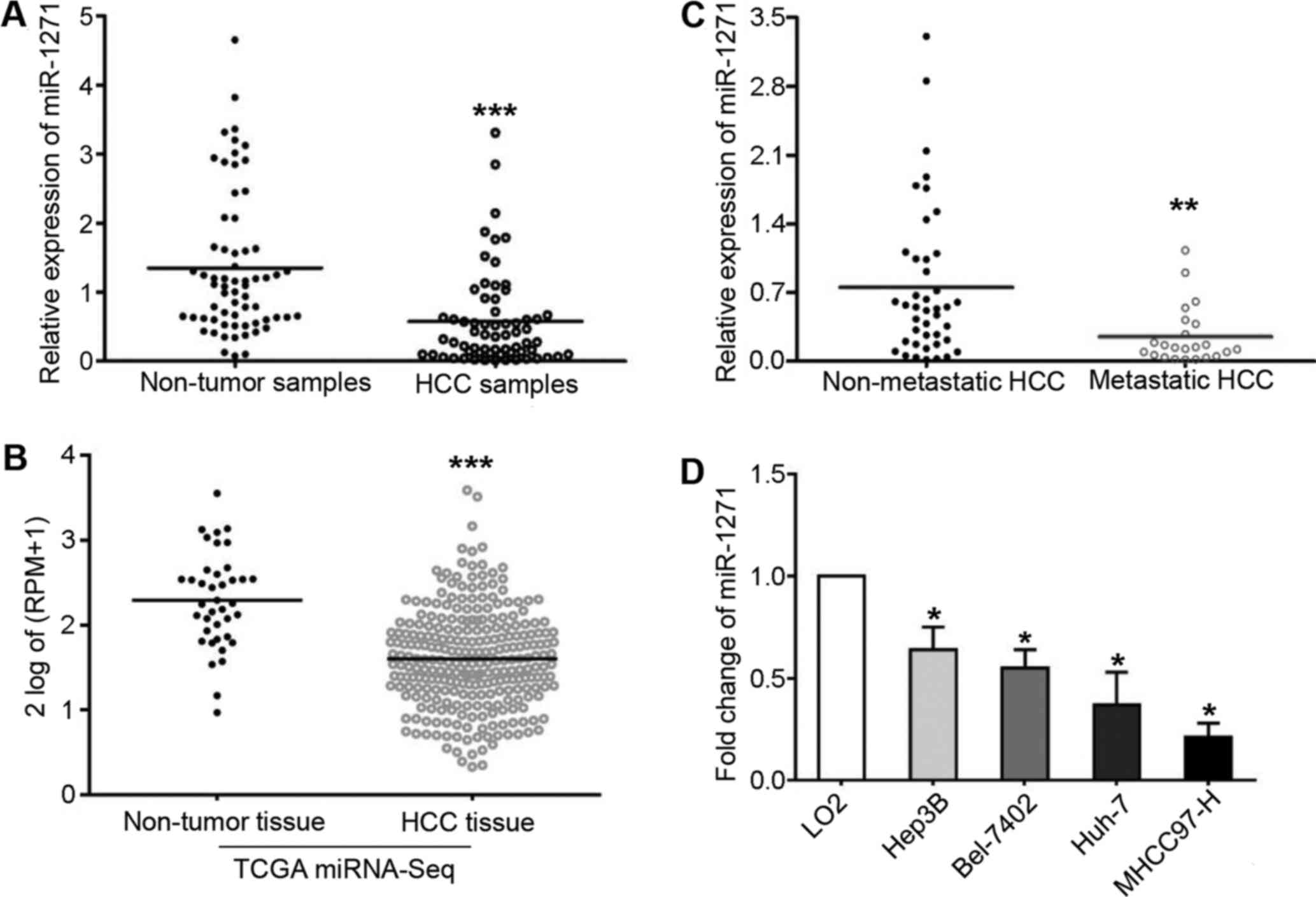
We analyzed miR-1271 expression by RT-qPCR and found
that it was frequently downregulated in HCC tissues (P=0.007;
Fig. 1A). Consistent with our
findings, miR-1271 was also discovered to be markedly downregulated
in 271 HCC tissues obtained from the TCGA miRNA-Seq data set
(P<0.001; Fig. 1B). These
findings encouraged us to further demonstrate a lower expression of
miR-1271 in metastatic HCC tissues compared with non-metastatic
tissues (P=0.004; Fig. 1C).
Moreover, we determined the expression levels of miR-1271 in 4 HCC
cell lines and LO2 cells. As shown in Fig. 1D, a significant change was
confirmed by one-way ANOVA analysis in these 5 cell lines
(P<0.05). Furthermore, all 4 HCC cell lines expressed decreased
levels of miR-1271 compared with the LO2 cells (P<0.05,
respectively).
Inhibitory effects of miR-1271 on
metastasis and EMT in HCC
After examining the expression of miR-1271 in
different HCC cell lines, we overexpressed miR-1271 in MHCC97-H
cells by transfection with miR-1271 lentivirus, and knocked down
its expression levels in Hep3B cells by transfection with
anti-miR-1271 lentivirus (data not shown). Wound healing assays
were used to demonstrate that miR-1271 over-expression inhibited
the migration of the MHCC97-H cells (P=0.021; Fig. 2A), whereas miR-1271 knockdown
promoted the migration of the Hep3B cells (P=0.031; Fig. 2B). The inhibitory effect of
miR-1271 on HCC cell migration was also be confirmed by a Transwell
chamber model (P=0.030 and P=0.034; Fig. 2C and D). Similarly, in the
Transwell chamber invasion assays, the upregulation of miR-1271
decreased the invasion of the MHCC97-H cells (P=0.009; Fig. 2E) and the downregulation of
miR-1271 increased the invasion of the Hep3B cells (P=0.031;
Fig. 2F).
To further investigate the effects of miR-1271 on
tumor metastasis in vivo, we established a lung metastasis
model by tail vein injection of HCC cells, and the maximum number
of lung metastatic clusters number of a single lung we found was
measured and found to be 19. As shown in Fig. 3A, the total number of lung
metastatic clusters in the miR-1271 group was much lower than that
in the miR-ctrl group (P=0.007). By contrast, a promoting effect on
in vivo tumor metastasis induced by miR-1271 knockdown was
observed in nude mice injected with recombinant Hep3B cells
(P=0.029; Fig. 3B). Taken
together, these results demonstrated that miR-1271 significantly
inhibited the metastasis of HCC cells in vitro and in
vivo.
As already discussed above, EMT serves as a
micro-foundation for cancer metastasis. Thus, we evaluated whether
miR-1271 regulates the EMT phenotype of HCC cells. As shown in
Fig. 3C, the results from western
blot analysis revealed that the re-induction of miR-1271 in the
MHCC97-H cells increased the expression of epithelial markers
(E-cadherin, P<0.05) but decreased the expression levels of
mesenchymal markers (N-cadherin and vimentin; P<0.05,
respectively). On the contrary, miR-1271 knockdown induced EMT by
decreasing E-cadherin expression and increasing the expression of
two mesenchymal markers expression (Fig. 3D; P<0.05, respectively).
PTP4A1 is a direct downstream target of
miR-1271
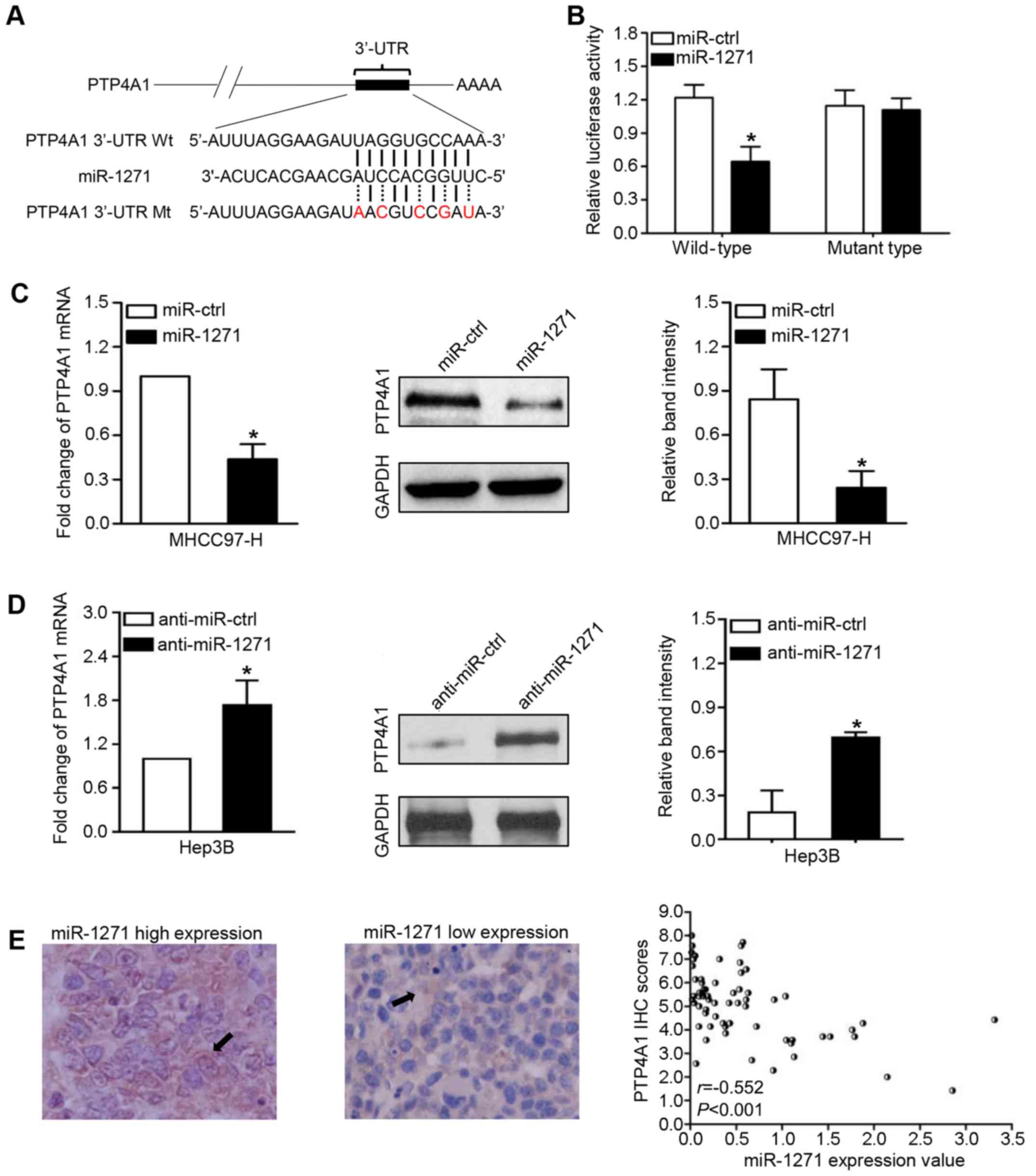
To investigate the targets through which miR-1271
suppresses HCC metastasis, we used two online databases
(TargetScan, http://www.targetscan.org/ and MiRanda http://www.microrna.org/) to predict that PTP4A1 was a
high potential target of miR-1271 in HCC (Context++ score
percentile, 99%; miRSVR score: −0.7645; Fig. 4A). To confirm this prediction, we
first used a Dual-Luciferase reporter system to demonstrate that
miR-1271, as we had expected, suppressed the luciferase activity of
PTP4A1 with a wild-type 3′-UTR, but did not suppress the activity
of PTP4A1 containing a mutant type 3′-UTR (P=0.035; Fig. 4B). In addition, the results from
RT-qPCR and western blot analysis revealed that miR-1271 negatively
modulated the mRNA and protein levels of PTP4A1 in the HCC cells
(P<0.05, respectively; Fig. 4C and
D). Importantly, our study revealed a negative correlation
between miR-1271 levels and PTP4A1 protein expression in 65 HCC
tissues (r=−0.552, P<0.001; Fig.
4E). On the whole, these results strongly indicate that PTP4A1
is a target of miR-1271 in HCC.
Alterations in PTP4A1 expression abrogate
the effects of miR-1271 on HCC cells
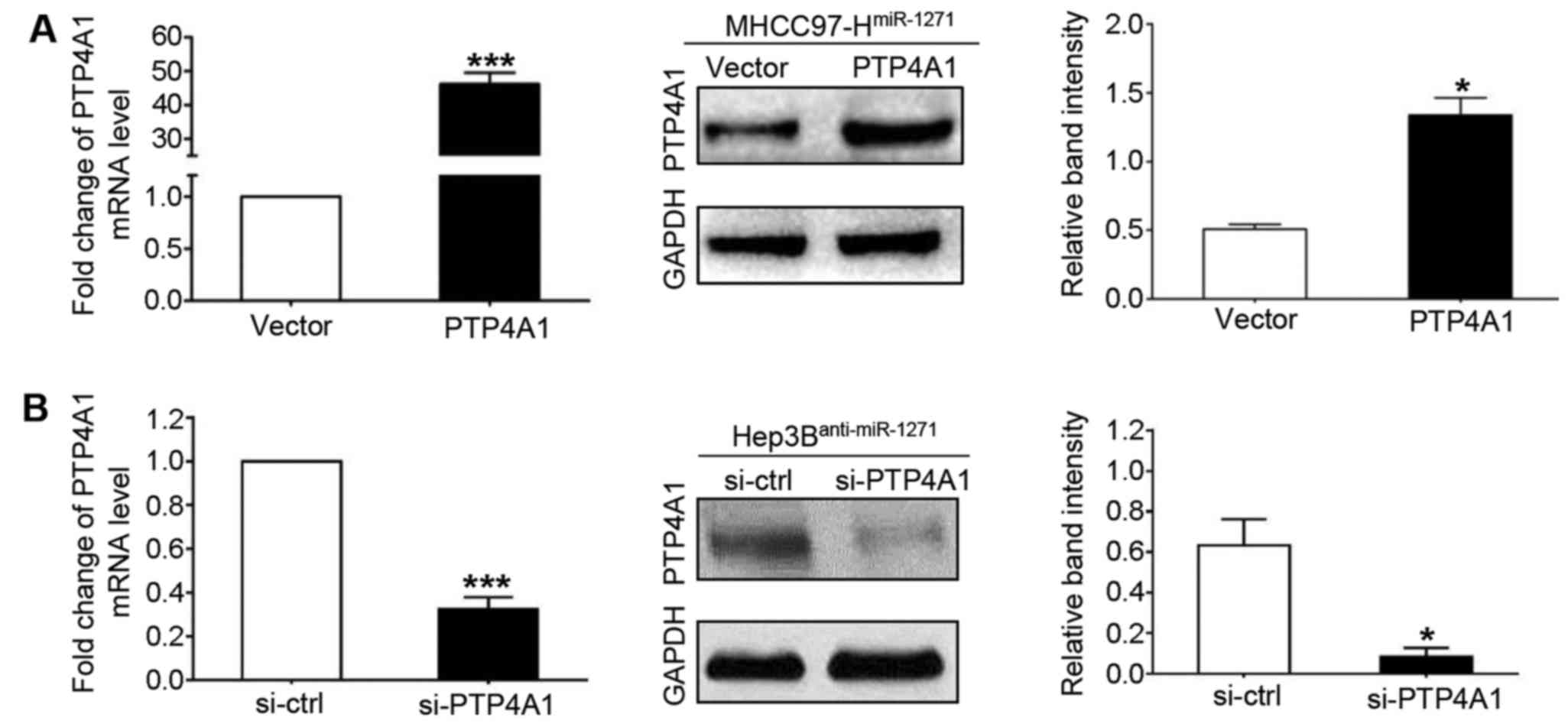
To further confirm these results, we repeated the
above-mentioned in vitro experiments using another method.
We transfected PTP4A1 vectors into the cells to increase PTP4A1
expression in the MHCC97-H cells that stably overexpressed miR-1271
(by transfection with miR-1271 expression vector; P<0.001 and
P=0.031, respectively; Fig. 5A),
and used PTP4A1 siRNA (si-PTP4A1) to knockdown PTP4A1 expression in
the Hep3B cells with a stably decreased expression of miR-1271 (by
transfection with miR-1271 inhibitor; P<0.001 and P= 0.016,
respectively; Fig. 5B). PTP4A1
overexpression increased the migration (P=0.007; Fig. 6A), invasion (P=0.024; Fig. 6B) and mesenchymal transition
(P<0.05, respectively; Fig. 6C)
in miR-1271-overexpressing MHCC97-H cells. Similarly, PTP4A1
knockdown decreased the migration (P=0.040; Fig. 6D) and invasion (P=0.033, Fig. 6E) and inhibited EMT (P<0.05,
respectively; Fig. 6F) in the
Hep3B cells transfected with anti-miR-1271.
miR-1271 prevents HCC metastasis by
inducing c-Src phosphorylation
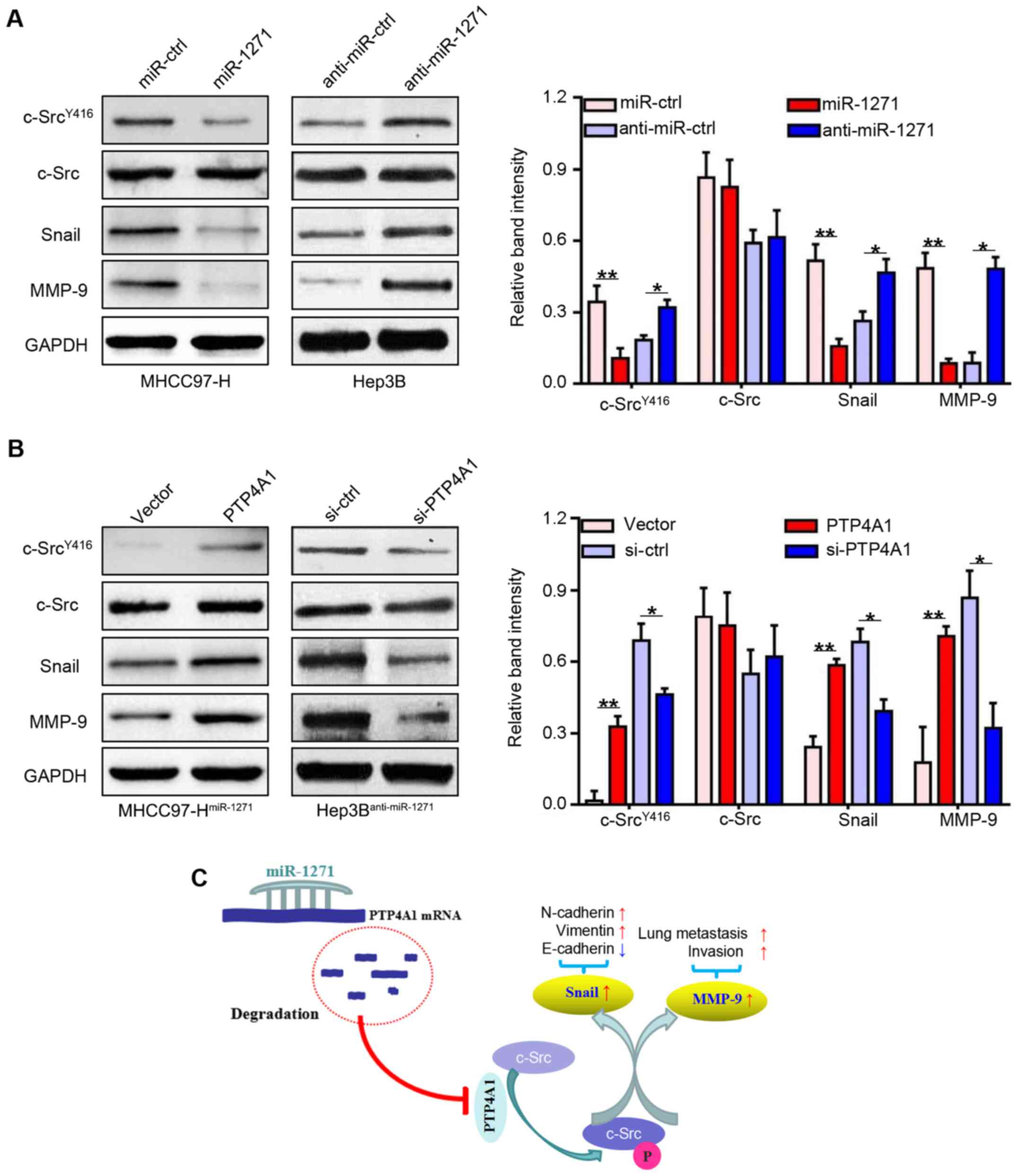
To explore the underlying mechanisms of PTP4A1 in
the regulation of HCC metastasis, we used western blot analysis to
detect the expression of total c-Src expression, the
phosphorylation of c-Src and two metastasis-associated c-Src
downstream targets (Snail and MMP-9). As we had expected, the
expression levels of c-SrcY416, Snail and MMP-9 were
downregulated in the MHCC97-H cells that stably over-expressed
miR-1271 (P<0.05 and P<0.01, respectively; Fig. 7A, left lanes), and were upregulated
in the Hep3B cells in which miR-1271 was inhibited (P<0.05 and
P<0.01, respectively; Fig. 7A,
right lanes). By contrast, the expression levels of these genes
were increased following transfection with PTP4A1 expression vector
(P<0.05 and P<0.01, respectively; Fig. 7B, left lanes). Moreover,
transfection with si-PTP4A1 decreased the expression levels of
these genes, which was similar to the effects induced by miR-1271
overexpression (P<0.05 and P<0.01, respectively; Fig. 7B, right lanes). These results thus
indicate that the multiple anticancer functions of miR-1271 are
attributed to the inhibition of the PTP4A1/c-Src signaling pathway
(Fig. 7C).
Clinical significance of miR-1271 and
PTP4A1 in human HCC
We divided the 65 HCC patients into 2 groups
according to the mean value of miR-1271 expression or PTP4A1 IHC
scores. As shown in Table I, a low
expression of miR-1271 was associated with tumor metastasis
(χ2=6.040, P=0.014) and an advanced TNM stage (III+IV
stage, χ2=5.298, P=0.021). Moreover, a high expression
of PTP4A1 was associated with a large tumor size (>5 cm,
χ2=6.023, P=0.014), tumor metastasis
(χ2=4.190, P=0.041) and an advanced TNM stage
(χ2=5.747, P=0.017). Kaplan-Meier survival curves
revealed that patients in the low miR-1271 expression group had a
poorer 3-year overall survival rate (HR=2.597, P=0.001; Fig. 8A) and tumor-free survival rate
(HR=2.864, P<0.001; Fig. 8B)
compared with patients in the high miR-1271 group. Furthermore, the
3-year overall survival rate (HR=2.172, P=0.008; Fig. 8C) and tumor-free survival rate
(HR=2.780, P<0.001; Fig. 8D)
were lower in the high PTP4A1 expression group than the low PTP4A1
expression group. On the whole, these data suggest that miR-1271
and PTP4A1 are prognostic indicators for patients with HCC.
 | Table ICorrelation between the
clinicopathological characteristics and miR-1271 and PTP4A1
expression in patients with HCC (n=65) |
Table I
Correlation between the
clinicopathological characteristics and miR-1271 and PTP4A1
expression in patients with HCC (n=65)
| Clinical
characteristics | miR-1271 expression
| χ2 | P-value | PTP4A1 expression
| χ2 | P-value |
|---|
| Low (n=44) | High (n=21) | Low (n=28) | High (n=37) |
|---|
| Sex | | | | | | | | |
| Male | 30 | 9 | 3.799 | 0.051 | 15 | 24 | 0.847 | 0.357 |
| Female | 14 | 12 | | | 13 | 13 | | |
| Age (years) | | | | | | | | |
| <50 | 17 | 11 | 1.095 | 0.295 | 10 | 18 | 1.087 | 0.297 |
| ≥50 | 27 | 10 | | | 18 | 19 | | |
| HBsAg | | | | | | | | |
| Negative | 11 | 4 | 0.047 | 0.828 | 7 | 8 | 0.102 | 0.749 |
| Positive | 33 | 17 | | | 21 | 29 | | |
| Liver
cirrhosis | | | | | | | | |
| Absent | 15 | 12 | 3.111 | 0.078 | 15 | 12 | 2.933 | 0.087 |
| Present | 29 | 9 | | | 13 | 25 | | |
| Tumor size
(cm) | | | | | | | | |
| ≤5 | 14 | 12 | 3.799 | 0.051 | 16 | 10 | 6.023 | 0.014a |
| >5 | 30 | 9 | | | 12 | 27 | | |
| Edmondson-Steiner
grading | | | | | | | | |
| I+II | 19 | 14 | 3.137 | 0.077 | 18 | 15 | 3.596 | 0.058 |
| III+IV | 25 | 7 | | | 10 | 22 | | |
| Metastasis | | | | | | | | |
| Absent | 24 | 18 | 6.040 | 0.014a | 22 | 20 | 4.190 | 0.041a |
| Present | 20 | 3 | | | 6 | 17 | | |
| TNM stage | | | | | | | | |
| I+II | 18 | 15 | 5.298 | 0.021a | 19 | 14 | 5.747 | 0.017a |
| III+IV | 26 | 6 | | | 9 | 23 | | |
Discussion
The dysregulation of miRNA expression is a common
incident in the development and progression of human cancers,
including HCC (20). In the
present study, we investigated the role of miR-1271 in HCC and
discovered the following: First, miR-1271 was identified as a
downregulated miRNA in HCC samples and cell lines. This finding was
still consistently observed in the 271 HCC tissues from the TCGA
database. We then found that the 23 HCC tissues from patients with
metastasis expressed lower miR-1271 levels than other 42 HCC
tissues. The intrahepatic and hematogenous metastasis of HCC
attributed to the poor prognosis of patients with HCC. miRNAs have
been recognized as important regulators of human cancer metastasis
(21). Hence, in the present
study, we analyzed our clinical data to discover that a low
expression of miR-1271 was associated with tumor metastasis and a
poor prognosis of patients with HCC. This prognostic value of
miR-1271 in HCC was consistent with a previous result in
neuroglioma (22). These results
strongly indicated that miR-1271 is associated with tumor
metastasis in HCC.
The biological functions of miR-1271 have been
partly uncovered in other types of cancer. miR-1271 has been
demonstrated to inhibit cell proliferation in oral squamous cell
carcinoma (23) and to sensitize
human gastric cancer cells to cisplatin treatment (24). In the present study, and to the
best of our knowledge, we report for the first time that miR-1271
is a critical inhibitor of cell migration and invasion in HCC
through gain- and loss-of-function experiments in vitro.
Moreover, the overexpression of miR-1271 also suppressed the
formation of lung metastatic clusters in mice. EMT is identified as
loss of epithelial marker (E-cadherin) expression and an increase
in mesenchymal marker (vimentin and N-cadherin) expression
(25). EMT is involved in numerous
liver disease states, such as HBV or HCV-associated viral
hepatitis, non-alcoholic fatty liver disease, liver fibrosis and
HCC (26). These diseases present
EMT in hepatocytes and cancer cells, leading to cell migration or
invasion. Therefore, EMT has been recognized as a fundamental
mechanism for HCC metastasis. In this study, we demonstrated that
the overexpression of miR-1271 increased E-cadherin expression, and
decreased N-cadherin and vimentin expression, whereas the
inhibition of miR-1271 exerted the opposite effects. These findings
suggest that miR-1271 has considerable potential for cancer
therapy.
PTP4A1 (also known as phosphatase of regenerating
liver 1) belongs to a subfamily of prenylated protein tyrosine
phosphatases (PTPs) (27). Even
the expression of PTP4A1 in human normal tissues has not been well
characterized, Wang et al (28) found that the PTP4A1 mRNA levels
were elevated in a number of tumor cell lines, including HeLa,
SK-Lu-1 and several melanoma cell lines. Our results also showed
that a high expression of PTP4A1 in HCC tissues was associated with
lower overall survival and disease-free rates. Although the
transcriptional regulation of PTP4A1 is not yet well understood,
post-transcriptional regulation by miRNAs has been widely reported.
PTP4A1 serves as a tumor suppressor target of miR-339-5p in
colorectal (29), and miR-601
(30) and miR-944 (31) in breast cancer. In this study,
PTP4A1 was demonstrated as an effective target of miR-1271 by the
following evidence: First, miR-1271 only decreased luciferase
activity in MHCC97-H cells carrying the wild-type 3′-UTR of PTP4A1.
Second, miR-1271 overexpression decreased the mRNA and protein
expression levels of PTP4A1 in MHCC97-H cells, whereas miR-1271
knockdown increased their expression levels in Hep3B cells. In
addition, a negative correlation between miR-1271 and PTP4A1
protein expression was confirmed in 65 HCC tissue samples. Finally,
altering PTP4A1 expression in HCC cells abrogated the effects
induced by miR-1271. These results suggest that PTP4A1 is a
downstream effector of miR-1271 functions in HCC.
The proto-oncogene, tyrosine-protein kinase Src
(simply c-Src), has been linked to the PTP family. Liang et
al (32,33) revealed that PTP4A3 promoted
proliferation and invasion by the reduction of C-terminal Src
kinase (Csk), leading to a decrease in Src-Tyr527. However, in
multiple myeloma cells, PTP4A3 has been demonstrated to activate
Src by increasing the phosphorylation at Tyr416, but not by
decreasing the phosphorylation at Tyr527 (34). In A549 lung cancer cells, the
inhibition of PTP4A1 directly decreased c-Src expression (35). Intriguingly, there were no
significant changes in total c-Src expression levels or its
phosphorylation status by PTP4A2 knockdown (36). Src activation induces Snail and
MMP-9 expression (37,38). In this study, miR-1271 inhibited
the phosphorylation of c-Src at Tyr416 (Y416), and Snail and MMP-9
expression. Moreover, PTP4A1 knockdown mimicked this function,
whereas PTP4A1 overexpression abrogated it. Even though the
detailed mechanisms underlying the activation of c-Src by PTP4A1
are not yet clear, it is unreasonable that PTP4A1 regulates Src
activation directly. First, our result showed that PTP4A1, a
phosphatase, increased c-Src phosphorylation at Y416. Second, Luo
et al (39) were unable to
obtain direct binding evidence by immunoprecipitation. These
results indicate that miR-1271-mediated PTP4A1 inhibition
deactivates c-Src through an indirect signaling pathway.
In conclusion, the present study demonstrates that
the downregulation of miR-1271 in HCC tissues is associated with
metastasis and a poor prognosis. miR-1271 is confirmed as a novel
tumor metastasis and EMT inhibitor in HCC. The multiple anticancer
functions of miR-1271 are attributed to the inhibition of the
PTP4A1/c-Src signaling pathway. Taken together, our findings
suggest that miR-1271 is a prospective therapeutic target for
HCC.
Acknowledgments
The present study was supported by a grant from the
National Natural Scientific Foundation of China (no. 81472247).
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Burkhart RA, Ronnekleiv-Kelly SM and
Pawlik TM: Personalized therapy in hepatocellular carcinoma:
Molecular markers of prognosis and therapeutic response. Surg
Oncol. 26:138–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695.
2017.PubMed/NCBI
|
|
4
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tu K, Li C, Zheng X, Yang W, Yao Y and Liu
Q: Prognostic significance of miR-218 in human hepatocellular
carcinoma and its role in cell growth. Oncol Rep. 32:1571–1577.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang X, Liang L, Zhang XF, Jia HL, Qin Y,
Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, et al: MicroRNA-26a
suppresses tumor growth and metastasis of human hepatocellular
carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology.
58:158–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC,
Huang CH, Lee YS, Yen TC and Hsieh SY: MicroRNA-214 downregulation
contributes to tumor angiogenesis by inducing secretion of the
hepatoma-derived growth factor in human hepatoma. J Hepatol.
57:584–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li DP, Fan J, Wu YJ, Xie YF, Zha JM and
Zhou XM: MiR-155 up-regulated by TGF-β promotes
epithelial-mesenchymal transition, invasion and metastasis of human
hepatocellular carcinoma cells in vitro. Am J Transl Res.
9:2956–2965. 2017.
|
|
9
|
Sakabe T, Azumi J, Umekita Y, Toriguchi K,
Hatano E, Hirooka Y and Shiota G: Prognostic relevance of miR-137
in patients with hepatocellular carcinoma. Liver Int. 37:271–279.
2017. View Article : Google Scholar
|
|
10
|
von Felden J, Heim D, Schulze K, Krech T,
Ewald F, Nashan B, Lohse AW and Wege H: High expression of micro
RNA-135A in hepatocellular carcinoma is associated with recurrence
within 12 months after resection. BMC Cancer. 17:602017. View Article : Google Scholar
|
|
11
|
Li L, Qu YW and Li YP: Over-expression of
miR-1271 inhibits endometrial cancer cells proliferation and
induces cell apoptosis by targeting CDK1. Eur Rev Med Pharmacol
Sci. 21:2816–2822. 2017.PubMed/NCBI
|
|
12
|
Zhong J, Liu Y, Xu Q, Yu J and Zhang M:
Inhibition of DIXDC1 by microRNA-1271 suppresses the proliferation
and invasion of prostate cancer cells. Biochem Biophys Res Commun.
484:794–800. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Z, Huang C and Hao D: MicroRNA-1271
inhibits proliferation and promotes apoptosis of multiple myeloma
cells through inhibiting smoothened-mediated Hedgehog signaling
pathway. Oncol Rep. 37:1261–1269. 2017. View Article : Google Scholar
|
|
14
|
Liu X, Ma L, Rao Q, Mao Y, Xin Y, Xu H, Li
C and Wang X: MiR-1271 inhibits ovarian cancer growth by targeting
cyclin G1. Med Sci Monit. 21:3152–3158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu H, Wang H, Liu X and Yu T: miR-1271
inhibits migration, invasion and epithelial-mesenchymal transition
by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem
Biophys Res Commun. 472:346–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Xu L and Jiang L: miR-1271
promotes non-small-cell lung cancer cell proliferation and invasion
via targeting HOXA5. Biochem Biophys Res Commun. 458:714–719. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Santamaria PG, Moreno-Bueno G, Portillo F
and Cano A: EMT: Present and future in clinical oncology. Mol
Oncol. 11:718–738. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jolly MK, Ware KE, Gilja S, Somarelli JA
and Levine H: EMT and MET: Necessary or permissive for metastasis?
Mol Oncol. 11:755–769. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li C, Yang W, Zhang J, Zheng X, Yao Y, Tu
K and Liu Q: SREBP-1 has a prognostic role and contributes to
invasion and metastasis in human hepatocellular carcinoma. Int J
Mol Sci. 15:7124–7138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Y, Bao Y and Yang W: Regulatory miRNAs
in colorectal carcinogenesis and metastasis. Int J Mol Sci.
18:182017. View Article : Google Scholar
|
|
22
|
Gong J, Wang ZX and Liu ZY: miRNA-1271
inhibits cell proliferation in neuroglioma by targeting fibronectin
1. Mol Med Rep. 16:143–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kong D, Zhang G, Ma H and Jiang G:
miR-1271 inhibits OSCC cell growth and metastasis by targeting ALK.
Neoplasma. 62:559–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang M, Shan X, Zhou X, Qiu T, Zhu W, Ding
Y, Shu Y and Liu P: miR-1271 regulates cisplatin resistance of
human gastric cancer cell lines by targeting IGF1R, IRS1, mTOR and
BCL2. Anticancer Agents Med Chem. 14:884–891. 2014. View Article : Google Scholar
|
|
25
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Serrano-Gomez SJ, Maziveyi M and Alahari
SK: Regulation of epithelial-mesenchymal transition through
epigenetic and post-translational modifications. Mol Cancer.
15:182016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bessette DC, Qiu D and Pallen CJ: PRL
PTPs: Mediators and markers of cancer progression. Cancer
Metastasis Rev. 27:231–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang J, Kirby CE and Herbst R: The
tyrosine phosphatase PRL-1 localizes to the endoplasmic reticulum
and the mitotic spindle and is required for normal mitosis. J Biol
Chem. 277:46659–46668. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou C, Liu G, Wang L, Lu Y, Yuan L, Zheng
L, Chen F, Peng F and Li X: MiR-339–5p regulates the growth, colony
formation and metastasis of colorectal cancer cells by targeting
PRL-1. PLoS One. 8:e631422013. View Article : Google Scholar
|
|
30
|
Hu JY, Yi W, Wei X, Zhang MY, Xu R, Zeng
LS, Huang ZJ and Chen JS: miR-601 is a prognostic marker and
suppresses cell growth and invasion by targeting PTP4A1 in breast
cancer. Biomed Pharmacother. 79:247–253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Flores-Pérez A, Marchat LA,
Rodríguez-Cuevas S, Bautista VP, Fuentes-Mera L, Romero-Zamora D,
Maciel-Dominguez A, de la Cruz OH, Fonseca-Sánchez M, Ruíz-García
E, et al: Suppression of cell migration is promoted by miR-944
through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC
Cancer. 16:3792016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liang F, Liang J, Wang WQ, Sun JP, Udho E
and Zhang ZY: PRL3 promotes cell invasion and proliferation by
down-regulation of Csk leading to Src activation. J Biol Chem.
282:5413–5419. 2007. View Article : Google Scholar
|
|
33
|
Liang F, Luo Y, Dong Y, Walls CD, Liang J,
Jiang HY, Sanford JR, Wek RC and Zhang ZY: Translational control of
C-terminal Src kinase (Csk) expression by PRL3 phosphatase. J Biol
Chem. 283:10339–10346. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Abdollahi P, Vandsemb EN, Hjort MA, Misund
K, Holien T, Sponaas AM, Rø TB, Slørdahl TS and Børset M: Src
Family kinases are regulated in multiple myeloma cells by
phosphatase of regenerating liver-3. Mol Cancer Res. 15:69–77.
2017. View Article : Google Scholar
|
|
35
|
Achiwa H and Lazo JS: PRL-1 tyrosine
phosphatase regulates c-Src levels, adherence, and invasion in
human lung cancer cells. Cancer Res. 67:643–650. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y and Lazo JS: Metastasis-associated
phosphatase PRL-2 regulates tumor cell migration and invasion.
Oncogene. 31:818–827. 2012. View Article : Google Scholar
|
|
37
|
Liu X and Feng R: Inhibition of epithelial
to mesenchymal transition in metastatic breast carcinoma cells by
c-Src suppression. Acta Biochim Biophys Sin (Shanghai). 42:496–501.
2010. View Article : Google Scholar
|
|
38
|
Mon NN, Senga T and Ito S: Interleukin-1β
activates focal adhesion kinase and Src to induce matrix
metalloproteinase-9 production and invasion of MCF-7 breast cancer
cells. Oncol Lett. 13:955–960. 2017.PubMed/NCBI
|
|
39
|
Luo Y, Liang F and Zhang ZY: PRL1 promotes
cell migration and invasion by increasing MMP2 and MMP9 expression
through Src and ERK1/2 pathways. Biochemistry. 48:1838–1846. 2009.
View Article : Google Scholar : PubMed/NCBI
|