Introduction
Gastric cancer (GC) as one of the most common
malignant tumors and remains a leading cause of cancer-related
mortality worldwide, and an estimated 1000,000 individuals are
diag-nosed with GC annually (1,2). The
activity imbalance between oncogenes and anti-oncogenes has been
implicated in gastric carcinogenesis (3,4).
Accumulating evidence has indicated that the targeting of oncogenes
or downstream genes associated with cancer progression may provide
effective strategies for the intervention of carcinogenesis
(5,6).
Interleukin (IL)-21, identified as a member of the
IL-2 family, is involved in biological activities in cancer and
autoimmunity by binding to its receptor, IL-21R (7), which is highly expressed in
hematological malignancies (8,9) and
enhances the aggressiveness of follicular lymphoma (10). IL-21/IL-21R axis results in the
pathogenesis of leukemia and large cell lymphoma through activation
of the JAK/STAT signaling pathway (11,12),
indicating that IL-21R may present a potential therapeutic target
for cancer therapy. However, knowledge about the functions and
regulatory mechanism of IL-21R in GC is limited.
Few genetic alterations and a low copy number gain
for IL-21R suggest that post-transcriptional modulation may play a
role in its upregulation in GC. In this study, using bioinformatic
analysis, we found that IL-21R expression was mainly regulated by
miR-125a (www.microrna.org), and it was listed in
the top rank. MicroRNAs (miRNAs or miRs), a subgroup of small
regulatory ncRNAs, which negatively modulate the protein-coding
genes by binding to their 3′ untranslated regions (3′UTRs)
(13). The aberrant expression of
miRNAs has been shown to be associated with various malignancies
(14-16) and miRNAs can serve as non-invasive
biomarkers for the prognosis of patients with GC (17,18).
miR-30a has been shown to suppress Th17 differentiation and
demyelination by targeting IL-21R (19), and integrated miRNA-mRNA profiling
has identified miR-583 as a negative regulator of IL2Rγ (20); however, the inhibition of
miR-3940-5p has been shown to facilitate T-cell activity by
targeting IL2Rγ (21). Increasing
evidence documents the potential role of long non-coding RNAs
(lncRNAs) as miRNA sponges in human cancer, and lncRNAs may emerge
as important players in the regulation of miRNAs and their target
genes (22,23).
In the present study, we investigated the clinical
significance of IL-21R expression in GC samples, and revealed the
functions of IL-21R and its regulation by miR-125a and lncRNA
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in
GC cells. The correlation of miR-125a with IL-21R or MALAT1
expression was analyzed in primary GC samples, and the association
of MALAT1 expression with the poor survival of and recurrence in
patients with GC was analyzed by Kaplan-Meier plotter analysis. We
aimed to uncover the molecular mechanisms through which IL-21R
upregulation contributes to gastric tumorigenesis and provide a
potential therapeutic marker for clinical intervention.
Materials and methods
Cell culture
Human GC cell lines, including the undifferentiated
HGC-27 cell line, the moderately differentiated SGC-7901 cell line,
the poorly differentiated AGS and BGC-823 cell lines, the lowly
differentiated MKN-45 cell line and the human gastric mucosa
epithelial GES-1 cells were stored at the Laboratory of the First
Affiliated Hospital of Zhengzhou University. The cells were
cultured in a humidified incubator with 5% CO2 at 37˚C
in RPMI-1640 medium or Dulbecco’s modified Eagle’s medium (DMEM;
Nanjing KeyGen Biotech. Co. Ltd., Nanjing, China) containing 10%
fetal bovine serum (FBS; Keygen).
Clinical samples and data
A tissue microarray (TMA) of gastric adenocarcinoma
(GAC) (Chip lot no. HSTM-Ade180Sur-01; array no. I11-020-1) was
provided by Outdo Biotech Co., Ltd. (Shanghai, China). The TMA was
prepared for immunohistochemical (IHC) analysis. Human GC tissues
and the corresponding adjacent non-tumor tissues (ANTT) were
obtained from the biopsy samples of 89 cases of GC admitted in the
First Affiliated Hospital of Zhengzhou University (Zhengzhou,
China) from January, 2007 to December, 2011. The baseline
characteristics of the patients prior to neo-adjuvant chemotherapy
are summarized in Table I. This
study was approved by Medical Ethics Committee of Zhengzhou
University, and written informed consent was obtained from the
patients prior to sample collection. Two pathologists respectively
reviewed all the cases. In addition, 386 cases of GAC patients and
35 ANTT, as well as the relative expression level of IL-21R were
downloaded from The Cancer Genome Atlas (TCGA)-GAC RNA sequencing
database (https://genome-cancer.ucsc.edu). Other cohorts
(GSE51105, GSE22377 and GSE15459) for the analysis of the
prognostic value of lncRNA NEAT1 in GAC were downloaded from the
Kaplan-Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=gastric).
 | Table IAssociation of IL-21R expression with
clinicopatho-logical factors of patients with gastric cancer. |
Table I
Association of IL-21R expression with
clinicopatho-logical factors of patients with gastric cancer.
| Parameters | Patients n (%) 89
(100) | IL-21R expression
| P-value |
|---|
| Low (%)59
(66.3) | High (%)30
(33.7) |
|---|
| Age (years) | | | | |
| ≥60 | 29 | 16 (55.2) | 13 (44.8) | |
| <60 | 60 | 43 (71.7) | 17 (28.3) | 0.125 |
| Sex | | | | |
| Female | 20 | 11(55.0) | 9 (45.0) | |
| Male | 69 | 48 (69.6) | 21 (30.4) | 0.228 |
| Tumor size
(cm) | | | | |
| ≥3.5 | 17 | 7 (41.2) | 10 (58.8) | |
| <3.5 | 72 | 52 (72.2) | 20 (27,8) | 0.015 |
| TNM stage | | | | |
| I/II | 32 | 18 (56.3) | 14 (43.7) | |
| III/IV | 57 | 41 (71.9) | 16 (28.1) | 0.135 |
|
Differentiation | | | | |
| Well/moderate | 27 | 16 (59.3) | 11 (40.7) | |
| Poor | 62 | 43 (69.4) | 19 (30.6) | 0.357 |
| T
classification | | | | |
| T1/T2 | 19 | 13 (68.4) | 6 (31.6) | |
| T3/T4 | 70 | 46 (65.7) | 24 (34.3) | 0.826 |
| N
classification | | | | |
| Negative | 66 | 48 (72.7) | 18 (27.3) | |
| Positive | 23 | 11 (47.8) | 12 (52.2) | 0.031 |
Bioinformatics analysis
The analysis of genomic alterations (copy number,
somatic mutation and mRNA upregulation) and the methylation level
of IL-21R in GAC (n=298) was conducted using a TCGA dataset and
software cBioPortal (www.cbioportal.org). The miRNAs that may target the
IL-21R gene were identified by using the prediction tool,
microRNA.org (http://www.microrna.org/), according to the mirSVR and
PhastCons scores, and the lncRNAs that may sponge miR-125a were
screened out using starBase v2.0 (http://starbase.sysu.edu.cn/).
IHC analysis
IL-21R antibody (rabbit, polyclonal antibody,
ab5980; Abcam, Cambridge, MA, USA) was used for the IHC detection
of protein expression in the TMA. IL-21R antibody was used at a
1:200 dilution. Endogenous peroxidase was inhibited by incubation
with freshly prepared 3% hydrogen peroxide with 0.1% sodium azide.
Non-specific staining was blocked with 0.5% casein and 5% normal
serum. The TMA samples were incubated with biotinylated antibodies
and horseradish peroxidase. Staining was developed with
diami-nobenzidine substrate and the sections were counterstained
with hematoxylin. PBS replaced IL-21R antibody in the negative
controls and its expression was quantified as previously described
(24).
Vector construction and transfection
Small interfering RNA (siRNA, 100 µM)
targeting the IL-21R gene [siIL-21R sequence,
CCTACACCTGCCACATGGATGTATT; or non-target negative control (NC)
sequence, CCTCCACGTCACGTATAGTG ACATT] were provided by GenePharma
(Shanghai, China). For MALAT1 overexpression, the full-length human
MALAT1 cDNA was amplified using the following primers: Forward,
5′-GGCGGTACCATGAAACAATTTGGAGAAG-3′ and reverse,
5′-GCGCTCGAGCTAAGTTTGTACATTTTGCC-3′; miR-125a mimic (forward,
5′-AGCTAAGCTTCTCTCTGTGT CTCTATTTCTGTCGTTT-3′ and reverse,
5′-ACTGCTCGAG GTCAGGTTTCAGTTGGTGGTCAAATG-3′) was provided by
GenePharma, and lentivirus-mediated IL-21R was synthesized by
Shanghai Genechem (Shanghai, China). The HGC-27 and MKN-45 cells
were transfected with these vectors and were then incubated at 5%
CO2 at 37˚C. The medium was refreshed, and the cells
were then cultured for a further 48 h. The cell transfection
efficiency was evaluated by reverse transcription-quantitative PCR
(RT-qPCR) and western blot analysis.
RT-qPCR
Total RNA was isolated from the GC cell lines using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s instructions. Complementary DNA (cDNA) was produced
from the RNA using the PrimeScript™ Reverse Transcription kit
(Takara, Tokyo, Japan) and its amplification was performed using
the SYBR-Green Master Mix kit (Takara) according to the
manufacturer’s instructions on an ABI 7500 System (Applied
Biosystems/Thermo Fisher Scientific, Waltham, MA, USA). The primers
specific for IL-21R and miR-125a were designed and synthesized by
Shanghai Sangon Biotech (Shanghai, China). Quantitative PCR for
miR-125a-3p and IL-21R was performed using the SYBR-Green Master
Mix kit and the specific primers as follows: Forward,
5′-TGACACAGGTGAGGTTCTTG-3′ and reverse,
5′-TATGGTTTTGACGACTGTGTGAT-3′; IL-21R forward,
5′-CCCGACCTCGTCTGCTACA-3′ and reverse, 5′-TGGTCTTGCCAGGTAAGGGT-3′;
U6 forward, 5′-CAAATTCGTGAAGCGTTCCATA-3′ and reverse,
5′-AGTGCAGGGTCCGAGGTATTC-3′ and; GAPDH forward,
5′-CCTGTACGCCAACACAGTGC-3′ and reverse, 5′-ATACTCCTGCTTGCTGATCC-3′.
The PCR cycling conditions were as follows: 94˚C for 30 sec, 56˚C
for 30 sec, and 72˚C for 90 sec, with 30 cycles, and a final
extension at 72˚C for 5 min. Thus, the analysis of the relative
gene expression data was conducted by using RT-qPCR and the
2-ΔΔCq method (25).
miR-125a expression was normalized to the U6 expression level and
GAPDH expression was used as an internal control for IL-21R. The
experiments were performed 3 times.
Western blot analysis
The GC cells were harvested and proteins were
extracted using lysis buffer (100 mM Tris-HCl, 2% SDS, 1 mM
Mercaptoethanol and 25% glycerol). The cell extracts were boiled in
loading buffer and equal amounts of cell extracts were separated on
15% SDS-PAGE gels. Separated protein bands were transferred into
polyvinylidene fluoride (PVDF) membranes. The primary antibodies
used were as follows: phosphorylated Janus kinase 2 (p-JAK2; rabbit
monoclonal antibody, #3776; Cell Signaling Technology, Danvers, MA,
USA), phosphorylated signal transducer and activator of
transcription 3 (p-STAT3; mouse monoclonal antibody, sc-8059; Santa
Cruz Biotechnology, Santa Cruz, CA, USA) and anti-GAPDH (rabbit
polyclonal antibody, ab153802, Abcam) and were diluted at a ratio
of 1:1,000 according to the manufacturer’s instructions and
incubated with the membranes overnight at 4˚C. Horseradish
peroxidase (HRP)-linked secondary antibodies (goat anti-mouse IgG,
ab205719 and goat anti-Rabbit IgG, ab205718; Abcam) were added at a
dilution ratio of 1:10,000, and incubated with the membranes at
room temperature for 1 h. The membranes were washed with PBS 3
times and the immunoreactive bands were visualized using ECL-PLUS
(GE Healthcare, Piscataway, NJ, USA) according to the
manufacturer’s instructions.
Cell viability assay
GC cells (2×103/well) were seeded in
96-well plates at 37˚C, 5% CO2. Following transfection
with siIL-21R or miR-125a mimic for 24, 48, 72, 96 and 120 h, CCK-8
solution (10 µl) was added to each well, followed by
incubation for 2 h. The optical densities at 492 nm were measured
using a microplate reader (Molecular Devices, Sunnyvale, CA,
USA).
Transwell invasion assay
Transwell 24 Well Permeable Supports (CLS3396) were
purchased from Sigma-Aldrich (St. Louis, MO, USA), and its filters
were coated with Matrigel on the upper surface of a polycarbonic
membrane. Following incubation at 37°C for 30 min, the Matrigel
solidified and served as the extracellular matrix for analysis of
tumor cell invasion. GC cells (1×105) in 100 µl
of serum-free DMEM were added to the upper compartment of the
chamber. A total of 200 µl conditioned medium was used as a
source of chemoattractant, and placed in the bottom compartment of
the chamber. Following 24 h of incubation at 37°C with 5%
CO2, the medium was removed from the upper chamber. The
non-invaded cells on the upper side of the chamber were scraped off
with a cotton swab. The cells that had migrated from the Matrigel
into the pores of the inserted filter were fixed with 100%
methanol, stained with hematoxylin (15°C, 10 min), and mounted and
dried at 80°C for 30 min. The number of GC cells invading through
the Matrigel was counted in 3 randomly selected visual fields from
the central and peripheral portion of the filter using an inverted
microscope (Olympus, Tokyo, Japan, ×200 magnification). Each assay
was repeated 3 times.
Dual-luciferase reporter assay
The GC cells were seeded into 24-well plates.
Following 24 h of incubation, 1 mg pmirGLO report vector (Promega
Corp., Madison, WI, USA) carrying the wild-type 3′UTR or mutated
3′UTR of the miR-125a target was co-transfected with miR-125a or
miR-NC into the GC cells. At 48 h following transfection, Firefly
and Renilla luciferase activities were examined using a
Dual-luciferase Reporter System (Promega). pmirGLO report vector
was used as a positive control.
In vivo tumor xenograft experiments
Six-week-old nude mice (BALB/c-nu) (n=15, female)
were bred at the Laboratory Animal facility of Zhengzhou
University, and were housed individually in microisolator
ventilated cages (temperature, 26-28°C; 40-60% humidity and
ventilation for 10-15 times/h) with free access to water and food.
All experimental procedures were performed according to the
regulations and internal biosafety and bioethics guidelines and the
use of animals was approved by the Ethics Review Commission of
Zhengzhou University. For the localized model, 2×107 GC
MKN-45 cells stably transfected with IL-21R, miR-125a and empty
vector were injected subcutaneously into the right flanks of the
6-week old female BALB/c nude mice, which were supplied by Shanghai
SLAC Laboratory Animal Co. Mice bearing tumors approximately 0.5 cm
in diameter were randomized into the miR-NC + Lv-NC, miR-125a +
Lv-IL-21R and miR-125a + Lv-NC groups (n=5 in each group). The
tumors were measured every 3 days and the tumor volume was
calculated according to the following formula: Length ×
width2/2.
Statistical analysis
Data are presented as the means ± SEM. The
Kruskal-Wallis H test, Mann Whitney U test with Bonferroni’s
correction and the Chi-square test were applied to analyze the
differential expression of IL-21R in GC and adjacent normal
tissues. Pearson’s correlation coefficient analysis was used to
observe the correlations between IL-21R or MALAT1 expression and
miR-125a in GC tissues. Gene expression, cell proliferation and
invasion were calculated using a Student’s t-test or one-way
analysis of variance (ANOVA) between groups. For the parental and
control groups, the LSD method of multiple comparisons was used
when the probability for ANOVA was statistically significant.
Survival and recurrence curves were analyzed with the Kaplan-Meier
method (www.kmplot.com) and log-rank test.
Differences were considered statistically significant at
P<0.05.
Results
Expression of IL-21R is upregulated in GC
samples
IL-21R has been reported to be upregulated in
diffuse large B-cell lymphoma (DLBCL) and to be associated with an
unfavorable prognosis in patients with DLBCL (10). However, to date, at least to the
best of our knowledge, little is known about the expression of
IL-21R in human GC. In this study, the expression of IL-21R was
examined by IHC analysis, and the results revealed that its
expression level was increased in the 89 cases of GC as compared
with the ANTT (65.17 vs. 47.19%, P=0.015) (Fig. 1A). The mRNA level of IL-21R was
then validated by the datamining of the RNA sequencing data from
GAC publicly available at The Cancer Genome Atlas database (TCGA),
which indicated that IL-21R expression was markedly increased in
the total GAC samples (n=386) or pair-matched tissues (n=32) as
compared with the adjacent normal tissues (Fig. 1B). Consistently, IL-21R expression
was upregu-lated in the GC cell lines as compared with the
immortalized GES-1 cells (Fig.
1C). To further explore the reasons for the upregulation of
IL-21R in GC, we examined the genomic alterations of IL-21R in the
TCGA cohort by cBioPortal (www.cbioportal.org) (26), including copy number, somatic
mutation and mRNA upregulation, indicating that only 6% of cases
(18/298) had the genetic alterations for IL-21R, of which its mRNA
upregulation accounted for 2.7% of the cases. Moreover, IL-21R mRNA
upregulation could not be explained by the copy number alterations
in the GC samples (Fig. 1D).
IL-21R overexpression is associated with
a poor prognosis of patients with GC
According to the results presented in Fig. 1A, the samples were divided
according to the IL-21R expression level into the IL-21R high
expression (intermediate/strong, n=30) and low expression
(negative/low, n=59) groups. As indicated in Table I, IL-21R high expression was
positively associated with tumor size (P=0.015) and lymphatic
metastasis (P=0.031) in the patients with GC, but had no
association with other clinical factors (each P>0.05).
Kaplan-Meier analysis revealed that the patients with GC with a
high IL-21R expression had a shorter survival time and a higher
recurrence rate compared with those with a low IL-21R expression
(Fig. 1E). To further confirm this
result, Kaplan Meier plotter analysis (www.kmplot.com) (27)
demonstrated that a high IL-21R expression was associated with a
poor survival and recurrence in patients with GC (Fig. 1F).
Knockdown of IL-21R suppresses cell
proliferation and invasion
As IL-21R was found to be expressed abundantly in
the MKN-45 and HGC-27 cell lines, we performed siRNA-mediated
knockdown experiments to investigate its function in these two cell
lines. Following transfection with siIL-21R or NC, both the mRNA
and protein levels of IL-21R were significantly decreased in these
cells (Fig. 2A). The knockdown of
IL-21R reduced the cell proliferative activities, as shown by CCK-8
assay (Fig. 2B). In addition, the
cell invasive ability was evidently weakened by siIL-21R
transfection as compared with the NC group (Fig. 2C and D).
IL-21R is a direct target of miR-125a in
GC
According to the binding sites with the IL-21R
3′UTR, miRNAs were identified by microRNA.org
(http://www.microrna.org/) with the mirSVR and
PhastCons score. We found that miR-125a-3p (miR-125a) was ranked at
the top of the miRNA list and had the potential to bind with IL-21R
3′UTR. Given the negative correlation of miRNAs with their target
genes, we used the TCGA cohort to analyze the correlation of IL-21R
with miR-125a expression in paired GC samples, indicating that
miR-125a expression was downregulated (Fig. 3A) and exhibited a negative
correlation with IL-21R expression in the GC samples (Fig. 3B). To confirm whether IL-21R is a
direct target of miR-125a, we performed a luciferase assay. The
binding site of miR-125a with IL-21R 3′UTR is shown in Fig. 3C. Transfection with miR-125a mimic
inhibited the luciferase activity of the wild-type (Wt) IL-21R
3′UTR, but had no effect on the mutant (Mut) sequence of the
binding site (Fig. 3D).
Additionally, both the mRNA and protein expression levels of IL-21R
were decreased in the HGC-27 and MKN-45 cells following
transfection with miR-125a mimic (Fig.
3E). These results suggested miR-125a regulates IL-21R
expression by targeting its 3′UTR.
IL-21R overexpression reverses the tumor
suppressive effects of miR-125a
Rescue experiments were further conducted to
determine whether IL-21R is a functional target of miR-125a in GC
cells. Following co-transfection with Lv-IL-21R and miR-125a mimic
for 72 h, a CCK-8 proliferation assay revealed that the
overexpression of IL-21R increased the cell proliferative activity
and partly attenuated the anti-proliferative effects of miR-125a on
the HGC-27 and MKN-45 cell lines (Fig.
4A). In addition, the enforced expression of IL-21R enhanced
the cell invasive potential and partly alleviated the inhibitory
effects induced by miR-125a in the HGC-27 and MKN-45 cell lines
(Fig. 4B-D). Moreover, our in
vivo experiments illustrated that the overexpression of IL-21R
partly abolished the anti-tumor effects of miR-125a (Fig. 4E and F), indicating that IL-21R was
a target of miR-125a in GC.
lncRNA MALAT1 functions as an endogenous
sponge of miR-125a
For miR-125a-3p, at a very high stringency, 3
lncRNAs including MALAT1, LINC00657 and CTC-444N24.11 were
identified to have a complementary sequence of miR-125a-3p using
starBase v2.0 (http://starbase.sysu.edu.cn/) (Fig. 5A), of which MALAT1 as a star
lncRNA, has been implicated in the tumorigenesis and progression of
multiple malignancies, including GC (28-30).
Thus, in this study, we first analyzed MALAT1 expression and its
correlation with miR-125a, and found that MALAT1 expression was
upregulated (Fig. 5B) and
negatively correlated with miR-125a expression in the GC samples
(Fig. 5C). To verify whether
MALAT1 can bind to miR-125a, we conducted a luciferase gene
reporter assay. The binding site of MALAT1 with miR-125a is shown
in Fig. 5D. Dual-luciferase assay
demonstrated a marked decrease in the luciferase activities
following co-transfection of the cells with miR-125a mimics and
Wt-MALAT1 expression vector, but not with the Mut-MALAT1 vector
(Fig. 5E). The restored expression
of MALAT1 decreased miR-125a expression (Fig. 5F), whereas it increased IL-21R
expression (Fig. 5G). Transfection
with miR-125a mimic not only inhibited the activities of the
IL-21R/JAK2/STAT3 pathway, but also partly attenuated the increased
activities of this pathway induced by MALAT1, as indicated by
western blot analysis (Fig. 5H).
The results of CCK-8 assay revealed that the increased
proliferative activities induced by MALAT1 were reduced by miR-125a
in the GC cells (Fig. 5I). These
results suggest that lncRNA MALAT1 may act as an endogenous sponge
of miR-125a to regulate IL-21R signaling in GC cells.
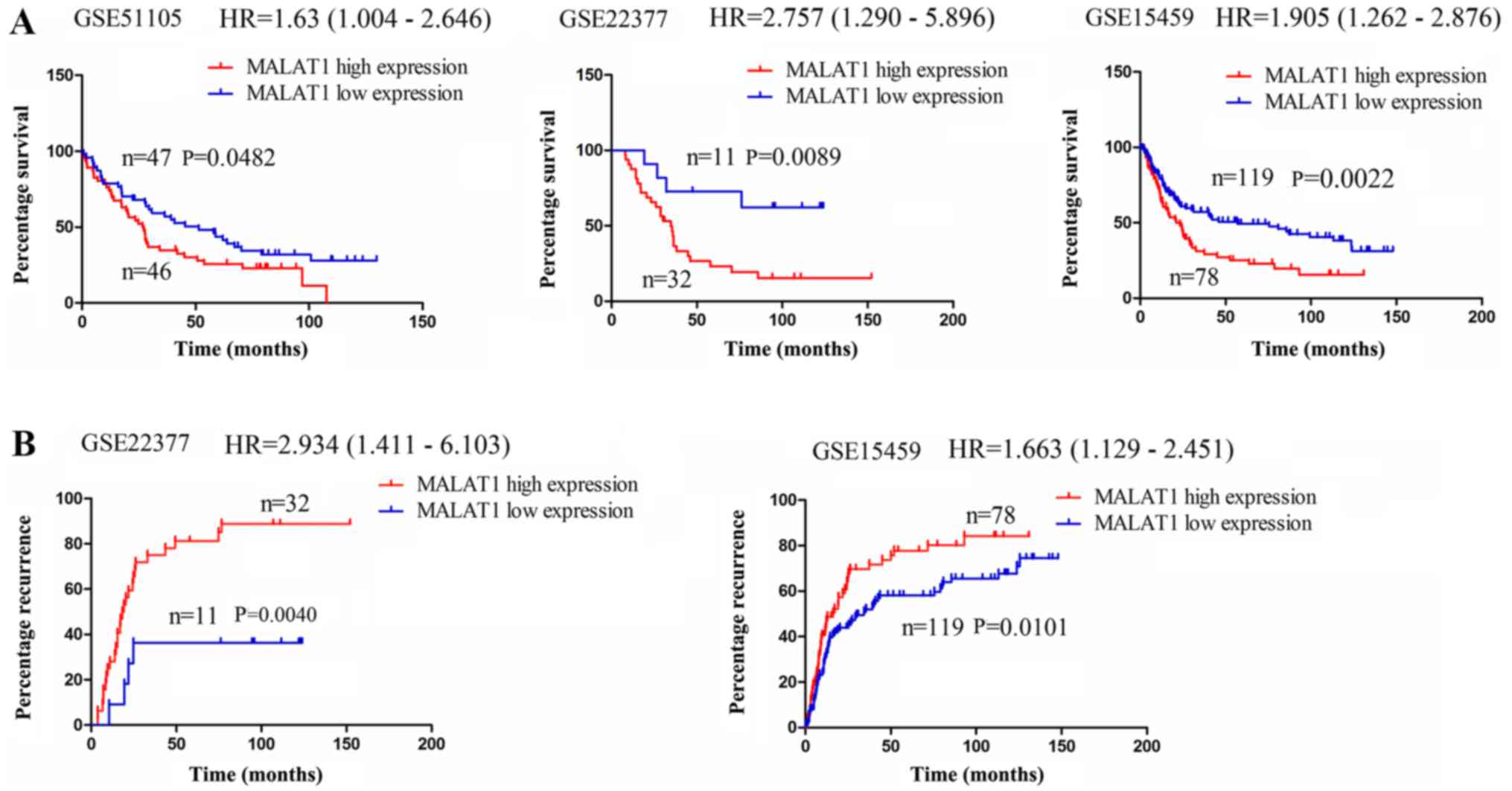
lncRNA MALAT1 is associated with a poor
survival of and recurrence in patients with GC
The expression of lncRNA MALAT1 has been shown to be
upregulated in GC (29,31). Thus, in this study, we investigated
whether it was associated with the survival of and recurrence in
patients with GC. Using the online Kaplan Meier plotter tools
(www.kmplot.com) (27), we found that in the cohorts in the
GSE51105, GSE22377 and GSE15459 databases, the patient with GC with
a high MALAT1 expression exhibited a lower survival rate compared
with those with a low MALAT1 expression (Fig. 6A); In the cohorts in the GSE22377
and GSE15459 databases, it was found that the patients with GC with
a high MALAT1 expression displayed a higher recurrence rate
compared with those with a low MALAT1 expression (Fig. 6B). These data thus indicated that
lncRNA MALAT1 may represent a risk factor for survival and
recurrence in patients with GC. According to all the results
obtained in vitro and in vivo, we found that lncRNA
MALAT1 may act as a sponge of miR-125a to regulate IL-21R
signaling, leading to gastric tumorigenicity (Fig. 7).
Discussion
IL-21 mainly originates from activated
CD4+ T cells and natural killer (NK) T cells and exerts
its function by interacting with IL-21R, which is expressed in
lymphoid tissues, as well as in epithelial cells (32). Recent studies have demonstrated
that the ectopic expression of IL-21R is found in hematological
malignancies, such as DLBCL (10),
adult T-cell leukemia (33) and
chronic lymphocytic leukemia (34). However, little knowledge is
available regarding the IL-21R expression level in other epithelial
tumors, particularly in GC, at least to the best of our knowledge.
In this study, we found that IL-21R exhibited an increased
expression in GC. The use of the TCGA cohort further verified this
result in 386 cases of GAC samples. Consistent with previous
reports (10,33,34),
a high IL-21R expression was found to be associated with tumor
size, lymphatic metastasis, poor survival and recurrence, acting as
an independent prognosis factor for the survival of and recurrence
in patients with GC. The results of this study indicated that
IL-21R may represent a potential biomarker for GC.
Functionally, IL21R, expressed by bone marrow
CD14+ cells, accelerates osteoclast formation in
multiple myeloma patients (33),
participates in the mediation of pro-apoptotic signals in CLL B
cells (35) and matrix
metalloproteinase signaling in breast cancer, and promotes tumor
proliferation, migration and invasion (36). In this study, we also found that
the knockdown of IL-21R suppressed GC cell proliferation and
invasion. These results indicate that IL-21R may play an oncogenic
role in GC cells.
To further explore the mechanisms of action of
IL-21R in GC, we investigated its genetic and copy number
alterations in GC and found these factors could not comprehensively
explain the reasons for IL-21R upregulation in GC. Thus, we focused
on its post-transcriptional regulation, such as miRNAs. By
bioinformatics analysis, we identified IL-21R as a direct target of
miR-125a, and IL-21R overexpression partly alleviated the tumor
suppressive effects of miR-125a in GC in vitro and in
vivo. Other studies have demonstrated that miR-125a functions
as a tumor suppressive factor in various types of cancer (37-39).
These data suggested the upregulation of IL-21R was partly due to
the downregulation of miR-125a in GC.
It has been recently demonstrated that lncRNAs
function as miRNAs sponges and inhibit their activities and
functions (40). Accumulating data
have indicated that lncRNA MALAT1 promotes cell proliferation,
migration and metastasis, and decreases cell apoptosis by sponging
multiple miRNAs in various malignancies, such as miR-143-3p and
miR-204 in hepatocellular carcinoma (41,42),
miR-101 in glioma (43), and
miR-429 in renal cell carcinoma (44). Of note, in this study, we also
identified MALAT1 as a negative regulator of miR-125a-3p in GC
samples, and confirmed that MALAT1 could bind to miR-125a, thereby
inhibiting its activity and increasing IL-21R expression. Rescue
experiments demonstrated that miR-125a mimic suppressed the
activation of the IL-21R/JAK2/STAT3 signaling pathway and partly
reversed the promoting effects of MALAT1 on this signaling pathway
and on the proliferation of GC cells. These data suggest lncRNA
MALAT1 functions as a sponge of miR-125a to regulate IL-21R
signaling in GC cells. The findings of this study further
documented that MALAT1 was a biomarker for GC metastasis and
represented a risk factor for the survival and recurrence of
GC.
In conclusion, the upregulation of IL-21R in GC was
associated with tumor size and lymphatic metastasis and acted as an
independent prognostic factor of a poor survival and recurrence in
GC. IL-21R functioned as an oncogenic factor and alleviated the
suppressive effects of miR-125a in GC cells. lncRNA MALAT1 acted as
a sponge of miR-125a to regulate IL-21R signaling, leading to
gastric tumorigenicity (Fig.
7).
Acknowledgments
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Henan Province (Grant no. 162300410284).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
LY and JZ contributed equally to this article. XWH
designed this article and supervised its quality. LY wrote this
paper and performed the experiments. JZ performed the experiments
and revised the paper. DG and JM were responsible for collecting
all the data, and SFS used the prediction tools and analyzed the
data in the study. XWH and JZ checked the manuscript and all
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by Medical Ethics Committee
of Zhengzhou University, and written informed consent was obtained
from the patients prior to sample collection. All experimental
procedures were performed according to the regulations and internal
biosafety and bioethics guidelines and the use of animals was
approved by the Ethics Review Commission of Zhengzhou
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hu XT and He C: Recent progress in the
study of methylated tumor suppressor genes in gastric cancer. Chin
J Cancer. 32:31–41. 2013. View Article : Google Scholar :
|
|
2
|
Yang XB, Zhao JJ, Huang CY, Wang QJ, Pan
K, Wang DD, Pan QZ, Jiang SS, Lv L, Gao X, et al: Decreased
expression of the FOXO3a gene is associated with poor prognosis in
primary gastric adenocarcinoma patients. PLoS One. 8:e781582013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wogan GN, Hecht SS, Felton JS, Conney AH
and Loeb LA: Environmental and chemical carcinogenesis. Semin
Cancer Biol. 14:473–486. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buffart TE, Tijssen M, El-Bchiri J, Duval
A, van de Wiel MA, Ylstra B, Meijer GA and Carvalho B: NMD
inhibition fails to identify tumour suppressor genes in
microsatellite stable gastric cancer cell lines. BMC Med Genomics.
2:392009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friday BB and Adjei AA: K-ras as a target
for cancer therapy. Biochim Biophys Acta. 1756:127–144.
2005.PubMed/NCBI
|
|
6
|
Bah N, Maillet L, Ryan J, Dubreil S,
Gautier F, Letai A, Juin P and Barillé-Nion S: Bcl-xL controls a
switch between cell death modes during mitotic arrest. Cell Death
Dis. 5:e12912014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan MJ and Wang T: Advances of the
interleukin-21 signaling pathway in immunity and angiogenesis.
Biomed Rep. 5:3–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cha Z, Gu H, Guo H, Tu X, Zang Y, Zhao C,
Hua M, Rechlic JR, Olasnova LM, Song H, et al: Effect of
interleukin 21 and its receptor on CD8+ T cells in the
pathogenesis of diffuse large B-cell lymphoma. Oncol Lett.
8:421–425. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Browning RL, Mo X, Muthusamy N and Byrd
JC: CpG oligo-deoxynucleotide CpG-685 upregulates functional
interleukin-21 receptor on chronic lymphocytic leukemia B cells
through an NF-κB mediated pathway. Oncotarget. 6:15931–15939. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wood B, Sikdar S, Choi SJ, Virk S,
Alhejaily A, Baetz T and LeBrun DP: Abundant expression of
interleukin-21 receptor in follicular lymphoma cells is associated
with more aggressive disease. Leuk Lymphoma. 54:1212–1220. 2013.
View Article : Google Scholar
|
|
11
|
Zhang M, Mathews Griner LA, Ju W, Duveau
DY, Guha R, Petrus MN, Wen B, Maeda M, Shinn P, Ferrer M, et al:
Selective targeting of JAK/STAT signaling is potentiated by Bcl-xL
blockade in IL-2-dependent adult T-cell leukemia. Proc Natl Acad
Sci USA. 112:12480–12485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hao Y, Chapuy B, Monti S, Sun HH, Rodig SJ
and Shipp MA: Selective JAK2 inhibition specifically decreases
Hodgkin lymphoma and mediastinal large B-cell lymphoma growth in
vitro and in vivo. Clin Cancer Res. 20:2674–2683. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang Z, Zhang L, Liao Q, Wang Y, Yu F,
Feng M, Xiang X and Xiong J: Regulation of TRIM24 by miR-511
modulates cell proliferation in gastric cancer. J Exp Clin Cancer
Res. 36:172017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ding L, Zhang S, Xu M, Zhang R, Sui P and
Yang Q: MicroRNA-27a contributes to the malignant behavior of
gastric cancer cells by directly targeting PH domain and
leucinerich repeat protein phosphatase 2. J Exp Clin Cancer Res.
36:452017. View Article : Google Scholar
|
|
16
|
Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He
Y, Chen G, Zhou Q, Wang W, Zhou X, et al: Radiation-induced
miR-208a increases the proliferation and radioresistance by
targeting p21 in human lung cancer cells. J Exp Clin Cancer Res.
35:72016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li
L, Xu H, Zhong J and Zeng X: Serum miR-20a is a promising biomarker
for gastric cancer. Biomed Rep. 6:429–434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Imaoka H, Toiyama Y, Okigami M, Yasuda H,
Saigusa S, Ohi M, Tanaka K, Inoue Y, Mohri Y and Kusunoki M:
Circulating microRNA-203 predicts metastases, early recurrence, and
poor prognosis in human gastric cancer. Gastric Cancer. 19:744–753.
2016. View Article : Google Scholar
|
|
19
|
Qu X, Zhou J, Wang T, Han J, Ma L, Yu H,
Geng D, Fan H, Zhang Q, Hua F, et al: MiR-30a inhibits Th17
differentiation and demyelination of EAE mice by targeting the
IL-21R. Brain Behav Immun. 57:193–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yun S, Lee SU, Kim JM, Lee HJ, Song HY,
Kim YK, Jung H, Park YJ, Yoon SR, Oh SR, et al: Integrated
mRNA-microRNA profiling of human NK cell differentiation identifies
MiR-583 as a negative regulator of IL2Rγ expression. PLoS One.
9:e1089132014. View Article : Google Scholar
|
|
21
|
Wang Y, Wang K, Dang N, Wang L and Zhang
M: Downregulation of miR-3940-5p promotes T-cell activity by
targeting the cytokine receptor IL-2R gamma on human cutaneous
T-cell lines. Immunobiology. 221:1378–1381. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Laneve P, Po A, Favia A, Legnini I, Alfano
V, Rea J, Di Carlo V, Bevilacqua V, Miele E, Mastronuzzi A, et al:
The long noncoding RNA linc-NeD125 controls the expression of
medulloblastoma driver genes by microRNA sponge activity.
Oncotarget. 8:31003–31015. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X,
Yang Y, Xiao B and Guo J: LncRNA-RMRP promotes carcinogenesis by
acting as a miR-206 sponge and is used as a novel biomarker for
gastric cancer. Oncotarget. 7:37812–37824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Wang G, Chu SJ, Zhu JS, Zhang R,
Lu WW, Xia LQ, Lu YM, Da W and Sun Q: Loss of large tumor
suppressor 1 promotes growth and metastasis of gastric cancer cells
through upregulation of the YAP signaling. Oncotarget.
7:16180–16193. 2016.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y
and Yang K: Up-regulation of long noncoding RNA MALAT1 contributes
to proliferation and metastasis in esophageal squamous cell
carcinoma. J Exp Clin Cancer Res. 34:72015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N,
Bin J, Liao Y and Liao W: Long non-coding RNA MALAT1 promotes
gastric cancer tumorigenicity and metastasis by regulating
vasculogenic mimicry and angiogenesis. Cancer Lett. 395:31–44.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jadaliha M, Zong X, Malakar P, Ray T,
Singh DK, Freier SM, Jensen T, Prasanth SG, Karni R, Ray PS, et al:
Functional and prognostic significance of long non-coding RNA
MALAT1 as a metastasis driver in ER negative lymph node negative
breast cancer. Oncotarget. 7:40418–40436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Deng QJ, Xie LQ and Li H: Overexpressed
MALAT1 promotes invasion and metastasis of gastric cancer cells via
increasing EGFL7 expression. Life Sci. 157:38–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Davis ID, Skak K, Smyth MJ, Kristjansen
PE, Miller DM and Sivakumar PV: Interleukin-21 signaling: Functions
in cancer and autoimmunity. Clin Cancer Res. 13:6926–6932. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ueda M, Imada K, Imura A, Koga H,
Hishizawa M and Uchiyama T: Expression of functional interleukin-21
receptor on adult T-cell leukaemia cells. Br J Haematol.
128:169–176. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bolzoni M, Ronchetti D, Storti P, Donofrio
G, Marchica V, Costa F, Agnelli L, Toscani D, Vescovini R, Todoerti
K, et al: IL21R expressing CD14+CD16+ monocytes expand in multiple
myeloma patients leading to increased osteoclasts. Haematologica.
102:773–784. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
de Totero D, Meazza R, Zupo S, Cutrona G,
Matis S, Colombo M, Balleari E, Pierri I, Fabbi M, Capaia M, et al:
Interleukin-21 receptor (IL-21R) is up-regulated by CD40 triggering
and mediates proapoptotic signals in chronic lymphocytic leukemia B
cells. Blood. 107:3708–3715. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang LN, Cui YX, Ruge F and Jiang WG:
Interleukin 21 and its receptor play a role in proliferation,
migration and invasion of breast cancer cells. Cancer Genomics
Proteomics. 12:211–221. 2015.PubMed/NCBI
|
|
37
|
Yin F, Zhang JN, Wang SW, Zhou CH, Zhao
MM, Fan WH, Fan M and Liu S: MiR-125a-3p regulates glioma apoptosis
and invasion by regulating Nrg1. PLoS One. 10:e01167592015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pan L, Zhou L, Yin W, Bai J and Liu R:
miR-125a induces apoptosis, metabolism disorder and
migrationimpairment in pancreatic cancer cells by targeting
Mfn2-related mitochondrial fission. Int J Oncol. 53:124–136.
2018.PubMed/NCBI
|
|
39
|
Yan L, Yu MC, Gao GL, Liang HW, Zhou XY,
Zhu ZT, Zhang CY, Wang YB and Chen X: MiR-125a-5p functions as a
tumour suppressor in breast cancer by downregulating BAP1. J Cell
Biochem. Aug 4–2018.Epub ahead of print. View Article : Google Scholar
|
|
40
|
Bayoumi AS, Sayed A, Broskova Z, Teoh JP,
Wilson J, Su H, Tang YL and Kim IM: Crosstalk between Long
Noncoding RNAs and MicroRNAs in Health and Disease. Int J Mol Sci.
17:3562016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen L, Yao H, Wang K and Liu X: Long
non-coding RNA MALAT1 regulates ZEB1 expression by sponging
miR-143-3p and promotes hepatocellular carcinoma progression. J
Cell Biochem. 118:4836–4843. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J,
Tan D and Liu S: The long non-coding RNA MALAT1 promotes the
migration and invasion of hepatocellular carcinoma by sponging
miR-204 and releasing SIRT1. Tumour Biol. 39:10104283177181352017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li Z, Xu C, Ding B, Gao M, Wei X and Ji N:
Long non-coding RNA MALAT1 promotes proliferation and suppresses
apoptosis of glioma cells through derepressing Rap1B by sponging
miR-101. J Neurooncol. 134:19–28. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jiang LT, Wan CH, Guo QH, Yang SJ, Wu JD
and Cai J: Long noncoding RNA metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) promotes renal cell carcinoma
progression via sponging miRNA-429. Med Sci Monit. 24:1794–1801.
2018. View Article : Google Scholar : PubMed/NCBI
|