Introduction
Myelodysplastic syndrome (MDS) presents a group of
heterogeneous myeloid clones derived from haematopoietic stem cells
(HSCs), usually manifesting as ineffective haematopoietic,
refractory haematopoietic reduction and failure. With approximately
40% of patients with MDS developing acute myeloid leukaemia (AML),
allogeneic HSC transplantation (HSCT) is the most commonly used
treatment (1). Recent studies have
greatly illuminated the genomic landscape of MDS (2); however, there is limited information
on MDS pathogenesis and the developmental mechanisms. Thus, these
aspects of MDS warrant clarification in order to improve patient
prognosis. Accumulating evidence has indicated that the
dysregulation of microRNAs (miRNAs or miRs) is involved in cancer
proliferation, differentiation, apoptosis and metastasis, with
miRNAs functioning as oncogenes or tumour suppressors, providing a
promising method for the management of cancer (3,4).
miR-143 expression changes, including both up- and downregulation,
is frequently observed in various types of cancer. Whilst the
downregulation of miR-143 causes it to act as a tumour suppressor
in various types of tumour, including colorectal cancer (5-7),
prostate cancer (8,9), gastric cancer (10,11),
cervical cancer (12,13), nasopharyngeal carcinoma (14,15)
and human osteosarcoma (16,17),
miR-143 may also act as an anti-oncogene in T-cell leukaemia Jurkat
cells (18). Various studies have
demonstrated that miR-143 expression is downregulated in MDS
(19-21); however the expression of miR-143
warrants further investigation by cohort studies. In addition, the
molecular mechanisms of action of miR-143 in MDS/AML need to be
further elucidated.
In the present cohort study, bone marrow (BM)
samples from 33 patients with MDS and 11 healthy individuals were
collected, with the healthy samples serving as the controls. To
explore the molecular mechanisms of the disease, a series of
experiments were performed in which miR-143 was either
overexpressed or downregulated in in vitro recombinant
lentiviral transfections of SKM-1 cells, a cell line used to
transform MDS to AML harboring multiple karyotypic abnormalities,
whilst lacking the 5q deletion. The effects of miR-143 on the
proliferation, differentiation and apoptosis of SKM-1 cells were
investigated and the in vivo vs. the in vitro effect
of miR-143 was verified.
Materials and methods
Samples and cell lines
Upon receiving written informed consent from all
participants, a total of 44 BM samples (33 MDS/AML patient samples
and 11 samples from healthy individuals) were obtained between
October, 2012 to April, 2014 from the First Affiliated Hospital of
Chongqing Medical University, Chongqing, China. The demographic
characteristics of the patients included in this study are
presented in Table I.
 | Table IDemographic and clinical
characteristic of the patients studied. |
Table I
Demographic and clinical
characteristic of the patients studied.
| Number of
patients | 33 |
| Sex
(male/female) | 20/13 |
| Median (range) | |
| Age (years) | 50.2 (17-79) |
| BM blasts (%) | 22.4 (2-88) |
| Type, N (%) | |
| RAEB-1 | 7 (21.2%) |
| RAEB-2 | 11 (33.3%) |
| Secondary AML | 15 (45.5%) |
|
Cytogenetic/FISH | |
| 5q- | 12 |
| 8+ | 2 |
| Normal | 19 |
The SKM-1 cells [kindly provided by Professor
Jianfeng Zhou of Tongji Medical College of Huazhong University of
Science and Technology (Wuhan, China)] were cultured in RPMI-1640
(Gibco/Thermo Fisher Scientific, Waltham, MA, USA) with 10%
heat-inactivated foetal bovine serum (FBS) (Gibco/Thermo Fisher
Scientific) and were maintained in a humidified atmosphere of 5%
CO2 at 37˚C (22,23).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Mononuclear cells (MNCs) were isolated from the BM
samples using Ficoll-Paque (GE Healthcare, MA, USA), and total RNA
was extracted using RNAiso Plus (Takara, Dalian, China) according
to the manufacturer's instructions. A commercially available cDNA
synthesis kit (Takara) was used for cDNA synthesis. Quantitative
PCR (qPCR) was performed using a CFX96 Touch™ Real-Time PCR
Detection System (Bio-Rad, Hercules, CA, USA). The total reaction
volume was 20 µl and was prepared as follows: SYBR Premix Ex Taq
(10 µl), 0.8 µl of each primer (10 µmol/l), 2 µl of cDNA template
and ddH2O (6.4 µl). The RT-qPCR conditions were as
follows: 95°C for 3 min; 95°C for 20 sec, followed by 62°C for 50
sec repeated over 40 cycles. All primers were designed and
synthesised by Novland (Novland Co., Ltd., Shanghai, China) and are
presented in Table II. U6 was
amplified as a reference for normalization and the results were
analysed using the 2−ΔΔCq method (24).
 | Table IISequences of primers using in
RT-qPCR. |
Table II
Sequences of primers using in
RT-qPCR.
| Genes | Forward and reverse
primers |
|---|
| miR-143 | F:
5'-TGTGACACTGAGATGAAGCACTG-3' |
| R:
5'-TATGGTTGTTCTGCTCTCTGTCTC-3' |
| U6 | F:
5'-ATTGGAACGATACAGAGAAGATT-3' |
| R:
5'-GGAACGCTTCACGAATTTG-3' |
| FasL | F:
5'-TCCCATCCTCCTGACCAC-3' |
| R:
5'-TCGTAAACCGCTTCCCTC-3' |
| Caspase-8 | F:
5'-GGAGTCCATTATCCGTAGT-3' |
| R:
5'-AGTTTGAGGGTCTGCTTT-3' |
| Caspase-3 | F:
5'-ATGACATCTCGGTCTGGT-3' |
| R:
5'-AGAAACATCACGCATCAA-3' |
| Caspase-9 | F:
5'-CCAAGCCTCTTCTTACTTCACC-3' |
| R:
5'-CATCGTTCTGCCATCACTCA-3' |
| Actin | F:
5'-CCACGAAACTACCTTCAACTAA-3' |
| R:
5'-GTGATCTCCTTCTGCATCCTGT-3' |
Construction of recombinant lentiviral
vectors carrying hsa-miR-143 and hsa-miR-143-inhibitor
The miR-143 gene overexpression and knockdown
lentiviral vector (LV) system contained GV217, pHelper 1.0 and
pHelper 2.0 before packaging (GeneChem Co., Shanghai, China). The
target sequences for the miR-143 overexpression lentiviral vector
system were as follows: 5'-CGGCCGCGACTCTAGCAGAGCTGGAGAGGTGGAG-3'
and 5'-ATAAGCTTGATATCGCAGGAAGGACAGAGTGTTTCC-3'. The primers for the
miR-143-3p inhibition lentiviral vector were as follows:
5'-TGAGATGAAGCACTGTAGCTC-3' and 5'-GAGCTACAGTGCTTCATCTCA-3'. The
production, purification and titration of the lenti-virus were
performed as previously described by Tiscornia et al
(25). Three vectors for
overexpression and downregulation were co-transfected into 293T
cells [kindly provided by Professor Jianfeng Zhou of Tongji Medical
College of Huazhong University of Science and Technology (Wuhan,
China)] in serum-free media using Lipofectamine 2000 (Invitrogen
Inc., Carlsbad, CA, USA). Following 8 h of incubation, the medium
was changed to complete medium containing RPMI-1640 and 10%
heat-inactivated FBS. The high-titre recombinant lentiviral
vectors, LV-hsa-miR-143 and LV-hsa-miR-143-inhibitors, were
harvested after 2 days of transfection.
RNA interference
The SKM-1 cells were seeded (2×105
cells/ml) in 6-well plates and infected with the lentivirus at a
multiplicity of infection (MOI) of 10. The medium was replaced with
fresh basic medium after 10 h. The infection efficiency was
evaluated using a fluorescence microscope (SN:3832002173, Zeiss,
Oberkochen, Germany) and the infection rate was detected by a flow
cytometer (CytoFLEX, Beckman Coulter Corp., Tokyo, Japan) after 4
days.
Cell proliferation assays
Cell proliferation was determined using a Cell
Counting kit-8 (CCK-8) assay (7Sea Biotech, Shanghai, China).
Briefly, approximately 3x103 cells/100 µl were seeded in
96-well plates and cultured at 37°C for 1 day prior to
transfection. The cells were then stably transfected with the
lentivirus at an MOI of 10 for 10 h. The medium was replaced with
fresh basic medium, and the cells were further incubated at 37°C
for 24, 48 and 72 h. Following incubation, CCK-8 reagent (10 µl)
was added to each well, and the cells were incubated for an
additional 2 h. Finally, the absorbance at 450 nm was measured by
spectrophotometer (SN:1510-03523, Type:1510, Thermo Fisher
Scientific, Vantaa, Finland).
Cell apoptosis and cell cycle assays
SKM-1 cell apoptosis was evaluated with an Annexin
V-FITC/PI Apoptosis Detection kit (BD Biosciences, Heidelberg,
Germany) according to the manufacturer's instructions using a flow
cytometer (BD Biosciences, San Jose, CA, USA). For the cell cycle
assay, the cells were harvested and fixed with 70% anhydrous
alcohol for 4 h at 4°C. The cells were then incubated with 100
µg/ml of propidium iodide (PI) for 30 min at room temperature. The
cell cycle profiles were analysed using Multicycle software
(Phoenix Flow Systems, San Diego, CA, USA).
Western blot analysis
Total protein from the SKM-1 cells and tumour
tissues was harvested using RIPA lysis buffer supplemented with 1
µM PMSF (Beyotime, Shanghai, China), and the protein (40 µg) was
loaded and separated on a 10% SDS-polyacrylamide gradient gel. The
proteins were transferred onto polyvinylidene fluoride (PVDF)
membranes with glycine transfer buffer and blocked for 1 h with 5%
non-fat milk in PBS containing 0.1% Tween-20 (PBS-T). The blots
were then incubated overnight at 4°C with each primary antibody.
The following primary antibodies were used in this study: Rabbit
anti-human FasL (cat. no. YT0785), cleaved caspase-8 (cat. no.
YT0011), cleaved caspase-3 (cat. no. YT0004) and cleaved caspase-9
(cat. no. YT0013) (Immunoway Biotechnology, Newark, DE, USA), each
at a dilution of 1:500. Horseradish peroxidase-conjugated goat
anti-rabbit IgG (cat. no. ZB-2306, ZSGB-BIO, Beijing, China) was
used as the secondary antibody and GAPDH (cat. no. #5174, CST
(Shanghai) Biological Reagents Co., Ltd., Shanghai, China) served
as a loading control. The membranes were then washed 3 times with
PBS containing 0.1% Tween-20 and incubated with the secondary
antibody at a dilution of 1:7,000 for 2 h at room temperature.
After washing with PBS-T (3×10 ml, 5 min each), the protein bands
were visualised with an ECL kit (Beyotime), and the band intensity
was analysed using Quantity One software (Bio-Rad).
Firefly dual-luciferase reporter
assay
The TargetScan (http://www.targetscan.org/) website was used to screen
out the downstream target genes that may be associated with miR-143
and to select the one we were interested in for validation. The
wild-type and mutated 3'-UTR fragment from MLLT3/AF9 mRNA was
cloned into dual luciferase reporter vectors (Promega, Madison, WI,
USA). 293T cells were seeded into 24-well plates and were
transfected with negative control vectors, miR-143 mimics (miR-143
inhibitors or a precursor control) and pMIR-MLLT3-WT (or a mutated
version of pMIR-MLLT3-Mut), together with the luciferase-reporter
vectors. After 2 days of transfection, the Dual-Luciferase Reporter
Assay system (Promega) was used to detect the lucif-erase activity
according to the manufacturer's instructions.
SKM-1 xenograft tumour assay
Non-obese diabetic severely compromised
immunodeficient (NOD/SCID) mice are ideal models for in vivo
experiments of blood system tumours, as they have a wide range of
immunodeficiencies, such as a lack of functional lymphocytes, low
levels of serum immunoglobulin, reduced natural killer (NK) cell
activity, dysfunction of antigen-presenting cells, a lack of
circulating complement, less rejection and graft anti-host disease
(26). NOD/SCID mice are generated
by backcrossing SCID mice with non-obese diabetic mice (NOD/Lt).
Currently, subcutaneous injections (27) are commonly used for the
establishment of blood tumours.
Following approval from the Ethics Committee of the
First Affiliated Hospital of Chongqing Medical University, 5- to
6-week-old female NOD/SCID mice (21.57±0.89 g) were purchased from
Beijing HFK Bioscience and bred at the Experimental Animal Centre
at Chongqing Medical University. Briefly, 15 mice were randomly
divided into 3 groups (5 mice per group) as follows:
LV-miR-143-treated SKM-1 cells, LV-control-treated SKM-1 cells and
untreated SKM-1 cells. To generate subcutaneous xenografts,
2×107 SKM-1 cells suspended in a total volume of 200 µl
(PBS pre-chilled to 4°C) were injected into the right flanks of
each mouse. The tumour volume was measured with a calliper every 3
days using the following formula: Volume (mm3) =L x
W2/2 (L represents the largest diameter, while W is the
smallest diameter of the tumour). At the end of the 28-day
observation period, the mice were sacrificed. The weight of the
mice upon sacrifice was 24.2±1.49 g, the maximum tumour diameter of
a single tumour was 1.5 cm and no mouse developed multiple tumours
in this study. The tumour tissues were harvested and preserved in
4% paraformaldehyde buffer for histological analyses (28).
Histological analyses
The xenograft tumour tissues were fixed in 4%
paraformaldehyde solution, paraffin-embedded and cut into
3-µm-thick sections. The paraffin sections were then stained with
haematoxylin and eosin (H&E). Immunohistochemistry for FasL and
caspase-3 was performed on two sets of xenograft tissues from each
group using a standard technique (29). One set of slides was incubated with
the FasL antibody (cat. no. 13098-1-AP, 1:500 dilution, rabbit
polyclonal; Proteintech Group, Inc., Chicago, IL, USA), whilst the
other set was incubated with the caspase-3 antibody (cat. no.
66470-2-lg, 1:500 dilution, rabbit polyclonal; Proteintech) in
Normal Antibody Diluent (ScyTek, phosphate-buffered). The reaction
was developed using the protocol of Histostain™-Plus kits
(ZSGB-BIO) and counterstained with haematoxylin. Apoptosis was also
assessed in situ using the terminal deoxynucleotidyl transferase
(TdT) dUTP Nick-End Labeling (TUNEL) apoptosis detection kit (cat.
no. C1089, Beyotime) according to the manufacturer's instructions.
Cell counting were performed in 3 random fields of each section and
cells with dark brown cores were identified as apoptotic cells. The
results showed as a percentage of apoptotic cells.
Statistical analysis
All statistical analyses were conducted using SPSS
20.0 software (IBM, Chicago, IL, USA). The results are presented as
the means ± standard deviation (SD). A two-tailed Student's t-test
was carried out to determine significance when only 2 groups were
compared. Variations between groups were analysed with a one-way
ANOVA, followed by Tukey's multiple comparison test. Values of
P<0.05 and P<0.01 were considered to indicate statistically
significant differences and highly statistically significant
differences, respectively.
Results
miR-143 expression levels are decreased
in MDS/AML samples and cell lines
The expression levels of miR-143 in 33 patients with
MDS/AML and SKM-1 were compared to those of 11 healthy controls.
Following normalization and data calculations, miR-143 expression
was found to be significantly decreased in MDSand secondary AML
(sAML) samples and cell lines compared to the healthy controls
(Fig. 1A and B). Compared to the
low-risk MDS group, the expression of miR-143 was evidently
decreased in the high-risk MDS group (Fig. 1C). Moreover, the miR-143 expression
levels in patients with MDS with the 5q deletion were significantly
lower than the normal cytogenetic type (Fig. 1D).
Recombinant lentiviral vectors lead to
variations in the miR-143 levels in SKM-1 cells
To clarify the role of miR-143 in MDS, the SKM-1
cells were transfected with LV-hsa-miR-143 and
LV-hsa-miR-143-inhibitors, respectively. The infection efficiency
was evaluated using a fluorescence microscope (Fig. 2A) and flow cytometry, with >60%
of the SKM-1 cells being positive for green fluorescent protein
(GFP) (Fig. 2B). miR-143
expression was then detected by RT-qPCR. Compared to the LV-control
and the normal control (SKM-1 cells), LV-hsa-miR-143 induced the
upregulation of miR-143 expression 9-fold, whereas
LV-hsa-miR-143-inhibitors downregulated the expression of miR-143
by approximately a quarter (Fig.
2C).
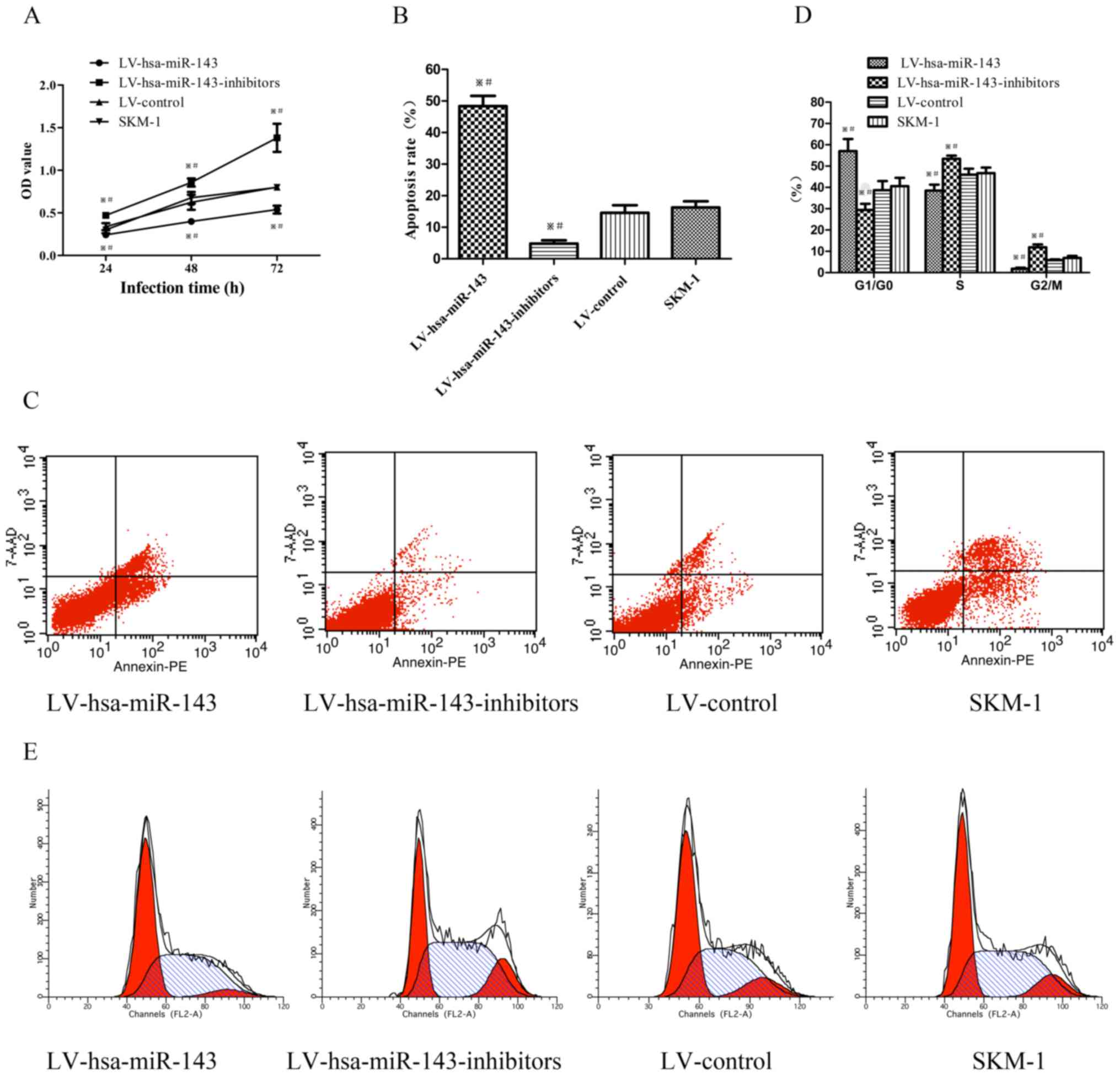
miR-143 affects the proliferation,
apoptosis and cell cycle of SKM-1 cells
To assess the role of miR-143 in the SKM-1 cells
following transfection, a CCK-8 assay was used to evaluate the
effects of miR-143 on the proliferation of SKM-1 cells. The optical
density (OD) value of the cells transfected with LV-hsa-mir-143 was
significantly lower than that of the negative control and SKM-1
cells, whereas the OD of cells transfected with
LV-hsa-miR-143-inhibitors was higher than control groups (Fig. 3A). The apoptotic rate of the SKM-1
cells trans-fected with LV-hsa-mir-143 demonstrated a significant
3-fold increase, while the apoptotic rate of the cells transfected
with LV-hsa-miR-143-inhibitors was decreased by more than half,
compared to the control groups (Fig.
3B and C). Furthermore, the cell cycle profile was examined by
flow cytometry. The number of SKM-1 cells transfected with
LV-hsa-miR-143 arrested in the G1/G0 phase increased by >16%,
while that of the cells transfected with LV-hsa-miR-143-inhibitors
decreased by 10% compared to the control groups (Fig. 3D and E). Taken together, these
results suggest that miR-143 inhibits the proliferation, and
induces the apoptosis and cell cycle arrest of SKM-1 cells.
The overexpression of miR-143 activates
the Fas/FasL apop- totic pathway
To further examine the specific mechanisms of the
miR-143-induced apoptosis in SKM-1 cells, RT-qPCR (Fig. 4A) and western blot analysis
(Fig. 4B and C) were employed for
the detection of FasL, cleaved caspase-8, -9 and -3. An increased
expression of the apoptotic factors, caspase-8, -3 and -9 and FasL
at both the mRNA and protein level compared to the LV-control and
SKM-1 cells was observed, confirming that miR-143 overexpression
activates the Fas/FasL apoptotic pathway.
LV-hsa-miR-143 binds to the 3'-UTR of AF9
and inhibits its expression
To further elucidate the possible molecular
mechanisms involved, the target prediction website, Targetscan was
utilised (30). Bioinformatics
analyses indicated that the MLLT3/AF9 gene was a putative target of
miR-143 (Fig. 5A). We conducted a
Firefly luciferase reporter assay to clarify whether AF9 is a
direct target of miR-143. The luciferase reporter containing the
AF9 3'-UTR was signifi-cantly suppressed by miR-143, whereas the
mutated reporter was not affected (Fig. 5B). Subsequently, miR-143 inhibitors
and pMIR-MLLT3-WT vector were transfected into the SKM-1 cells,
with results demonstrating that miR-143 inhibitors can increase
luciferase activity, hence indicating that LV-hsa-miR-143 binds to
the 3'-UTR of AF9 and inhibits its expression (Fig. 5C). Furthermore, western blot
analysis was performed to determine whether the knockdown miR-143
led to an upregulation of MLLT3/AF9 expression (Fig. 5D and E). The results revealed that
AF9 was a target of and regulated by miR-143.
Overexpression of miR-143 inhibits tumour
growth and promotes apoptosis in vivo
To verify the function of miR-143 in tumour growth
in the complex microenvironment, NOD/SCID mice were subcutaneously
injected with LV-hsa-miR-143 SKM-1 cells, LV-control SKM-1 cells or
untreated SKM-1 cells. Consistent with the in vitro
findings, tumours derived from SKM-1 cells overexpressing miR-143
exhibited significantly reduced growth (Fig. 6A-C). Western blot analysis was
employed for the detection of FasL and cleaved caspase-3 in the
tumour samples, with the results demonstrating that the
overexpression of miR-143 enhanced the expression of FasL and
cleaved caspase-3 compared to that in the SKM-1 cell group
(Fig. 6D and E). H&E staining
of the xenograft tumours revealed unsystematic and irregular tumour
cell arrangements and an increased the nuclear-cytoplasmic ratio,
which is consistent with the pathological characteristics of
malignancy (Fig. 6F). The results
of immunohistochemical staining revealed that both FasL and
caspase-3 were strongly positive in the LV-hsa-miR-143 group
(Fig. 6G). Moreover, a TUNEL assay
was performed, which revealed that the proportion of apoptotic
cells was higher in the LV-hsa-miR-143 group (31.71±2.78%) than in
the LV-control group (15.32±3.91%, P<0.001) or the untreated
SKM-1 cell group (13.29±4.15%, P<0.001, Fig. 6I and J). Taken together, these
results confirm that the overexpression of miR-143 activates the
FasL apop-totic pathway.
Discussion
Recent studies have demonstrated that miR-143 plays
an inhibitory role in different tumours types and has a significant
effect on the haematopoietic system (31,32).
To further support these findings, the results of the present study
suggested that the overexpression of miR-143 significantly
inhibited AML cell proliferation. Additionally, we also
demonstrated that the down-regulation of miR-143 significantly
promoted cell proliferation, suggesting that miR-143 is likely an
anticancer factor for MDS/AML. To further elucidate the mechanisms
through which miR-143 inhibits SKM-1 cell proliferation, flow
cytometric analysis was performed to assess the cell cycle profile,
with an increased cell number of cells in the LV-hsa-miR-143 group
in the G0/G1 phase and a decreased number in the S phase being
observed. By contrast, the LV-hsa-miR-143-inhibitors group
exhibited favorable conditions for cell apoptosis in the G0/G1
phase. Therefore, it can be concluded that miR-143 inhibits AML
cell proliferation, promotes apoptosis and arrests cell growth
cycle at the G0/G1 phase.
To verify whether the in vitro effects of
miR-143 on MDS reflect the in vivo response, experiments
using NOD/SCID mice which were subcutaneously injected with SKM-1
cells stably expressing miR-143 were performed. The results
indicated that the upregulation of miR-143 expression increased
apoptosis through the activation of the FasL pathway, thereby
attenuating the growth of the tumour xenografts. Furthermore, these
findings provide evidence that the overexpression of miR-143
promotes the apoptosis of SKM-1 cells by activating the Fas/FasL
signalling pathway. To further explore the mechanisms through which
miR-143 overexpression promotes apoptosis, the MLLT3/AF9 gene was
selected using the TargetScan online bioinformatics tool, which
predicts biological targets of miRNAs by searching for the presence
of conserved 8mer, 7mer and 6mer sites matching the seed region of
each miRNA (33). MLLT3/AF9 is a
proto-oncogene capable of relieving the expression of key genes in
leukaemia, and it may be a key regulator of lymphoma cell
proliferation or carcinogenesis (34-36).
As described herein, both a wild-type and mutant vector of the
MLLT3/AF9 3'-UTR were constructed and inhibition was verified with
a Firefly luciferase reporter gene assay. The results demonstrated
that miR-143 analogues inhibited wild-type MLLT3/AF9, whereas the
mutant remaind unaffected, thus confirming the status of the
MLLT3/AF9 gene as a target gene of miR-143.
Taken together, the findings of this study suggest
that miR-143 overexpression inhibits the proliferation of SKM-1
cells both in vitro and in vivo. This may be due to
miR-143 affecting the apoptotic pathway of FasL, hence supporting
the use of MLLT3/AF9 inhibitors as potential antitumour therapy for
MDS, whose effects need to be extensively confirmed by clinical
trials.
Acknowledgments
The authors would like to thank Mr. Peng etc. from
The Chongqing Key Laboratory of Translational Medicine in Major
Metabolic Diseases for providing assistance with the experiments.
The MDS/AML cell line SKM-1 cells were kindly provided by Professor
Jianfeng Zhou of Tongji Medical College of Huazhong University of
Science and Technology (Wuhan, China).
Funding
This study was supported by the Chongqing Education
Commission Foundation (KJ1702017) and the National Natural Science
Foundation of China (grant nos. 30971277 and 81250034).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CW and CP performed the molecular experiments and
drafted the manuscript. JC and LD performed the in vivo
assays. XK took part in data collection and processing. ZZ was
responsible for the preparation of pathological sections and
immunohistochemistry. JC and LW were responsible for the conception
and design of the study and manuscript revision. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
All clinical samples were collected by the informed
consent signed by the patients and approved by the Institutional
Review Board of The First Affiliated Hospital of Chongqing Medical
University. Animal handling and procedures were approved by the
Ethics Committee of the First Affiliated Hospital of Chongqing
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
de Witte T, Bowen D, Robin M, Malcovati L,
Niederwieser D, Yakoub-Agha I, Mufti GJ, Fenaux P, Sanz G, Martino
R, et al: Allogeneic hematopoietic stem cell transplantation for
MDS and CMML: Recommendations from an international expert panel.
Blood. 129:1753–1762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pellagatti A and Boultwood J: The
molecular pathogenesis of the myelodysplastic syndromes. Eur J
Haematol. 95:3–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mandal N: Bibliometric analysis of global
publication output and collaboration structure study in microRNA
research. Scientometrics. 98:2011–2037. 2014. View Article : Google Scholar
|
|
4
|
Kuang X, Chi J and Wang L: Deregulated
microRNA expression and its pathogenetic implications for
myelodysplastic syndromes. Hematology. 21:593–602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Simmer F, Venderbosch S, Dijkstra JR,
Vink-Börger EM, Faber C, Mekenkamp LJ, Koopman M, De Haan AF, Punt
CJ and Nagtegaal ID: MicroRNA-143 is a putative predictive factor
for the response to fluoropyrimidine-based chemotherapy in patients
with metastatic colorectal cancer. Oncotarget. 6:22996–23007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu Y, Ma Z, He Y, Liu W, Su Y and Tang Z:
PART-1 functions as a competitive endogenous RNA for promoting
tumor progression by sponging miR-143 in colorectal cancer. Biochem
Biophys Res Commun. 490:317–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu B, Liu X and Chang H: MicroRNA-143
inhibits colorectal cancer cell proliferation by targeting MMP7.
Minerva Med. 108:13–19. 2017.
|
|
8
|
Zhou P, Chen WG and Li XW: MicroRNA-143
acts as a tumor suppressor by targeting hexokinase 2 in human
prostate cancer. Am J Cancer Res. 5:2056–2063. 2015.PubMed/NCBI
|
|
9
|
Rodríguez M, Bajo-Santos C, Hessvik NP,
Lorenz S, Fromm B, Berge V, Sandvig K, Linē A and Llorente A:
Identification of non-invasive miRNAs biomarkers for prostate
cancer by deep sequencing analysis of urinary exosomes. Mol Cancer.
16:1562017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang F, Liu J, Zou Y, Jiao Y, Huang Y, Fan
L, Li X, Yu H, He C, Wei W, et al: MicroRNA-143-3p, up-regulated in
H. pylori-positive gastric cancer, suppresses tumor growth,
migration and invasion by directly targeting AKT2. Oncotarget.
8:28711–28724. 2017.PubMed/NCBI
|
|
11
|
Du F, Feng Y, Fang J and Yang M:
MicroRNA-143 enhances chemosensitivity of Quercetin through
autophagy inhibition via target GABARAPL1 in gastric cancer cells.
Biomed Pharmacother. 74:169–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Wang X and Zhang Y and Zhang Y: E3
ubiquitin ligase isolated by differential display regulates
cervical cancer growth in vitro and in vivo via microRNA-143. Exp
Ther Med. 12:676–682. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong P, Xiong Y, Hanley SJB, Yue J and
Watari H: Musashi-2, a novel oncoprotein promoting cervical cancer
cell growth and invasion, is negatively regulated by p53-induced
miR-143 and miR-107 activation. J Exp Clin Cancer Res. 36:1502017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen JH, Yang R, Zhang W and Wang YP:
Functions of microRNA-143 in the apoptosis, invasion and migration
of naso-pharyngeal carcinoma. Exp Ther Med. 12:3749–3755. 2016.
View Article : Google Scholar
|
|
15
|
He B, Xu Z, Chen J, Zheng D, Li A and
Zhang LS: Upregulated microRNA-143 inhibits cell proliferation in
human nasopha-ryngeal carcinoma. Oncol Lett. 12:5023–5028. 2016.
View Article : Google Scholar
|
|
16
|
Li WH, Wu HJ, Li YX, Pan HG, Meng T and
Wang X: MicroRNA-143 promotes apoptosis of osteosarcoma cells by
caspase-3 activation via targeting Bcl-2. Biomed Pharmacother.
80:8–15. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Wang G, Ding C, Liu P, Wang R,
Ding W, Tong D, Wu D, Li C, Wei Q, et al: Increased circular RNA
UBAP2 acts as a sponge of miR-143 to promote osteosarcoma
progression. Oncotarget. 8:61687–61697. 2017.PubMed/NCBI
|
|
18
|
Akao Y, Nakagawa Y, Iio A and Naoe T: Role
of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia
Jurkat cells. Leuk Res. 33:1530–1538. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ozdogan H, Gur Dedeoglu B, Oztemur
Islakoglu Y, Aydos A, Kose S, Atalay A, Yegin ZA, Avcu F, Uckan
Cetinkaya D and Ilhan O: DICER1 gene and miRNA dysregulation in
mesen-chymal stem cells of patients with myelodysplastic syndrome
and acute myeloblastic leukemia. Leuk Res. 63:62–71. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Votavova H, Grmanova M, Dostalova
Merkerova M, Belickova M, Vasikova A, Neuwirtova R and Cermak J:
Differential expression of microRNAs in CD34+ cells of
5q- syndrome. J Hematol Oncol. 4:12011. View Article : Google Scholar
|
|
21
|
Dostalova Merkerova M, Krejcik Z, Votavova
H, Belickova M, Vasikova A and Cermak J: Distinctive microRNA
expression profiles in CD34+ bone marrow cells from
patients with myelo-dysplastic syndrome. Eur J Hum Genet.
19:313–319. 2011. View Article : Google Scholar
|
|
22
|
Wang L, Luo J, Nian Q, Xiao Q, Yang Z and
Liu L: Ribosomal protein S14 silencing inhibits growth of acute
myeloid leukemia transformed from myelodysplastic syndromes via
activating p53. Hematology. 19:225–231. 2014. View Article : Google Scholar
|
|
23
|
Liu Z, Ding K, Li L, Liu H, Wang Y, Liu C
and Fu R: A novel histone deacetylase inhibitor Chidamide induces
G0/G1 arrest and apoptosis in myelodysplastic syndromes. Biomed
Pharmacother. 83:1032–1037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Tiscornia G, Singer O and Verma IM:
Production and purification of lentiviral vectors. Nat Protoc.
1:241–245. 2006. View Article : Google Scholar
|
|
26
|
Proetzel G and Wiles MV: Mouse Models for
Drug Discovery: Methods and Protocols. Humana Press; Totowa, NJ:
pp. 2010
|
|
27
|
Wu L, Li X, Su J, He Q, Zhang X, Chang C
and Pu Q: Efficacy and safety of CHG regimen (low-dose cytarabine,
homoharringtonine with G-CSF priming) as induction chemotherapy for
elderly patients with high-risk MDS or AML transformed from MDS. J
Cancer Res Clin Oncol. 137:1563–1569. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pramanik D, Campbell NR, Karikari C,
Chivukula R, Kent OA, Mendell JT and Maitra A: Restitution of tumor
suppressor microRNAs using a systemic nanovector inhibits
pancreatic cancer growth in mice. Mol Cancer Ther. 10:1470–1480.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guerenne L, Beurlet S, Said M, Gorombei P,
Le Pogam C, Guidez F, de la Grange P, Omidvar N, Vanneaux V, Mills
K, et al: GEP analysis validates high risk MDS and acute myeloid
leukemia post MDS mice models and highlights novel dysregulated
pathways. J Hematol Oncol. 9:52016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
31
|
Zhou J, Chaudhry H, Zhong Y, Ali MM,
Perkins LA, Owens WB, Morales JE, McGuire FR, Zumbrun EE, Zhang J,
et al: Dysregulation in microRNA expression in peripheral blood
mononuclear cells of sepsis patients is associated with
immunopathology. Cytokine. 71:89–100. 2015. View Article : Google Scholar
|
|
32
|
Shen JZ, Zhang YY, Fu HY, Wu DS and Zhou
HR: Overexpression of microRNA-143 inhibits growth and induces
apoptosis in human leukemia cells. Oncol Rep. 31:2035–2042. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen X, Clark J, Wunderlich M, Fan C,
Davis A, Chen S, Guan JL, Mulloy JC, Kumar A and Zheng Y: Autophagy
is dispensable for Kmt2a/Mll-Mllt3/Af9 AML maintenance and
anti-leukemic effect of chloroquine. Autophagy. 13:955–966. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang T, Luo Y, Wang T and Yang JY:
MicroRNA-297b-5p/3p target Mllt3/Af9 to suppress lymphoma cell
proliferation, migration and invasion in vitro and tumor growth in
nude mice. Leuk Lymphoma. 53:2033–2040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vogel T and Gruss P: Expression of
leukaemia associated transcription factor Af9/Mllt3 in the cerebral
cortex of the mouse. Gene Expr Patterns. 9:83–93. 2009. View Article : Google Scholar
|