Introduction
Thyroid cancer, which is derived from follicular
thyroid cells, is the most common endocrine malignancy, which
accounts for ~90% of neuroendocrine tumors (1,2). The
global incidence of thyroid cancer has markedly increased in recent
decades (3), with an estimated
300,000 new cases diagnosed annually and 40,000 cases of thyroid
cancer-associated morality occurring globally (4). Based on different pathological
features, thyroid cancer can be divided into five subtypes:
Papillary thyroid cancer (PTC), follicular thyroid cancer, poorly
differentiated thyroid cancer, anaplastic thyroid cancer and
thyroid squamous cell carcinoma (5). PTC, which accounts for 85-90% of all
thyroid cancer cases, is the most prevalent histological subtype of
thyroid cancer (6). Despite
significant advances in diagnosis and therapy, recurrence and/or
metastasis occurs in ~10% of patients with PTC, which heralds a
poor prognosis (7). Therefore, an
in-depth understanding of the molecular mechanism underlying the
initiation and progression of PTC will aid in the development of
novel diagnostic biomarkers and effective therapeutic strategies
for patients with PTC.
MicroRNAs (miRNAs/miRs) are an abundant class of
17-24 nucleotide long, non-coding RNAs (8). miRNAs negatively modulate gene
expression through direct binding to the 3-untranslated regions
(3'-UTRs) of target genes, thus leading to translational inhibition
and/or mRNA degradation (9).
miRNAs not only serve crucial roles in regulating various
fundamental cellular processes but are also closely associated with
tumorigenesis and tumor development (10,11).
Previous studies have reported that miRNAs are aberrantly expressed
in almost all types of human cancer, including PTC (12), colorectal cancer (13), lung cancer (14), glioblastoma (15) and bladder cancer (16). Increasing evidence has revealed
that various miRNAs are differentially expressed in PTC, and their
dysregulation has been implicated in the regulation of PTC
occurrence and development (17-19).
Furthermore, miRNAs may have tumor suppressive or oncogenic roles
in the progression of PTC and are able to regulate various
cancer-associated biological processes, including cell
proliferation, cell cycle progression, apoptosis, invasion,
metastasis and epithelial-mesenchymal transition (20-22).
Therefore, dysregulated miRNAs require further investigation, in
order to identify potential therapeutic targets for the treatment
of patients with PTC.
miR-766 has been well studied in numerous types of
human cancer, including renal cell carcinoma (23), lung adenocarcinoma (24) and colorectal cancer (25). However, the expression status,
specific roles and regulatory mechanisms of miR-766 in PTC remain
largely unclear. Therefore, the aim of the present study was to
detect miR-766 expression in PTC tissues and cell lines, to clarify
the clinical significance of miR-766 in patients with PTC, and to
explore the biological roles of miR-766 in the malignant biological
behaviors of PTC cells. This study also aimed to determine the
underlying mechanism of action of miR-766 in PTC cells.
Materials and methods
Clinical specimens
This study was approved by the Ethics Committee of
Daping Hospital (Chongqing, China), in accordance with the
principles expressed in the Declaration of Helsinki. Written
informed consent was obtained from all participants prior to
surgery. In total, 47 pairs of PTC tissues and matched adjacent
normal tissues were collected from patients who had received
surgical resection at Daping Hospital between June 2015 and May
2017. None of the patients had undergone chemotherapy or
radiotherapy prior to surgery. All tissues were quickly frozen in
liquid nitrogen after being excised and were stored at −80°C until
further use.
Cell lines and culture conditions
A normal human thyroid cell line (HT-ori3), two
human PTC cell lines (HTH83 and TPC-1) and a thyroid cancer cell
line (BCPAP) were purchased from American Type Culture Collection
(Manassas, VA, USA). Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% v/v heat-inactivated fetal bovine serum (FBS;
both from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
and 1% v/v penicillin-streptomycin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) were used to culture the cells. Cells were
grown at 37°C in a humidified atmosphere containing 5%
CO2.
Transfection
miR-766 mimics and a corresponding negative control
(miR-NC) were purchased from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The sequences were as follows: miR-766 mimics,
5'-ACUCCAGCCCCACAGCCUCAGC-3'; miR-NC, 5'-UUCUCCGAACGUGUCACGUTT-3'.
Small interfering RNA (siRNA) targeting insulin receptor substrate
2 (IRS2) expression (IRS2 siRNA) and a negative control siRNA (NC
siRNA) were acquired from Shanghai GenePharma Co., Ltd. (Shanghai,
China). The sequences were as follows: IRS2 siRNA,
5'-AAUAGCUGCAAGAGCGAUGAC-3'; NC siRNA, 5'-UUCUCCGAACGUGUCACGUTT-3'.
An IRS2 overexpression plasmid was chemically synthesized using
pcDNA3.1(+) basic vectors at the Chinese Academy of Sciences
(Changchun, China). HTH83 and TPC-1 cells were seeded into 6-well
plates at a density of 8×105 cells/well. Cells were transfected
with mimics (100 pmol), siRNAs (100 pmol) or plasmids (4 µg)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Cells
were incubated at 37°C in a humidified atmosphere containing 5%
CO2. After 48 h, reverse transcription-quantitative
polymerase chain reaction (RT-qPCR), flow cytometric analysis, and
cell migration and invasion assays were performed. MTT and colony
formation assays were conducted at 24 h post-transfection. Western
blot analysis was conducted 72 h post-transfection.
RT-qPCR analysis
Total RNA was isolated from cultured cells or
homogenized tissues using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
For the detection of miR-766 expression, RT was performed using a
TaqMan MicroRNA RT kit, according to the manufacturer's protocol,
followed by qPCR with a TaqMan MicroRNA PCR kit (both from Applied
Biosystems; Thermo Fisher Scientific, Inc.). The cycling conditions
for qPCR were as follows: 50°C for 2 min and 95°C for 10 min,
followed by 40 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 60 sec, and a final extension step
at 4°C for 5 min. U6 small nuclear RNA served as an internal
control for miR-766 expression. For IRS2 mRNA quantification, first
strand cDNA was synthesized using a PrimeScript RT Reagent kit
(Takara Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's protocol. qPCR was then performed using a SYBR
Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.). The cycling
conditions for qPCR were as follows: 5 min at 95°C, followed by 40
cycles at 95°C for 30 sec and 65°C for 45 sec, and a final
extension step at 40°C for 30 sec. GAPDH was used for IRS2 mRNA
normalization. The primers were designed as follows: miR-766,
forward 5'-TCG AGTACTTGAGATGGAGTTTT-3', reverse
5'-GGCCGCGTTGCAGTGAGCCGAG-3'; U6, forward
5'-GCTTCGGCAGCACATATACTAAAAT-3', reverse
5'-CGCTTCACGAATTTGCGTGTCAT-3'; IRS2, forward,
5'-CACAATTCCAAGCGCCACAA-3', reverse 5'-ATCAAAGCTCCAGGCTGACC-3'; and
GAPDH, forward 5'-ACCCACTCCTCCACCTTTG-3' and reverse
5'-CTCTTGTGCTCTTGCTGGG-3'. Relative gene expression was calculated
using the 2−ΔΔCq method (26).
MTT assay
After 24 h of incubation, transfected cells in the
exponential growth stage were collected and seeded into 96-well
plates at a density of 3,000 cells/well. At 0, 24, 48 and 72 h
following inoculation, the MTT assay was performed to determine
cell proliferation. Briefly, 20 µl 5 mg/ml MTT solution
(Sigma-Aldrich; Merck KGaA) was added to each well for 4 h at 37°C
and 5% CO2. Subsequently, the culture medium was gently
removed and formazan precipitates were dissolved in 100 µl
dimethyl sulfoxide (Beyotime Institute of Biotechnology,, Inc.,
Shanghai, China). The absorbance was detected at a wavelength of
490 nm using a microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Colony formation assay
Logarithmically growing transfected cells were
harvested after 24 h of incubation and were plated into 6-well
plates at a density of 1,000 cells/well. Cells were then maintained
at 37°C in a humidified atmosphere containing 5% CO2 for
2 weeks. Subsequently, cells were washed with PBS (Gibco; Thermo
Fisher Scientific, Inc.), fixed with 100% methanol at room
temperature for 20 min and stained with methyl violet (Beyotime
Institute of Biotechnology) at room temperature for 20 min. The
number of colonies (>50 cells/colony) was counted under an
inverted light microscope (IX71; Olympus Corporation, Tokyo,
Japan).
Flow cytometric analysis of cell
apoptosis
An Annexin V-fluorescein isothiocyanate (FITC)
Apoptosis Detection kit (Biolegend, Inc., San Diego, CA, USA) was
used to evaluate the percentage of apoptotic cells. Briefly,
transfected cells were seeded into 6-well plates (1×106
cells/well), incubated for 48 h at 37°C and 5% CO2,
harvested, washed three times with PBS, and suspended in 100
µl binding buffer. Subsequently, 5 µl Annexin V-FITC
and 5 µl propidium iodide were added and the transfected
cells were incubated for 15 min at room temperature in the dark.
Finally, flow cytometry (FACScan; BD Biosciences, Franklin Lakes,
NJ, USA) was used to measure the rate of apoptosis. The data were
analyzed with CellQuest version 5.1 (BD Biosciences).
Cell migration and invasion assays
The migratory ability of PTC cells was evaluated
using Transwell chambers (BD Biosciences) with an 8-µm pore
polycarbonate membrane. A total of 48 h post-transfection,
5×104 cells were suspended in FBS-free DMEM and were
inoculated into the upper chamber. The lower chambers were filled
with 500 µl DMEM supplemented with 20% FBS. After 24 h at
37°C, the non-migrated cells were removed using a cotton swab,
whereas the migrated cells were fixed with 100% methanol and
stained with 0.5% crystal violet (Beyotime Institute of
Biotechnology). Images of the cells were captured and migratory
ability was quantified by counting the number of migrated cells in
five randomly selected visual fields from each chamber using an
inverted light microscope. The cell invasion assay was conducted in
a similar manner to the migration assay; however, the Transwell
chambers were initially coated with Matrigel (BD Biosciences).
In vivo xenograft experiment
BALB/c nude mice (female; age, 4 weeks; weight, 20
g) were purchased from the Shanghai Laboratory Animal Center
(Chinese Academy of Sciences, Shanghai, China) and were divided
into two groups (n=4/group), which were subcutaneously injected
with TPC-1 cells transfected with miR-766 mimics or miR-NC,
respectively. The animals were maintained under specific
pathogen-free conditions (25°C, 50% humidity, 10-h light/14-h dark
cycle). The mice received free access to normal rodent food and
water, which was autoclaved. Tumor length and width were measured
every 2 days. BALB/c nude mice were sacrificed 30 days following
implantation, and the tumor xenografts were excised and weighed.
The tumor volumes were analyzed using the following formula: Tumor
volume = 1/2 × tumor length × tumor width. Experimental procedures
and protocols were approved by the Institutional Animal Care and
Use Committee of Daping Hospital.
Bioinformatics analysis and luciferase
reporter assay
The putative miR-766 target genes were predicted
using TargetScan software (http://www.targetscan.org) and miRDB software
(http://mirdb.org/). The wild-type (wt) and mutant
(mut) 3'-UTRs of IRS2 were created by Shanghai GenePharma Co.,
Ltd., and were inserted downstream of the psiCHECK2 luciferase
reporter vector (Promega Corporation, Madison, WI, USA), in order
to generate psiCHECK2-IRS2-3'-UTR wt and psiCHECK2-IRS2-3'-UTR mut,
respectively. Cells were plated into 24-well plates 12 h prior to
transfection at a density of 1.5×105 cells/well.
Co-transfection with the constructed luciferase reporter plasmids
(0.2 µg) and miR-766 mimics (50 pmol) or miR-NC (50 pmol)
was performed using Lipofectamine® 2000, according to
the manufacturer's protocol. A total of 48 h post-transfection,
luciferase activity was determined using a dual-luciferase reporter
assay system (Promega Corporation), according to the manufacturer's
protocol. The activity of firefly luciferase was normalized to that
of Renilla luciferase.
Western blot analysis
Total protein was isolated from tissues or cells
using radioimmunoprecipitation assay buffer (Sigma-Aldrich; Merck
KGaA) and was quantified using a bicinchoninic acid protein assay
kit (Beyotime Institute of Biotechnology). Equal amounts of protein
(30 µg) were separated by 10% SDS-PAGE and were transferred
to polyvinylidene fluoride membranes (Beyotime Institute of
Biotechnology), followed by blocking at room temperature for 1 h
with 5% w/v dried skimmed milk diluted in Tris-buffered saline with
0.1% Tween-20 (TBST). The membranes were then incubated overnight
at 4°C with primary antibodies against IRS2 (cat. no. ab52606;
1:1,000 dilution; Abcam, Cambridge, UK), phosphorylated
(p)-phosphoinositide 3-kinase (PI3K; cat. no. ab182651; 1:1,000
dilution; Abcam), PI3K (cat. no. ab191606; 1:1000 dilution; Abcam),
p-protein kinase B (Akt; cat. no. sc-81433; 1:1,000 dilution; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA), Akt (cat. no. sc-56878;
1:1,000 dilution; Santa Cruz Biotechnology, Inc.), or GAPDH (cat.
no. ab128915; 1:1,000 dilution; Abcam). After three washes with
TBST, the membranes were incubated with corresponding horseradish
peroxidase-conjugated secondary antibodies (cat. nos. ab205719 or
ab6721; 1:5,000 dilutions; Abcam) for 2 h at room temperature and
were subjected to visualization using an enhanced chemiluminescence
detection kit (Pierce; Thermo Fisher Scientific, Inc.). Protein
expression was semi-quantified using Quantity One software version
4.62 (Bio-Rad Laboratories, Inc.).
Statistical analyses
SPSS (version 17.0; SPSS, Inc., Chicago, IL, USA)
was used for statistical analysis. Data are presented as the means
± standard deviation from at least three independent experiments,
and the differences between groups were compared using Student's
t-test or one-way analysis of variance (ANOVA).
Student-Newman-Keuls test was used as a post hoc test following
ANOVA. The association between miR-766 and clinicopathological
characteristics of patients with PTC was analyzed using the
χ2 test. Spearman correlation analysis was employed to
investigate the association between miR-766 and IRS2 mRNA
expression in PTC tissues. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-766 expression is decreased in PTC
tissues and cell lines
To uncover the expression pattern of miR-766 in PTC,
RT-qPCR analysis was performed to detect miR-766 expression in 47
pairs of PTC tissues and matched adjacent normal tissues. PTC
tissues exhibited significantly lower miR-766 expression compared
with in the matched adjacent normal tissues (Fig. 1A; P<0.05). Subsequently, the
clinical significance of miR-766 in PTC was investigated. Briefly,
all patients with PTC were divided into miR-766 low (n=24) or
miR-766 high (n=23) expression groups, based on the median value of
miR-766 expression. Low miR-766 expression was significantly
associated with TNM stage (P=0.008) and lymph node metastasis
(P=0.039), whereas no obvious association was observed between
miR-766 and age, sex or tumor size (all P>0.05; Table I). In addition, miR-766 expression
was measured in two PTC cell lines and in a general thyroid cancer
cell line. The data obtained from RT-qPCR indicated that miR-766
was downregulated in the two PTC cell lines (HTH83 and TPC-1) and
the thyroid cancer cell line (BCPAP) compared with in the normal
human thyroid cell line (HT-ori3) (Fig. 1B; P<0.05). These results
indicated that downregulation of miR-766 may have a critical role
in PTC tumorigenesis and development.
 | Table IAssociation between miR-766
expression and clinico-pathological features in patients with
papillary thyroid cancer. |
Table I
Association between miR-766
expression and clinico-pathological features in patients with
papillary thyroid cancer.
| Variable | miR-766 expression
| P-value |
|---|
| Low | High |
|---|
| Age | | | 0.556 |
| <60 years | 8 | 10 | |
| ≥60 years | 16 | 13 | |
| Sex | | | 0.561 |
| Male | 12 | 7 | |
| Female | 12 | 16 | |
| Tumor size | | | 0.238 |
| <5 cm | 15 | 12 | |
| ≥5 cm | 9 | 11 | |
| Lymph node
metastasis | | | 0.039a |
| Negative | 6 | 13 | |
| Positive | 18 | 10 | |
| TNM stage | | | 0.008a |
| I-II | 6 | 15 | |
| III-IV | 18 | 8 | |
miR-766 is involved in the regulation of
cell proliferation, apoptosis, migration and invasion in PTC
To clarify the roles of miR-766 in PTC progression,
HTH83 and TPC-1 cell lines, which exhibited lower miR-766
expression among the three thyroid cancer cell lines, were selected
for functional assays and were transfected with miR-766 mimics or
miR-NC. Post-transfection, miR-766 was markedly overexpressed in
miR-766 mimics-transfected HTH83 and TPC-1 cells compared with in
cells transfected with miR-NC (Fig.
2A; P<0.05). The role of miR-766 overexpression in the
proliferation of PTC cells was determined by MTT and colony
formation assays. Upregulation of miR-766 significantly inhibited
the proliferation (Fig. 2B,
P<0.05) and colony formation (Fig.
2C, P<0.05) of HTH83 and TPC-1 cells. Since an alteration in
cell proliferation is often accompanied with changes in the rate of
apoptosis, the effects of miR-766 on cell apoptosis were assessed
using flow cytometry. Transfection of miR-766 mimics markedly
promoted the apoptosis of HTH83 and TPC-1 cells (Fig. 2D; P<0.05). Cell migration and
invasion assays were used to determine whether miR-766 was
implicated in the regulation of PTC cell metastasis. Ectopic
miR-766 expression markedly restricted the migration (Fig. 2E; P<0.05) and invasion (Fig. 2F; P<0.05) of HTH83 and TPC-1
cells. These findings suggested that miR-766 may exert tumor
suppressor activity in PTC cells.
IRS2 is a direct target gene of miR-766
in PTC cells
Based on bioinformatics analysis, the present study
revealed that the 3'-UTR of IRS2 matched the seed sequence of
miR-766 (Fig. 3A). IRS2 was
selected for further experimental identification because this gene
has previously been implicated in the initiation and progression of
PTC (27). A luciferase reporter
assay was performed to test this hypothesis. The assay demonstrated
that ectopic miR-766 expression significantly reduced the
luciferase activity of psiCHECK2-IRS2-3'-UTR wt in HTH83 and TPC-1
cells (Fig. 3B, P<0.05), but
not that of psiCHECK2-IRS2-3'-UTR mut. These findings indicated
that the 3'-UTR of IRS2 may be directly targeted by miR-766 in PTC
cells. To further clarify the association between miR-766 and IRS2
in PTC, IRS2 mRNA expression was detected in 47 pairs of PTC
tissues and matched adjacent normal tissues using RT-qPCR. The mRNA
expression levels of IRS2 were significantly increased in PTC
tissues (Fig. 3C; P<0.05) and
were inversely correlated with the expression levels of miR-766
(Fig. 3D; r=-0.5143, P=0.0002), as
determined by Spearman correlation analysis. To investigate whether
endogenous IRS2 expression could be regulated by miR-766, RT-qPCR
and western blot analysis were carried out to detect IRS2 mRNA and
protein expression in HTH83 and TPC-1 cells upon miR-766
overexpression. Compared with in the miR-NC group, HTH83 and TPC-1
cells transfected with miR-766 mimics exhibited significantly
decreased levels of IRS2 mRNA (Fig.
3E; P<0.05) and protein (Fig.
3F, P<0.05). Taken together, these results supported the
hypothesis that IRS2 is a direct target gene of miR-766 in PTC
cells.
 | Figure 3IRS2 is a direct target gene of
miR-766 in PTC cells. (A) Wt and mut binding sequences in the
3'-UTR of IRS2. (B) Relative luciferase activity was determined in
HTH83 and TPC-1 cells co-transfected with psiCHECK2-IRS2-3'-UTR wt
or psiCHECK2-IRS2-3'-UTR mut reporter plasmids, and miR-766 mimics
or miR-NC. *P<0.05 vs. miR-NC. (C) mRNA expression
levels of IRS2 in 47 pairs of PTC tissues and matched adjacent
normal tissues, as analyzed by reverse transcription-quantitative
polymerase chain reaction. *P<0.05 vs. normal
tissues. (D) Inverse correlation between miR-766 and IRS2 mRNA
expression in the same set of PTC tissues was validated through
Spearman correlation analysis. r=-0.5143, P=0.0002. (E and F) mRNA
and protein expression levels of IRS2 were detected in HTH83 and
TPC-1 cells transfected with miR-766 mimics or miR-NC.
*P<0.05 vs. miR-NC. 3'-UTR, 3'-untranslated region;
IRS2, insulin receptor substrate 2; miR-766, microRNA-766; mut,
mutant; NC, negative control; PTC, papillary thyroid cancer; wt,
wild-type. |
Knockdown of IRS2 simulates tumor
suppressor activity of miR-766 in PTC cells
IRS2 was validated as a direct target gene of
miR-766 in PTC cells. Therefore, it was hypothesized that the
tumor-suppressing roles of miR-766 in PTC cells may be achieved by
IRS2 knockdown. To evaluate this hypothesis, HTH83 and TPC-1 cells
were transfected with IRS2 siRNA to knockdown endogenous IRS2
expression and examine the function of IRS2 in PTC cells. Western
blot analysis indicated that IRS2 protein expression was
significantly decreased in IRS2 siRNA-transfected HTH83 and TPC-1
cells (Fig. 4A; P<0.05).
Inhibition of IRS2 significantly restricted HTH83 and TPC-1 cell
proliferation (Fig. 4B; P<0.05)
and colony formation (Fig. 4C;
P<0.05), and markedly enhanced cell apoptosis (Fig. 4D; P<0.05). In addition, as shown
in Fig. 4E and F, IRS2 knockdown
significantly attenuated the migratory (P<0.05) and invasive
(P<0.05) abilities of HTH83 and TPC-1 cells compared with in NC
siRNA-transfected cells. These results demonstrated that the
effects of IRS2 knockdown on PTC cells were similar to the effects
of miR-766 overexpression, further suggesting that IRS2 is a
functional downstream target of miR-766 in PTC cells.
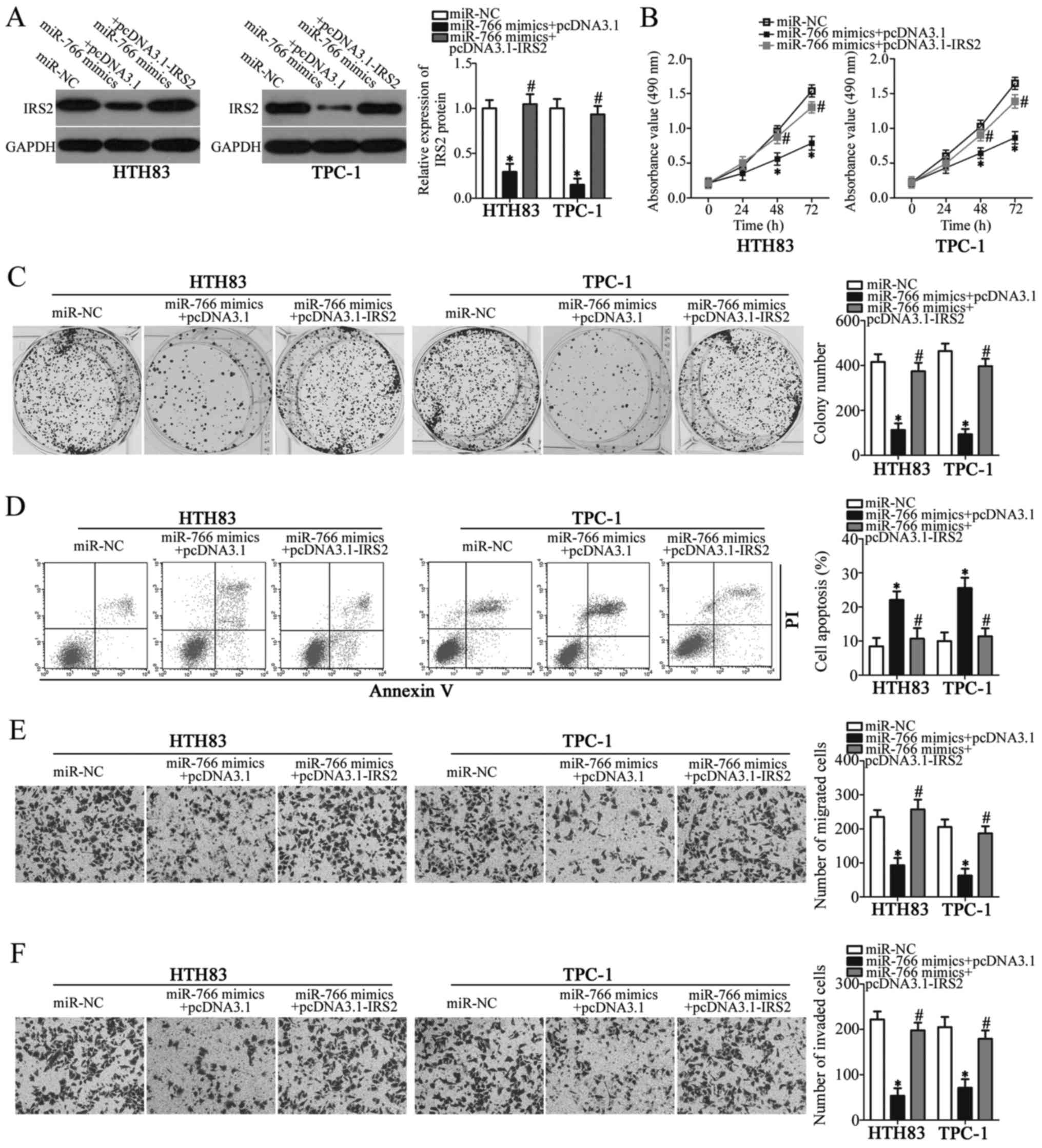
Reintroduction of IRS2 rescues the
effects of miR-766 on the malignant behaviors of PTC cells
Rescue experiments were performed to determine
whether miR-766 exerts tumor-suppressing roles in PTC cells by
inhibiting IRS2 expression. HTH83 and TPC-1 cells were
co-transfected with miR-766 mimics and empty pcDNA3.1 plasmid or
pcDNA3.1-IRS2 lacking the 3'-UTR. Western blot analysis was used to
detect the protein expression levels of IRS2 in the rescue
experiment. miR-766 overexpression-induced down-regulation of IRS2
was significantly recovered in HTH83 and TPC-1 cells following
co-transfection with miR-766 mimics and pcDNA3.1-IRS2 (Fig. 5A; P<0.05). In addition,
functional analyses indicated that restored IRS2 expression could
reverse the tumor suppressive roles of miR-766 overexpression on
the proliferation (Fig. 5B;
P<0.05), colony formation (Fig.
5C; P<0.05), apoptosis (Fig.
5D; P<0.05), migration (Fig.
5E; P<0.05) and invasion (Fig.
5F; P<0.05) of HTH83 and TPC-1 cells. These results
collectively indicated that miR-766 may inhibit the development of
PTC cells, at least partially, through the downregulation of IRS2
expression.
miR-766 suppresses activation of the
PI3K/Akt pathway in PTC cells by directly targeting IRS2
IRS2 has previously been reported to participate in
regulation of the PI3K/Akt pathway (28-30).
Therefore, it was hypothesized that miR-766 may target IRS2 to
inhibit the activation of PI3K/Akt signaling in PTC cells. To
confirm this hypothesis, HTH83 and TPC-1 cells were co-transfected
with miR-766 mimics and pcDNA3.1 or pcDNA3.1-IRS2, and western blot
analysis was performed 72 h post-transfection to determine the
expression of molecules associated with the PI3K/Akt pathway. A
marked decrease in p-PI3K and p-Akt was observed in HTH83 and TPC-1
cells upon miR-766 overexpression; however, this did not affect
total PI3K and Akt expression (Fig.
6). Furthermore, recovered IRS2 expression rescued the cellular
levels of p-PI3K and p-Akt, which were downregulated by miR-766
overexpression. These results suggested that miR-766 may inhibit
the PI3K/Akt signaling in PTC cells via regulation of IRS2.
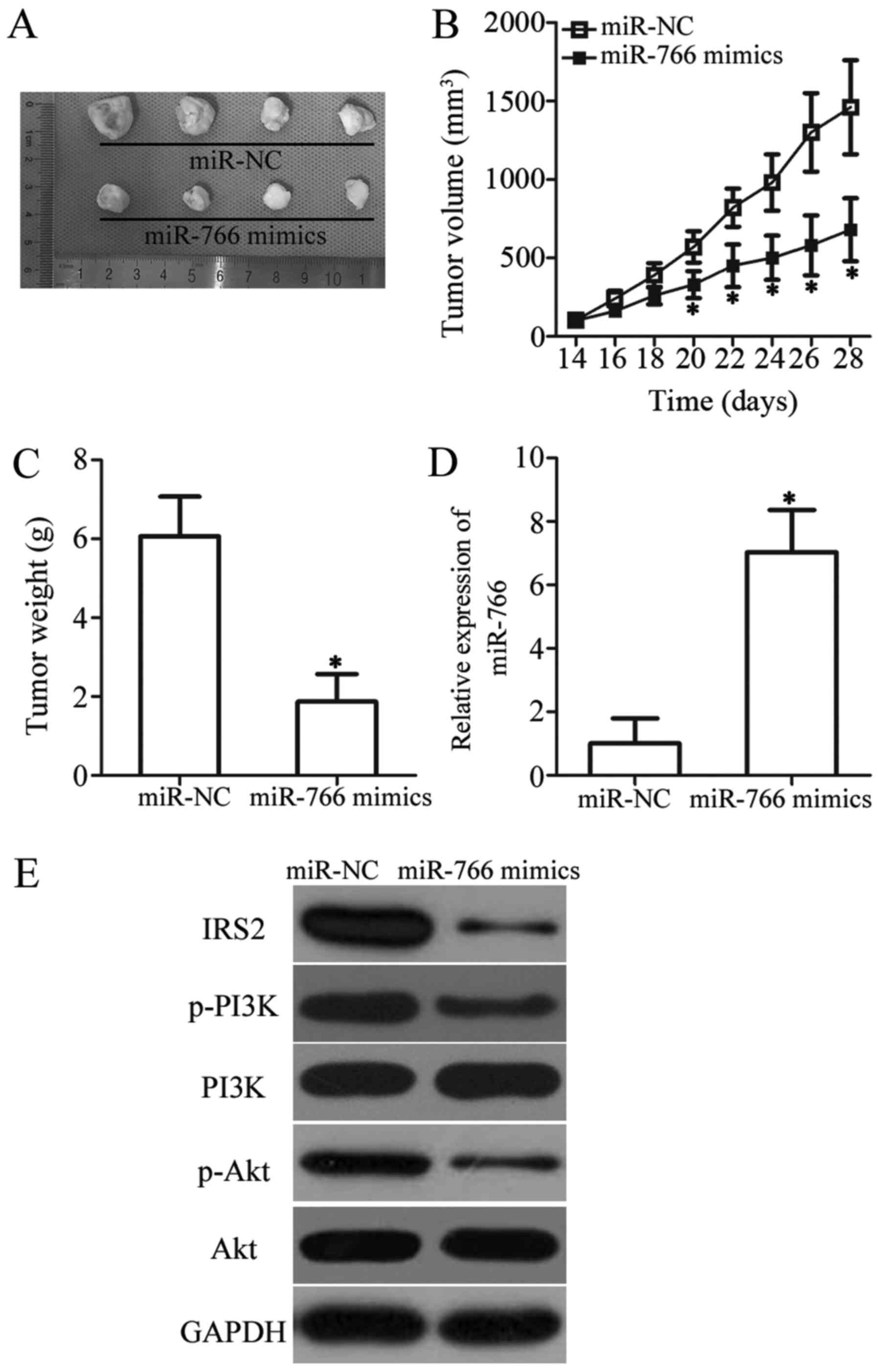
miR-766 hinders tumor growth in PTC in
vivo
To explore the precise role of miR-766 in PTC in
vivo, xenograft experiments were performed by subcutaneously
injecting miR-766-overexpressing TPC-1 cells into nude mice. The
miR-766-overexpressing tumor xenografts exhibited significant
suppression of tumor volume compared with in the miR-NC group
(Fig. 7A and B; P<0.05).
Subsequently, tumor xenografts were weighed; upregulation of
miR-766 decreased tumor weight in vivo (Fig. 7C; P<0.05). In addition, RT-qPCR
analysis revealed that miR-766 was markedly upregulated in tumor
xenografts that were generated using miR-766 mimics-transfected
cells (Fig. 7D; P<0.05).
Subsequently, the protein expression levels of IRS2 and molecules
associated with the PI3K/Akt pathway were detected in tumor
xenografts using western blot analysis. The protein expression
levels of IRS2, p-PI3K and p-Akt were downregulated in the tumor
xenografts from the miR-766 mimics groups compared with in those
from the miR-NC group (Fig. 7E).
Taken together, these results suggested that miR-766 may hinder PTC
tumor growth in vivo by suppressing IRS2 and activation of
the PI3K/Akt pathway.
Discussion
miRNAs are widely dysregulated in PTC, and
dysregulated miRNAs, together with their target genes, comprise a
complex network that has been implicated in the regulation of PTC
pathogenesis (25,31,32).
Notably, miRNAs have been proposed as novel diagnostic biomarkers
and potential therapeutic targets for anticancer treatment
(33). Therefore, further
exploration of the functional roles of aberrantly expressed miRNAs
in PTC and the underlying molecular mechanisms may aid in the
identification of novel therapeutic targets. The present study is
the first, to the best of our knowledge, to detect miR-766
expression in PTC, to clarify the clinical significance of miR-766
in PTC, and to examine the detailed roles of miR-766 in PTC
progression. Notably, the molecular mechanisms underlying the
activity of miR-766 in PTC cells were explored.
Expression of miR-766 is decreased in renal cell
carcinoma, and low miR-766 expression is significantly correlated
with the clinical stage. In addition, patients with renal cell
carcinoma and low miR-766 expression exhibit poorer prognosis
compared with those patients with high miR-766 expression (23). Conversely, miR-766 is overexpressed
in lung adenocarcinoma (24) and
colorectal cancer (25). miR-766
has been identified as an independent prognostic biomarker
predicting the clinical outcomes of patients with lung
adenocarcinoma (24). These
inconsistent observations indicate that the expression pattern of
miR-766 displays tissue specificity; however, the expression status
of miR-766 in PTC remains unknown. The present study demonstrated
that miR-766 was markedly downregulated in PTC tissues and cell
lines. In addition, the expression levels of miR-766 in PTC tissues
were associated with TNM stage and lymph node metastasis. These
findings implicated miR-766 as an attractive diagnostic and
prognosis biomarker for patients with PTC.
Dysregulation of miR-766 is involved in the
modulation of tumorigenesis and tumor development of numerous types
of human cancer. Restoration of miR-766 expression inhibits renal
cell carcinoma cell growth and promotes cell cycle arrest in
vitro, as well as decreasing tumor growth in vivo
(23). In breast cancer,
upregulation of miR-766 suppresses tumor sphere formation and
invasion in vitro, and attenuates in vivo lung
metastasis (34). Conversely,
miR-766 acts as an oncogene in colorectal cancer and increases cell
growth (35,36). Nevertheless, the biological roles
of miR-766 in PTC remain largely unidentified. In the present
study, functional assays revealed that miR-766 may have tumor
suppressor activity in PTC by affecting cell proliferation, colony
formation, apoptosis, migration and invasion in vitro, and
tumor growth in vivo. Therefore, miR-766 may represent a
valuable therapeutic target for the treatment of patients with
PTC.
miRNAs regulate biological behaviors associated with
carcinogenesis and cancer progression by directly binding to the
3'-UTRs of their target genes. Several genes, including splicing
factor SF2 (23) in renal cell
carcinoma, sex determining region Y-box 6 (35) and DNA methyl-transferase 3B
(36) in colorectal cancer, have
previously been demonstrated to be direct target genes of miR-766.
To illustrate the mechanisms underlying the cellular response to
miR-766, this study aimed to determine whether IRS2 is a direct
target gene of miR-766 in PTC. Bioinformatics analysis predicted
that the 3'-UTR of IRS2 matched the seed sequence of miR-766.
Luciferase reporter assays, RT-qPCR and western blot analysis
revealed that miR-766 could directly bind to the 3'-UTR of IRS2 and
decrease endogenous IRS2 expression in PTC cells. IRS2 was
upregulated in PTC tissues, and the IRS2 expression was inversely
correlated with miR-766 expression. The effects of IRS2 inhibition
on PTC cells were similar to the effects of miR-766 overexpression.
Furthermore, IRS2 reintroduction could abrogate the effects of
miR-766 on the malignant behaviors of PTC cells. These collective
results provided sufficient evidence to indicate that IRS2 may be a
direct and functional downstream target of miR-766 in PTC
cells.
IRS2 is a member of the IRS family, which mainly
interacts with SH2 domain-containing proteins to serve as adaptor
proteins for additional surface receptors (37). IRS2 is overexpressed in several
types of human cancer, including colorectal cancer (38), renal cell carcinoma (39), lung cancer (40) and breast cancer (41). It is a multifunctional oncogene
that has been implicated in the regulation of numerous biological
behaviors, such as cell proliferation, cell cycle, apoptosis,
invasion and epithelial-mesenchymal transition (42-44).
A previous study also demonstrated that IRS2 is upregulated in PTC
and serves oncogenic roles in the progression of PTC (27). In this study, miR-766 directly
targeted IRS2 to inhibit the malignancy of PTC cells. These prior
data suggested that the newly identified miR-766/IRS2 axis may
provide an effective therapeutic target for the management of
patients with PTC.
In conclusion, miR-766 was revealed to possess
antitumor properties and restrict the malignant biological
behaviors of PTC cells, at least in part by inhibiting IRS2 and
activation of the PI3K/Akt pathway. However, this study did not
explore the association between miR-766 and prognosis of patients
with PTC; this is a limitation, which will be resolved in future
studies.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ and JZ designed the study. JZ, ZL and YC
conducted RT-qPCR, MTT assay and colony formation assay. SZ, LG and
BG performed flow cytometry, migration and invasion assays, and
luciferase reporter assay. YJ, WT and SH carried out the in
vivo xenograft experiment and western blot analysis. They have
read and approved the final draft.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Daping Hospital, and was performed in accordance with
the Declaration of Helsinki and the guidelines of the Ethics
Committee of Daping Hospital. Written informed consent was obtained
from all patients for the use of their clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Xiang D, Xie L, Xu Y, Li Z, Hong Y and
Wang P: Papillary thyroid microcarcinomas located at the middle
part of the middle third of the thyroid gland correlates with the
presence of neck metastasis. Surgery. 157:526–533. 2015. View Article : Google Scholar
|
|
2
|
Kim HY, Park WY, Lee KE, Park WS, Chung
YS, Cho SJ and Youn YK: Comparative analysis of gene expression
profiles of papillary thyroid microcarcinoma and papillary thyroid
carcinoma. J Cancer Res Ther. 6:452–457. 2010. View Article : Google Scholar
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ye Y, Zhuang J, Wang G, He S, Ni J and Xia
W: MicroRNA-139 targets fibronectin 1 to inhibit papillary thyroid
carcinoma progression. Oncol Lett. 14:7799–7806. 2017.PubMed/NCBI
|
|
5
|
Catalano MG, Poli R, Pugliese M, Fortunati
N and Boccuzzi G: Emerging molecular therapies of advanced thyroid
cancer. Mol Aspects Med. 31:215–226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng XG, Chen ZF, Zhang KJ, Wang PG, Liu
ZM, Chen ZJ, Hou GY and Niu M: VEGF Trapon inhibits tumor growth in
papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 19:235–240.
2015.PubMed/NCBI
|
|
7
|
Fagin JA and Wells SA Jr: Biologic and
clinical perspectives on thyroid cancer. N Engl J Med.
375:23072016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dragomir M, Mafra ACP, Dias SMG, Vasilescu
C and Calin GA: Using microRNA Networks to understand cancer. Int J
Mol Sci. 19:192018. View Article : Google Scholar
|
|
11
|
Iorio MV, Casalini P, Tagliabue E, Ménard
S and Croce CM: MicroRNA profiling as a tool to understand
prognosis, therapy response and resistance in breast cancer. Eur J
Cancer. 44:2753–2759. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aragon Han P, Weng CH, Khawaja HT,
Nagarajan N, Schneider EB, Umbricht CB, Witwer KW and Zeiger MA:
MicroRNA expression and association with clinicopathologic features
in papillary thyroid cancer: A systematic review. Thyroid.
25:1322–1329. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yi R, Li Y, Wang FL, Miao G, Qi RM and
Zhao YY: MicroRNAs as diagnostic and prognostic biomarkers in
colorectal cancer. World J Gastrointest Oncol. 8:330–340. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Uddin A and Chakraborty S: Role of miRNAs
in lung cancer. J Cell Physiol. Apr 20–2018.Epub ahead of print.
View Article : Google Scholar
|
|
15
|
Ahir BK, Ozer H, Engelhard HH and Lakka
SS: MicroRNAs in glioblastoma pathogenesis and therapy: A
comprehensive review. Crit Rev Oncol Hematol. 120:22–33. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Homami A and Ghazi F: MicroRNAs as
biomarkers associated with bladder cancer. Med J Islam Repub Iran.
30:4752016.
|
|
17
|
Mutalib NS, Yusof AM, Mokhtar NM, Harun R,
Muhammad R and Jamal R: MicroRNAs and lymph node metastasis in
papillary thyroid cancers. Asian Pac J Cancer Prev. 17:25–35. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee JC, Gundara JS, Glover A, Serpell J
and Sidhu SB: MicroRNA expression profiles in the management of
papillary thyroid cancer. Oncologist. 19:1141–1147. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu G, Xie L and Miller D: Expression of
microRNAs in thyroid carcinoma. Methods Mol Biol. 1617:261–280.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu YT, Zheng HB, Zhang DQ, Zhou L and Sun
H: MicroRNA-1266 suppresses papillary thyroid carcinoma cell
metastasis and growth via targeting FGFR2. Eur Rev Med Pharmacol
Sci. 22:3430–3438. 2018.PubMed/NCBI
|
|
21
|
Wang R, Ma Q, Ji L, Yao Y, Ma M and Wen Q:
miR-622 suppresses tumor formation by directly targeting VEGFA in
papillary thyroid carcinoma. OncoTargets Ther. 11:1501–1509. 2018.
View Article : Google Scholar
|
|
22
|
Ma S, Jia W and Ni S: miR-199a-5p inhibits
the progression of papillary thyroid carcinoma by targeting SNAI1.
Biochem Biophys Res Commun. 497:181–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen C, Xue S, Zhang J, Chen W, Gong D,
Zheng J, Ma J, Xue W, Chen Y, Zhai W, et al:
DNA-methylation-mediated repression of miR-766-3p promotes cell
proliferation via targeting SF2 expression in renal cell carcinoma.
Int J Cancer. 141:1867–1878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Shi Y, Yin Z, Xue X and Zhou B: An
eight-miRNA signature as a potential biomarker for predicting
survival in lung adenocarcinoma. J Transl Med. 12:1592014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suresh R, Sethi S, Ali S, Giorgadze T and
Sarkar FH: Differential expression of microRNAs in papillary
thyroid carcinoma and their Role in Racial Disparity. J Cancer Sci
Ther. 7:145–154. 2015.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Dong S, Meng X, Xue S, Yan Z, Ren P and
Liu J: microRNA-141 inhibits thyroid cancer cell growth and
metastasis by targeting insulin receptor substrate 2. Am J Transl
Res. 8:1471–1481. 2016.PubMed/NCBI
|
|
28
|
Landis J and Shaw LM: Insulin receptor
substrate 2-mediated phosphatidylinositol 3-kinase signaling
selectively inhibits glycogen synthase kinase 3β to regulate
aerobic glycolysis. J Biol Chem. 289:18603–18613. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mercado-Matos J, Clark JL, Piper AJ,
Janusis J and Shaw LM: Differential involvement of the microtubule
cytoskeleton in insulin receptor substrate 1 (IRS-1) and IRS-2
signaling to AKT determines the response to microtubule disruption
in breast carcinoma cells. J Biol Chem. 292:7806–7816. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jeong SH and Lim DS: Insulin receptor
substrate 2: A bridge between Hippo and AKT pathways. BMB Rep.
51:209–210. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu J, Li C, Liu C, Zhao S, Wang Y and Fu
Z: Expressions of miRNAs in papillary thyroid carcinoma and their
associations with the clinical characteristics of PTC. Cancer
Biomark. 18:87–94. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yoruker EE, Terzioglu D, Teksoz S, Uslu
FE, Gezer U and Dalay N: MicroRNA expression profiles in papillary
thyroid carcinoma, benign thyroid nodules and healthy controls. J
Cancer. 7:803–809. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu Y, Wang H, Chen E, Xu Z, Chen B and Lu
G: Candidate microRNAs as biomarkers of thyroid carcinoma: A
systematic review, meta-analysis, and experimental validation.
Cancer Med. 5:2602–2614. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oh K and Lee DS: In vivo validation of
metastasis-regulating microRNA-766 in human triple-negative breast
cancer cells. Lab Anim Res. 33:256–263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li YC, Li CF, Chen LB, Li DD, Yang L, Jin
JP and Zhang B: MicroRNA-766 targeting regulation of SOX6
expression promoted cell proliferation of human colorectal cancer.
OncoTargets Ther. 8:2981–2988. 2015. View Article : Google Scholar
|
|
36
|
Afgar A, Fard-Esfahani P, Mehrtash A,
Azadmanesh K, Khodarahmi F, Ghadir M and Teimoori-Toolabi L:
miR-339 and especially miR-766 reactivate the expression of tumor
suppressor genes in colorectal cancer cell lines through DNA
methyltransferase 3B gene inhibition. Cancer Biol Ther.
17:1126–1138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
White MF: IRS proteins and the common path
to diabetes. Am J Physiol Endocrinol Metab. 283:E413–E422. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Day E, Poulogiannis G, McCaughan F,
Mulholland S, Arends MJ, Ibrahim AE and Dear PH: IRS2 is a
candidate driver oncogene on 13q34 in colorectal cancer. Int J Exp
Pathol. 94:203–211. 2013.PubMed/NCBI
|
|
39
|
Ma Y, Zhang H, He X, Song H, Qiang Y, Li
Y, Gao J and Wang Z: miR-106a* inhibits the
proliferation of renal carcinoma cells by targeting IRS-2. Tumour
Biol. 36:8389–8398. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu H, Lee MS, Tsai PY, Adler AS, Curry NL,
Challa S, Freinkman E, Hitchcock DS, Copps KD, White MF, et al:
Ablation of insulin receptor substrates 1 and 2 suppresses
Kras-driven lung tumorigenesis. Proc Natl Acad Sci USA.
115:4228–4233. 2018. View Article : Google Scholar
|
|
41
|
Porter HA, Perry A, Kingsley C, Tran NL
and Keegan AD: IRS1 is highly expressed in localized breast tumors
and regulates the sensitivity of breast cancer cells to
chemotherapy, while IRS2 is highly expressed in invasive breast
tumors. Cancer Lett. 338:239–248. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang P, Shao G, Lin X, Liu Y and Yang Z:
miR-338-3p inhibits the growth and invasion of non-small cell lung
cancer cells by targeting IRS2. Am J Cancer Res. 7:53–63.
2017.PubMed/NCBI
|
|
43
|
de Melo Campos P, Machado-Neto JA, Eide
CA, Savage SL, Scopim-Ribeiro R, da Silva Souza Duarte, Favaro PA,
Lorand-Metze I, Costa FF, Tognon CE, et al: IRS2 silencing
increases apoptosis and potentiates the effects of ruxolitinib in
JAK2V617F-positive myeloproliferative neoplasms. Oncotarget.
7:6948–6959. 2016.PubMed/NCBI
|
|
44
|
Liu H, Ren G, Zhu L, Liu X and He X: The
upregulation of miRNA-146a inhibited biological behaviors of ESCC
through inhibition of IRS2. Tumour Biol. 37:4641–4647. 2016.
View Article : Google Scholar
|