Introduction
Owing to differentiated stages and morphological
characteristics, lung cancer may be classified into small cell lung
cancer and non-small cell lung cancer (NSCLC) (1). NSCLC cases account for <85% of all
lung cancer cases, and are currently treated primarily by surgery
and chemotherapy (2,3). However, the typically late diagnosis
of lung cancer has led to a relatively low 5-year survival rate of
15%, therefore identification of novel biomarkers for diagnosing
early-stage lung cancer is of high importance (4). In addition, development of biomarkers
also satisfies the requirements for personalized treatment, which
might assist in specific diagnosis and treatment of NSCLC (5).
With the progress of human genome sequencing, it was
observed that merely 2% of total RNA could be translated into
proteins, and other RNAs that were not transcribed were classified
as non-coding RNA (ncRNA) (6,7).
ncRNAs may be divided into housekeeping ncRNAs and regulatory RNAs,
and the latter are further subcategorized into small ncRNAs
(<200 nt) and long non-coding RNAs (lncRNAs; ≥200 nt) (8). The reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) is the standard technique for
detecting lncRNA expression, owing to its high sensitivity and
precision (9). The aberrantly
expressed lncRNAs were extrapolated to be associated with human
disorders (10), since they were
involved with a number of biological processes, including
X-chromosome inactivation, reactivation of pluripotent stem cells,
differentiation of myocytes, cell apoptosis and cell invasion
(11-13). Of note, it has been identified that
development of lung cancer was accompanied by varied expression of
lncRNAs, and certain lncRNAs have been identified as principal
biomarkers for lung cancer development (14,15).
For instance, expression of lncRNA H19 was increased with increased
tumor size and advanced tumor-node-metastasis (TNM) staging when
NSCLC tissues were investigated (16). Additionally, H19 promoted
epithelial-mesenchymal transition (EMT) and metastasis of lung
cancer cells by decreasing E-cadherin expression and facilitating
Slug expression (17,18). These previous studies all indicate
that H19 is involved in the etiology of lung cancer, including lung
adenocarcinoma.
In addition, lncRNAs were hypothesized to act on
microRNAs (miRNAs/miRs) in various ways (19,20).
For example, H19 was documented to sponge miR-200b/c in mediating
EMT of breast cancer cells, whereas the reverse transition was
triggered when H19 sponged let-7b (21). H19 could also directly target
miR-29b-3p to boost metastasis of bladder cancer cells by
downregulating DNA methyltransferase 3B expression (22). Of note, miR-29b-3p was identified
to antagonize the aggravation of NSCLC (23), and signal transducer and activator
of transcription 3 (STAT3), the downstream target molecule of
miR-29b-3p, exhibited an increase in expression in lung cancer
cells (24). However, few studies
have stated clearly whether H19 could function on miR-29b-3p and
STAT3 to modify the pathogenesis of lung adenocarcinoma.
Thus, the aim of the present study was to
preliminarily elucidate the underlying mechanism between
H19/miR-29b-3p/STAT3 axis and the development of lung
adenocarcinoma, which may assist in identifying potential
biomarkers for diagnosing and treating lung adenocarcinoma.
Materials and methods
Chip analysis
starBase software (version 2.0; starbase. sysu.edu.cn) (25)
was applied to compare H19 expression between cancer tissues and
normal tissues, and to predict the downstream miRNA of H19. Using
this software, the expression data of H19 and miR-29b-3p within
lung adenocarcinoma tissues were drawn from The Cancer Genome Atlas
project (cancergenome.nih.gov) by way of the
Genomic Data Commons Data Portal (portal.gdc.cancer.gov). Furthermore, the potential
target sites of H19 and miR-29b-3p were predicted using miRanda
(www.miranda.org), and were investigated further
on basis of consideration of Argonaut (Ago) cross-linking
immunoprecipitation clusters. The Ago protein could greatly
influence miRNA processing and miRNA-derived cleavage within
animals (26,27), thereby affecting the target sites
between lncRNAs and miRNAs.
Collection of samples
A total of 305 lung adenocarcinoma tissues were
collected from patients who underwent surgical excision in The
First Affiliated Hospital of Jinzhou Medical University (Jinzhou,
China) between March 2016 and January 2017. The lung adenocarcinoma
subjects were diagnosed according to the standards of the 2015
World Health Organization classification (28), in which the definition of
adenocarcinoma has been modified, and thereby the probability of
NSCLCs diagnosed as adenocarcinoma was increased. The normal
pulmonary tissues obtained as the control group were located >3
cm from the lung adenocarcinoma tissues. The present study was
approved by the ethics committee of The First Affiliated Hospital
of Jinzhou Medical University and all patients provided written
informed consent.
Cell culture
Human lung adenocarcinoma cell lines (Calu-3,
NCI-H1975, A549 and NCI-H23) and normal lung cell line (HLF-a) were
purchased from the American Type Culture Collection (Manassas, VA,
USA). Cells were seeded in Dulbecco′s modified Eagle′s medium
(HyClone; GE Healthcare, Logan, UT, USA) containing 10% fetal
bovine serum, 1×105 U/l penicillin and 1×105
U/l streptomycin. The cells were cultured in 5% CO2 and
90% humidity at 37°C.
RT-qPCR
Total RNA was extracted from all cell lines and
tissues using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and its purity and
concentration were determined spectrophotometrically. Extracted
RNAs were reverse-transcribed into cDNAs using a Superscript II
reverse transcription kit (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer′s protocol, with reaction
conditions of: i) 42°C for 60 min, ii) 95°C for 5 min, and iii) 4°C
for 10 min. The cDNAs were subjected to PCR using a SYBR-Green
master kit (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the manufacturer′s protocol. The PCR conditions for
H19 and GAPDH were: i) Pre-denaturation at 95°C for 2 min, and ii)
40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C for
30 sec and extension at 72°C for 15 sec. Furthermore, the PCR
conditions for miR-29b-3p and U6 were: i) Pre-denaturation at 95°C
for 30 sec, and ii) 40 cycles of denaturation at 95°C for 10 sec,
annealing at 60°C for 30 sec and extension at 70°C for 5 sec. GAPDH
was used as the internal reference for H19, and U6 was used as the
internal reference for miR-29b-3p. The relative expression of H19
and miR-29b-3p were calculated according to the 2−ΔΔCq
method (29). Primers (Table I) were designed using Primer
Express software (version 2.0; Applied Biosystems; Thermo Fisher
Scientific, Inc.), and were synthesized by Sangon Biotech Co., Ltd.
(Shanghai, China).
 | Table IPrimer sequences for miR-29b-3p,
lncRNA H19, STAT3, U6 and GAPDH. |
Table I
Primer sequences for miR-29b-3p,
lncRNA H19, STAT3, U6 and GAPDH.
| RNA | Primer
sequence |
|---|
| miR-29b-3p | F:
5′-ACACTCCAGCTGGGTAGCACCATT TGAAATC-3′ |
| R:
5′-TGGTGTCGTGGAGTCG-3′ |
| U6 | F:
5′-CGCCCCACCCCTCCAG-3′ |
| R:
5′-CCGCCCAGACCCTCAGACT-3′ |
| lncRNA H19 | F:
5′-CTGTAACCGGCGCCAGAA-3′ |
| R:
5′-TGCATGGGAGAGCCCAGA-3′ |
| GAPDH | F:
5′-TGTGGGCATCAATGGATTTGG-3′ |
| R:
5′-ACACCATGTATTCCGGGTCAAT-3′ |
Western blotting
Total protein from tissues and cells was extracted
using radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) that contained proteinase inhibitor
(Roche Diagnostics, Indianapolis, IN, USA), and its concentration
was determined using a Bicinchoninic Acid protein assay kit (Thermo
Fisher Scientific, Inc.), according to the manufacturer′s protocol.
Subsequently, the protein extracts (30 µg for each sample)
were separated by SDS-PAGE (6 or 10% polyacrylamide gel), and
transferred onto polyvinylidene difluoride membranes. The membranes
were blocked with 5% skimmed milk in Tris-buffered saline
containing 0.25% Tween-20 at room temperature for 1 h before
overnight incubation at 4°C with rabbit anti-human monoclonal
antibodies against STAT3 (1:1,000; cat. no. ab68153; Abcam,
Cambridge, MA, USA), epithelial (E-)cadherin (1:1,000; cat. no.
20874-1-AP), vimentin (1:1,000; cat. no. 10366-1-AP), Snail
(1:1,000; cat. no. 13099-1-AP) (all from ProteinTech Group, Inc.,
Chicago, IL, USA), Slug (1:1,000; cat. no. sc-166476; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), β-actin (1:1,000; cat. no.
ab8227; Abcam) and GAPDH (1:1,000; cat. no. 5174; Cell Signaling
Technology, Inc., Danvers, MA, USA). Corresponding horseradish
peroxidase-conjugated mouse anti-rabbit secondary antibodies
(1:10,000; cat. no. 93702; Cell Signaling Technology, Inc.) were
added for another 1 h at room temperature. Enhanced
chemiluminescence detection reagent (EMD Millipore, Billerica, MA,
USA) was applied for development, and the band images were analyzed
utilizing a Gene Genius gel imaging system (Syngene Europe,
Cambridge, UK).
Cell transfection
Short interfering RNA (siRNA) for H19 (si-H19; 50
nM), negative control siRNA (si-NC; 50 nM), H19-overexpression
plasmid (pcDNA3.1-H19; 2 µg), pcDNA3.1 (2 µg),
miR-29b-3p mimic (50 nM), miR-29b-3p inhibitor (50 nM),
STAT3-overexpression plasmid (pcDNA3.1-STAT3; 2 µg), STAT3
siRNA (si-STAT3, 2 µg) and negative control (NC) were all
designed and synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou,
China). The sequences of H19-siRNA1#, H19-siRNA2# and H19-siRNA3#
were 5′-CCGUAAUUC ACUUAGAAGAdTdT-3′, 5′-CACAUAGAAAGGCAGGA UAdTdT-3′
and 5′-CCUUCUAAACGAAGGUUUAdTdT-3′, respectively. The sequence of
si-NC was 5′-UUCUCCGAAC GUGUCACGUTT-3′. The sequences for
miR-29b-3p mimic, miR-29b-3p inhibitor and miR-NC were
5′-UAGCACCAUUU GAAAUCAGUGUU-3′, 5′-AACACUGAUUUCAAAUGGUG CUA-3′ and
5′-UUCUCCGAACGUGUCACGUTT-3′, respectively. The forward and reverse
primers for si-STAT3 were 5′-AAGCAGCAGCTGAACAACATGTTCAAGAGACATGT
TGTTCAGCTGCTGCTT-3′ and 5′-AAGCAGCAGCTGAAC
AACATGTCTCTTGAACATGTTGTTCAGCTGCTGCTT-3′, respectively.
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for cell transfection, according to the
manufacturer′s protocol. After 48 h of transfection, Calu-3 and
NCI-H1975 cell lines were used for subsequent experiments.
Colony formation assay
Calu-3 and NCI-H1975 cell lines were inoculated into
6-well plates at a density of 1,000 cells/well. Following
continuous culture for 10 days, the cells were fixed with methanol
for 20 min. Finally, the cells were stained with crystal violet
(Sigma-Aldrich; Merck KGaA) for 15 min, and the number of colonies
that included >50 cells was determined using an inverted light
microscope (Leica Microsystems GmbH, Wetzlar, Germany) at ×100
magnification.
Cell proliferation assay
Calu-3 and NCI-H1975 cell lines were seeded in
96-well plates at a density of 2,500 cells/well. At 24, 48, 72 and
96 h after inoculation, Cell Counting Kit-8 (CCK-8) solution was
added to each well (Abmole Bioscience Inc., Houston, TX, USA) at 10
µl/well. Following incubation at 37°C for 1 h, the
absorbance at 450 nm was determined to calculate the cell
proliferation rate.
Cell apoptosis assay
Calu-3 and NCI-H1975 cell lines were resuspended in
500 µl binding buffer, and were stained with 5 µl
Annexin V-fluorescein isothiocyanate (FITC) and 10 µl
propidium iodide (PI) in the dark for 15 min, using an Annexin
V-FITC cell apoptosis kit (Beyotime Institute of Biotechnology,
Haimen, China), according to the manufacturer′s protocol.
Subsequently, flow cytometry (model, ELITE; laser wavelength, 488
nm; power, 15 mW; Beckman Coulter, Inc., Brea, CA, USA) was used to
analyze cell apoptosis. On the flow cytometry scattergrams, cells
in the lower-left quadrant labeled as
FITC-/PI- and the upper-left quadrant labeled
as FITC-/PI+ were designated as viable cells.
In contrast, cells in the upper-right quadrant tagged as
FITC+/PI+ were designated as necrotic cells,
and those in the lower-right quadrant labeled as
FITC+/PI- were designated as early-apoptotic
cells.
Dual-luciferase reporter gene assay
The H19 fragments that contained specific
miR-29b-3p-binding sites were cloned into the pmirGLO
dual-luciferase expression vector (Promega Corporation, Madison,
WI, USA), through which pmirGLO-H19-Wt was formed. Furthermore, the
same binding sites of miR-29b-3p in H19 were mutated to construct
pmirGLO-H19-Mut. Using a similar approach, pmirGLO-STAT3-Wt that
contained miR-29b-3p-binding sites and pmirGLO-STAT3-Mut vectors
were constructed. Cells that had been transfected with
pmirGLO-H19-Wt, pmirGLO-H19-Mut, pmirGLO-STAT3-Wt,
pmirGLO-STAT3-Mut or pmirGLO vector were, respectively, transfected
with miR-29b-3p mimic and miR-NC. At ~48 h after transfection,
luciferase activity was determined using the Dual-Luciferase
Reporter assay system (Promega Corporation).
Statistical analysis
All statistical analyses were performed using SPSS
software (version 17.0; SPSS, Inc., Chicago, IL, USA). The
measurement data that conformed to normal distribution are
expressed as the mean ± standard deviation, and the enumeration
data are represented as the frequency or percentage. Inter-group
comparisons among the measurement data were performed using
Student′s t test or analysis of variance with a Bonferroni post hoc
test, whereas the enumeration data were compared using a
χ2 test. The correlations between H19 expression and
miR-29b-3p expression, as well as between miR-29b-3p expression and
STAT3 expression, were determined by performing Spearman′s rank
correlation test. Furthermore, the Kaplan-Meier method was utilized
to calculate the accumulative survival rate of patients with lung
adenocarcinoma, with the log-rank test adopted for univariate
analysis of prognostic factors. The Cox regression model was used
for the multivariate analysis, to determine the association of
H19/miR-29b-3p expression with clinicopathological features and
overall survival of patients with lung adenocarcinoma. P<0.05
was considered to indicate a statistically significant
difference.
Results
Comparison of H19 and miR-29b-3p
expression between lung adenocarcinoma tissues/cells and normal
tissues/cells
The chip analysis results collected from The Cancer
Genome Atlas starBase (version 2.0) data portal indicated that H19
expression in lung cancer tissues was significantly increased
compared with in paracarcinoma tissues (P<0.05) and that there
was a negative correlation between H19 expression and miR-29b-3p
(Fig. 1). It was also identified
that H19 expression was significantly increased in the lung
adenocarcinoma tissues collected compared with in adjacent
paracancerous tissues (P<0.05), and miR-29b-3p expression in
lung adenocarcinoma was significantly decreased compared with in
adjacent paracancerous tissues (P<0.05) (Fig. 2A). Furthermore, H19 expression in
Calu-3, NCI-H1975, A549 and NCI-H23 cell lines were also increased
compared with that in the HLF-a cell line (P<0.05); in contrast,
the expression of miR-29b-3p was significantly decreased in the
cancer cell lines compared with in the HLF-a cell line (P<0.05)
(Fig. 2B). Since the expressions
of H19 and miR-29b-3p were altered most significantly within Calu-3
and NCI-H1975 cell lines when compared with HLF-a cell line, these
cell lines were selected for the subsequent cell experiments. In
addition, Spearman′s rank correlation analysis identified that
there was a negative correlation between miR-29b-3p expression and
H19 expression among the lung adenocarcinoma tissues investigated
(P<0.05; Fig. 2C).
Clinicopathological analyses identified that increased H19
expression and decreased miR-29b-3p were associated with longer
tumor diameter, more advanced TNM stage and invasive lung carcinoma
(P<0.05; Table II, Fig. 2D and E). The results of
multivariate analyses also indicated that H19 expression,
miR-29b-3p expression, tumor diameter and TNM staging could serve
as the independent predictors for poor survival rate of patients
with lung carcinoma patients (all P<0.05; Table III).
 | Figure 1Relevance of H19 to miR-29b-3p on the
basis of The Cancer Genome Atlas starBase (version 2.0) data
portal. (A) H19 expression was compared between cancer and normal
tissues. (B) H19 expression was negatively correlated with
miR-29b-3p expression in lung adenocarcinoma tissues. miR/miRNA,
microRNA; hsa, human; BLCA, bladder urothelial carcinoma; BRCA,
breast cancer; CRC, colorectal cancer; GBM, glioblastoma
multiforme; HNSC, head and neck squamous cell carcinoma; KICH,
kidney chromophobe; KIRC, kidney renal clear cell carcinoma; LAML,
acute myeloid leukemia; LUAD, lung adenocarcinoma; LUSC, lung
squamous cell carcinoma; OV, ovarian cancer; SKCM, skin cutaneous
melanoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial
carcinoma. |
 | Table IIAssociation between
clinicopathological characteristics and lncRNA H19/miR-29b-3p
expression in patients with lung adenocarcinoma. |
Table II
Association between
clinicopathological characteristics and lncRNA H19/miR-29b-3p
expression in patients with lung adenocarcinoma.
| Clinical
characteristic | lncRNA H19
expression
| miR-29b-3p
expression
|
|---|
| Low | High | P-value | Low | High | P-value |
|---|
| Age, years | | | | | | |
| >56 | 41 | 116 | 0.93 | 107 | 50 | 0.51 |
| ≤56 | 38 | 110 | | 106 | 42 | |
| Sex | | | | | | |
| Female | 39 | 104 | 0.608 | 100 | 43 | 0.973 |
| Male | 40 | 122 | | 113 | 49 | |
| History of
smoking | | | | | | |
| Yes | 45 | 84 | 0.002 | 84 | 45 | 0.124 |
| No | 34 | 142 | | 129 | 47 | |
| Tumor diameter,
cm | | | | | | |
| >1.56 | 46 | 162 | 0.027 | 153 | 55 | 0.038 |
| ≤1.56 | 33 | 64 | | 60 | 37 | |
| TNM staging | | | | | | |
| I+II | 41 | 85 | 0.026 | 76 | 50 | 0.002 |
| III+IV | 38 | 141 | | 137 | 42 | |
| Invasion | | | | | | |
| Yes | 53 | 178 | 0.037 | 170 | 61 | 0.012 |
| No | 26 | 48 | | 43 | 31 | |
| Histological
subtype | | | | | | |
| AIS | 46 | 28 | | 35 | 39 | |
| MIA | 23 | 18 | | 20 | 21 | |
| Lepidic
predominant | 10 | 19 | | 15 | 14 | |
| Acinar
predominant | 28 | 22 | | 23 | 27 | |
| Papillary
predominant | 17 | 26 | | 23 | 20 | |
| Micropapillary
predominant | 12 | 11 | | 10 | 13 | |
| Solid predominant
with mucin production | 14 | 17 | | 13 | 18 | |
| Invasive mucinous
adenocarcinoma | 6 | 8 | | 7 | 7 | |
 | Table IIIAssociation between clinical
characteristics and overall survival of patients with lung
adenocarcinoma patients. |
Table III
Association between clinical
characteristics and overall survival of patients with lung
adenocarcinoma patients.
| Clinical
characteristic | Univariate analysis
| Multivariate
analysis
|
|---|
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| lncRNA H19
expression | | | | | | |
| Low vs. high | 0.35 |
0.21-0.60 |
<0.001 | 0.45 |
0.25-0.81 | 0.007 |
| miR-29b-3p
expression | | | | | | |
| Low vs. high | 2.5 |
1.52-4.12 |
<0.001 | 1.77 |
1.03-3.07 | 0.04 |
| Age, years | | | | | | |
| >56 vs.
≤56 | 1.3 | 0.83-2.05 | 0.257 | 1.25 | 0.76-2.05 | 0.381 |
| Sex | | | | | | |
| Female vs.
male | 1 | 0.63-1.58 | 0.998 | 0.99 | 0.60-1.63 | 0.971 |
| History of
smoking | | | | | | |
| Yes vs. no | 0.9 | 0.57-1.43 | 0.662 | 1.13 | 0.68-1.88 | 0.648 |
| Tumor diameter,
cm | | | | | | |
| >1.56 vs.
≤1.56 | 2.58 |
1.57-4.23 |
<0.001 | 2.45 |
1.43-4.19 | 0.001 |
| TNM staging | | | | | | |
| I+II vs.
III+IV | 0.38 |
0.24-0.61 |
<0.001 | 0.43 |
0.25-0.72 | 0.001 |
| Invasion | | | | | | |
| Yes vs. no | 2.06 |
1.21-3.49 | 0.008 | 1.23 | 0.68-2.22 | 0.503 |
H19 and miR-29b-3p regulate
proliferation, viability and apoptosis of lung adenocarcinoma
cells
H19 expression in Calu-3 and NCI-H1975 cells was
significantly increased following transfection with pcDNA3.1-H19
(P<0.05), yet it was significantly decreased following
transfection with si-H19 (P<0.05) (Fig. 3A). Similarly, miR-29b-3p expression
was significantly increased and decreased following transfection
with miR-29b-3p mimic and miR-29b-3p inhibitor, respectively
(P<0.05; Fig. 3B). In
accordance with the results of CCK-8, flow cytometry and colony
formation assays, the proliferation, viability and survival of
Calu-3 and NCI-H1975 cell lines in the si-H19 group were
significantly decreased, compared with those in the si-NC group
(P<0.05; Fig. 4A). Conversely,
following transfection of pcDNA3.1-H19, the Calu-3 and NCI-H1975
cell lines exhibited significantly decreased apoptosis, along with
significantly increased proliferation and viability, when compared
with the pcDNA3.1 group (all P<0.05; Figs. 4 and 5A). In addition, with the NC group as the
reference, the proliferative capacity and viability of Calu-3 and
NCI-H1975 cell lines in the miR-29b-3p mimic group were
significantly decreased, and the apoptotic percentage of the cells
were significantly increased (P<0.05). By contrast, the
miR-29b-3-p inhibitor group exhibited significantly increased
proliferation and viability, as well as a significantly decreased
percentage of apoptotic cells in comparison with the control group
(P<0.05; Figs. 4 and 5A).
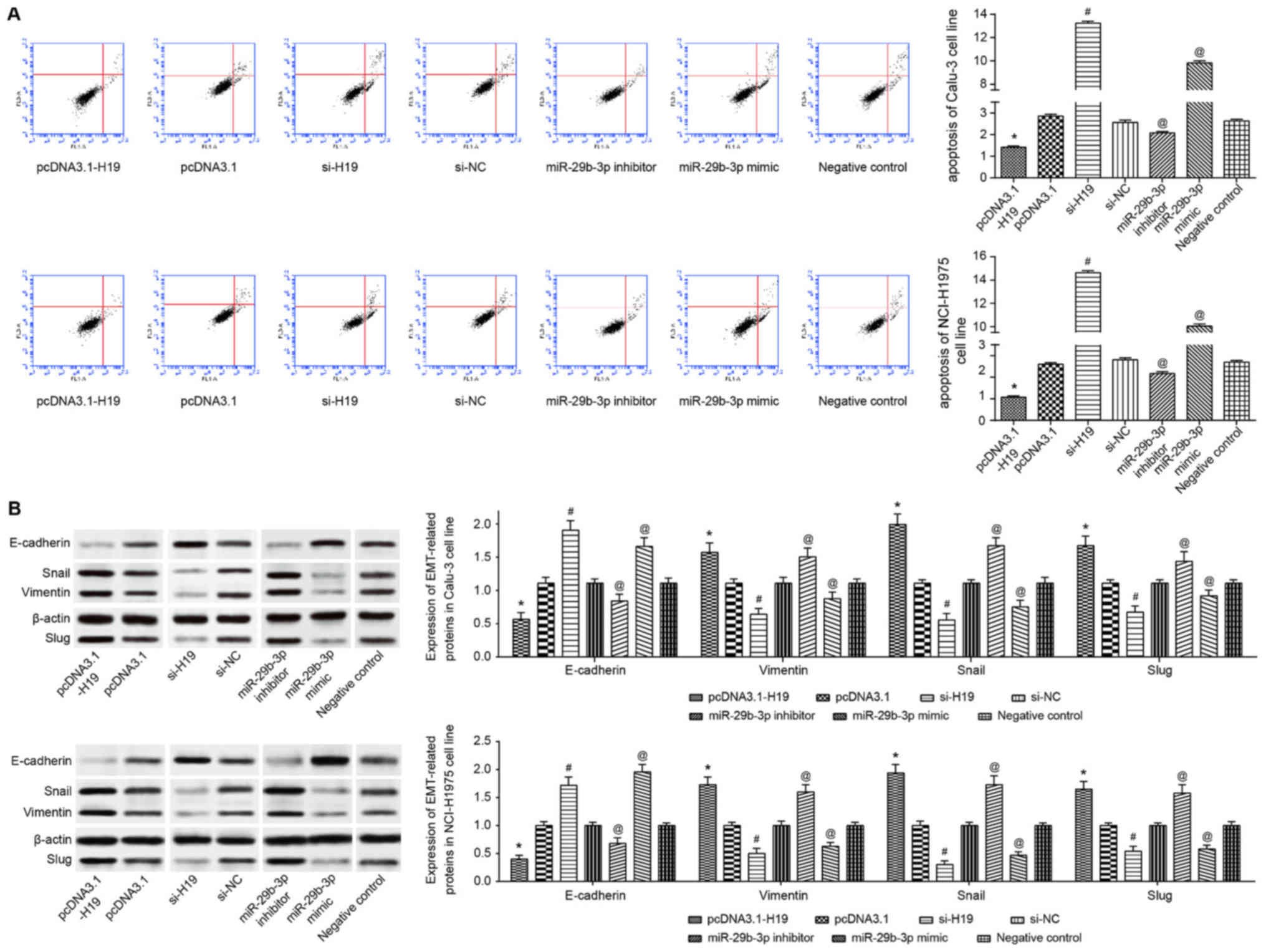 | Figure 5(A) Apoptotic status and (B) EMT
protein expression were compared among pcDNA3.1-H19-, pcDNA3.1-,
si-H19-, si-NC-, miR-29b-3p inhibitor-, miR-29b-3p mimic- and
negative control-transfected Calu-3 and NCI-H1975 cell lines.
*P<0.05 vs. pcDNA3.1; #P<0.05 vs.
si-NC; @P<0.05 vs. negative control. In (B), the
spaces between lanes marked by pcDNA3.1 and si-H19, as well as
between lanes marked by si-NC and miR-29b-3p inhibitor indicate
that the samples were run on different gels. Other spaces between
lanes and between rows indicate that irrelevant lanes have been
excised. EMT, epithelial-mesenchymal transition; si, short
interfering RNA; NC, negative control; miR, microRNA; E-cadherin,
epithelial cadherin. |
H19 and miR-29b-3p modify the expression
of EMT-specific proteins in lung adenocarcinoma cells
As the western blot results indicated, the
expression of epithelial marker (i.e. E-cadherin) in the miR-29b-3p
mimic group was increased significantly (P<0.05), whereas the
expression of interstitial markers, including vimentin, Snail and
Slug, was significantly decreased (P<0.05) in the Calu-3 and
NCI-H1975 cell lines (Fig. 5B).
Distinct from the miR-29b-3p mimic group, Calu-3 and NCI-H1975
cells of the miR-29b-3p inhibitor group were observed with
significantly increased vimentin, Snail and Slug expression, as
well as significantly decreased E-cadherin expression (P<0.05;
Fig. 5B). Furthermore, the
decrease in H19 expression in Calu-3 and NCI-H1975 cell lines
induced a significant decrease in vimentin, Snail and Slug
expression, and a significant increase in E-cadherin expression
(P<0.05; Fig. 5B).
H19 targets miR-29b-3p to decrease its
expression
It was predicted using starBase software that H19
could target miR-29b-3p at chr11: 2017218-2017240 and chr11:
2017218-2017320 (Fig. 6A).
Furthermore, the dual-luciferase reporter gene assay conducted
utilizing Calu-3 and NCI-H1975 cell lines indicated that
transfection of pmirGLO-H19-Wt and miR-29b-3p mimic could induce
significantly decreased luciferase activity compared with cells
transfected with pmirGLO-H19-Wt and miR-NC (P<0.05; Fig. 6B). No evident difference in
luciferase activity of the pmirGLO-H19-Mut+miR-29b-3p mimic group
from that of pmirGLO-H19-Wt+miR-NC group was observed. Furthermore,
RT-qPCR results indicated that increased H19 expression may
significantly decrease the expression level of miR-29b-3p in Calu-3
and NCI-H1975 cell lines (P<0.05; Fig. 6C). However, there was limited
effect on H19 expression, whether miR-29b-3p expression was
increased or decreased (P<0.05) (Fig. 6D).
STAT3 is modified by miR-29b-3p in lung
adenocarcinoma cells
STAT3 expression in lung adenocarcinoma cells was
positively correlated with H19 expression (P<0.05), yet it
appeared to be negatively correlated with miR-29b-3p expression
(P<0.05) (Fig. 7A). In
addition, overexpressed H19 and underexpressed miR-29b-3p could
contribute to abnormally overexpressed STAT3 in Calu-3 and
NCI-H1975 cell lines (P<0.05; Fig.
7B). Furthermore, the luciferase activity of miR-29b-3p mimic
binding to pmirGLO-STAT3-Wt was significantly increased compared
with that in cells co-transfected with miR-29b-3p mimic and
pmirGLO-STAT3-Mut (P<0.05), and the latter revealed lucif-erase
activity that was not significantly different from that of the
pmirGLO-STAT3-Wt+miR-NC group (Fig.
7C).
 | Figure 7Association between miR-29b-3p and
STAT3. (A) H19 expression was positively correlated with STAT3
expression in lung adenocarcinoma tissues. miR-29b-3p expression
was negatively correlated with STAT3 expression in lung
adenocarcinoma tissues. (B) STAT3 expression in pcDNA3.1-H19-,
pcDNA3.1-, si-H19-, si-NC-, miR-29b-3p inhibitor-, miR-29b-3p
mimic- and negative control-transfected Calu-3 and NCI-H1975 cell
lines. *P<0.05 vs. pcDNA3.1; #P<0.05
vs. si-NC; @P<0.05 vs. negative control. Spaces
between lanes marked by pcDNA3.1 and si-H19, as well as between
lanes marked by si-NC and miR-29b-3p inhibitor indicate that the
samples were run on different gels. Other spaces between lanes and
between rows indicate that irrelevant lanes have been excised. (C)
miR-29b-3p targeted STAT3 in certain sites. The luciferase
activities of Calu-3 and NCI-H1975 cell lines were compared between
miR-29b-3p mimic+STAT3-Wt and miR-29b-3p mimic+STAT3-Mut
groups.*P<0.05 vs. miR-29b-3p mimic+STAT3-Mut group.
miR/miRNA, microRNA; STAT3, signal transducer and activator of
transcription 3; si, short interfering; NC, negative control;
lncRNA, long non-coding RNA; hsa, human. |
STAT3 is modified by H19 and miR-29b-3p
in altering viability, proliferation and apoptosis of lung
adenocarcinoma cells, as well as the EMT-specific proteins in lung
adenocarcinoma cells
It was observed that the proliferation and viability
of Calu-3 and NCI-H1975 cell lines in the miR-NC+STAT3 group were
significantly increased compared with those of the miR-NC group
(P<0.05; Fig. 8), and the
apoptotic rate of the miR-NC+STAT3 group was below that of miR-NC
group (P<0.05; Fig. 9A).
Regarding the expression of EMT-specific proteins, it was
identified that the miR-NC+STAT3 group significantly increased
vimentin, Snail and Slug expression, as well as decreased
E-cadherin expressions, when compared with miR-NC group (P<0.05;
Fig. 9B). These results therefore
suggested that STAT3 inhibits the effect of miR-29b-3p on the
viability, proliferation, apoptosis and EMT of lung adenocarcinoma
cells.
Discussion
Primary bronchial carcinoma, also known as lung
cancer, is a major cause of mortality (30,31),
and the prevalence of its one pathological pattern (i.e. lung
adenocarcinoma) is increasing (32). Although traditional therapies for
lung adenocarcinoma, such as surgery, chemotherapy and
radiotherapy, have been performed, 5-year survival rate of patients
remains low at ~10% (2). Among
them, resistance to chemotherapies appears to be the factor
limiting the recovery of patients with lung adenocarcinoma
patients, and certain lncRNAs and miRNAs have been identified to
participate in the underlying molecular mechanism (33,34).
Notably, alterations in cell viability, survival, apoptosis and EMT
proteins may also guide the chemoresistance of tumor cells in a
different direction. Thus, in the present study, the role of lncRNA
H19 and miR-29b-3p in regulating EMT, viability and apoptosis of
lung carcinoma cells was investigated.
H19 was initially identified by Bartolomei et
al in 1991 (35), and it was
revealed to underlie the development process of bladder carcinoma,
hepatocellular carcinoma, breast cancer and lung cancer (35-38).
For example, H19 expressed in NSCLC tissues was increased ~2-fold
compared with in adjacent normal tissues (39,40).
The present study also revealed similar results, and it also
demonstrated that patients with lung adenocarcinoma with higher H19
expression exhibited an increased survival rate compared with those
with lower H19 expression. With regard to the in vitro
experiments, H19 was revealed to promote metastasis and
proliferation of lung adenocarcinoma cells, which led to decreased
sensitivity of the cells to cisplatin (40). Similar to this result, the present
study also revealed that upregulated H19 expression could increase
cell viability, cell proliferation and expression of EMT-specific
proteins in cells, as well as decrease cell apoptosis. Of note, a
previous study identified that the H19 promoter intensified by
c-myc could facilitate the proliferation of lung cancer cells by
increasing miR-107 expression (39). In the present study, it was
identified that H19 could suppress miR-29b-3p expression in lung
carcinoma cells, which also resulted in increased viability and
proliferation, along with decreased apoptosis of the neoplastic
cells. It was thus suggested that H19 may interact with various
miRNAs to modify the activity of lung adenocarcinoma cells, and
other downstream molecules require further investigation.
Nevertheless, the present study was limited by not establishing
mouse models to verify the effects of H19 and miR-29b-3p on the
progression of lung adenocarcinoma, as in a previous study
(41). One point that should be
underlined is how H19 acts on miR-29b-3p to regulate the
development of lung adenocarcinoma. Salmena et al (19) proposed a competing endogenous RNA
hypothesis that mRNAs, pseudogenes, lncRNAs and other endogenous
RNAs could competitively combine with the same miRNA with their
specific miRNA-binding sites, thereby limiting the inhibitory
effect of miRNA on the mRNA of target genes and increasing the
expression of target genes. Consistent with this hypothesis, the
present study also identified that H19 had a ′sponging′ function
(42), and H19 could target
miR-29b-3p to limit its expression. In addition, the aforementioned
miR-29b was previously identified to participate in the modulation
of cell apoptosis, the cell cycle and cell metastasis (43,44).
In particular, abnormally increased expression of miR-29b decreased
the proliferation, migration and invasion of lung cancer cells by
<30% (45). Furthermore,
miR-29b-3p expression was significantly decreased in pancreatic
carcinoma cells when compared with in normal cells, and
upregulation of miR-29b-3p expression could significantly limit
proliferation of the cells (46).
These results were verified in the present study, and it was
concluded that miR-29b-3p, which was regulated by H19, could
suppress proliferation, viability and EMT, and promote the
apoptosis of lung carcinoma cells.
In addition, the present study also indicated that
STAT3 was the target gene of miR-29b-3p, and miR-29b-3p could
directly regulate STAT3 expression. As a component of the Janus
kinase signaling pathway, STAT3 appears to be critical for cancer
onset and progression in the tumor microenvironment (47,48).
More specifically, activation of STAT3 usually either increased
cell proliferation and survival or decreased cell apoptosis
(49,50). Consistently, the present study
indicated that the STAT3 activated by H19 and miR-29b-3p allowed
increased proliferation and decreased apoptosis of the lung
adenocarcinoma cells, as well as activated EMT-specific protein
expression in the cells. As for whether H19, miR-29b-3p and STAT3
could alter metastasis of lung adenocarcinoma cells via induction
of the EMT process (51), further
investigation is required.
In summary, the H19/miR-29b-3p/STAT3 axis could
affect EMT, apoptosis, proliferation and viability of lung
carcinoma cells, which potentially reveals the underlying molecular
mechanism of lung adenocarcinoma. The results may provide a
foundation for developing a complete strategy for diagnosing and
treating lung carcinoma. However, the present study was limited by
the small size of the clinical samples analyzed, which may not be
applicable to the wider population. Furthermore, relevant animal
models to validate the study results were not established. Thirdly,
since the progression of lung adenocarcinoma resulted from mixed
effects of various genes, additional downstream and upstream genes
require investigation. In addition, although it is hypothesized
that H19 and miR-29b-3p may function to interfere with the
chemosensitivity of lung adenocarcinoma cells on the basis of their
molecular mechanisms, to the best of our knowledge, this has not
been investigated. Finally, since there are limited standards
formulated to unify the results of diverse methods, there may be
misunderstanding when experimental results that were focused on one
point were compared, such as detection methods for lncRNAs.
Therefore, further in-depth investigations are still required.
Funding
Not applicable.
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors′ contributions
LihL, LinL and SL conceived and designed the
experiments. LihL, LinL and SL performed the experiments. LihL and
LinL analyzed the data. SL drafted the manuscript. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Jinzhou Medical
University (Jinzhou, China) and all patients provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Wistuba II: Genetics of preneoplasia:
Lessons from lung cancer. Curr Mol Med. 7:3–14. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Verdecchia A, Francisci S, Brenner H,
Gatta G, Micheli A, Mangone L and Kunkler I; EUROCARE-4 Working
Group: Recent cancer survival in Europe: A 2000–02 period analysis
of EUROCARE-4 data. Lancet Oncol. 8:784–796. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Klebe S and Henderson DW: Facts and
fiction: Premalignant lesions of lung tissues. Pathology.
45:305–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W and Schlesinger
F: Landscape of transcription in human cells. Nature. 489:101–108.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gadgeel SM, Cote ML, Schwartz AG, Matherly
LH, Wozniak A and Bepler G: Parameters for individualizing systemic
therapy in non-small cell lung cancer. Drug Resist Updat.
13:196–204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Birney E, Stamatoyannopoulos JA, Dutta A,
Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis
ET and Thurman RE: Children′s Hospital Oakland Research Institute:
Identification and analysis of functional elements in 1% of the
human genome by the ENCODE pilot project. Nature. 447:799–816.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponting CP and Belgard TG: Transcribed
dark matter: Meaning or myth? Hum Mol Genet. 19:R162–R168. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inamura K: Major tumor suppressor and
oncogenic non-coding RNAs: Clinical relevance in lung cancer.
Cells. 6:62017. View Article : Google Scholar
|
|
9
|
Shi T, Gao G and Cao Y: Long noncoding
RNAs as novel biomarkers have a promising future in cancer
diagnostics. Dis Markers. 2016:90851952016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3:52015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gayen S, Maclary E, Buttigieg E, Hinten M
and Kalantry S: A primary role for the Tsix lncRNA in maintaining
random X-chromosome inactivation. Cell Reports. 11:1251–1265. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kawaguchi T and Hirose T: Chromatin
remodeling complexes in the assembly of long noncoding
RNA-dependent nuclear bodies. Nucleus. 6:462–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhuang W, Ge X, Yang S, Huang M, Zhuang W,
Chen P, Zhang X, Fu J, Qu J and Li B: Upregulation of lncRNA MEG3
promotes osteogenic differentiation of mesenchymal stem cells from
multiple myeloma patients by targeting BMP4 transcription. Stem
Cells. 33:1985–1997. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D and
Marra A: The long noncoding MALAT-1 RNA indicates a poor prognosis
in non-small cell lung cancer and induces migration and tumor
growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee YS and Dutta A: MicroRNAs: Small but
potent oncogenes or tumor suppressors. Curr Opin Investig Drugs.
7:560–564. 2006.PubMed/NCBI
|
|
16
|
Zhang E, Li W, Yin D, De W, Zhu L, Sun S
and Han L: c-Myc-regulated long non-coding RNA H19 indicates a poor
prognosis and affects cell proliferation in non-small-cell lung
cancer. Tumour Biol. 37:4007–4015. 2016. View Article : Google Scholar
|
|
17
|
Sadiq AA and Salgia R: MET as a possible
target for non-small-cell lung cancer. J Clin Oncol. 31:1089–1096.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Matouk IJ, Raveh E, Abu-lail R, Mezan S,
Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M,
et al: Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys
Acta. 1843:1414–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zaleska K: miRNA - Therapeutic tool in
breast cancer? Where are we now? Rep Pract Oncol Radiother.
20:79–86. 2014. View Article : Google Scholar
|
|
21
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal.
10:102017. View Article : Google Scholar
|
|
22
|
Lv M, Zhong Z, Huang M, Tian Q, Jiang R
and Chen J: lncRNA H19 regulates epithelial-mesenchymal transition
and metastasis of bladder cancer by miR-29b3p as competing
endogenous RNA. Biochim Biophys Acta Mol Cell Res. 1864:1887–1899.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Avasarala S, Van Scoyk M, Wang J, Sechler
M, Vandervest K, Brzezinski C, Weekes C, Edwards MG, Arcaroli J,
Davis RE, et al: hsa-miR29b, a critical downstream target of
non-canonical Wnt signaling, plays an anti-proliferative role in
non-small cell lung cancer cells via targeting MDM2 expression.
Biol Open. 2:675–685. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Weerasinghe P, Garcia GE, Zhu Q, Yuan P,
Feng L, Mao L and Jing N: Inhibition of Stat3 activation and tumor
growth suppression of non-small cell lung cancer by G-quartet
oligonucleotides. Int J Oncol. 31:129–136. 2007.PubMed/NCBI
|
|
25
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar
|
|
26
|
Shin C, Nam JW, Farh KK, Chiang HR,
Shkumatava A and Bartel DP: Expanding the microRNA targeting code:
Functional sites with centered pairing. Mol Cell. 38:789–802. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karginov FV, Cheloufi S, Chong MM, Stark
A, Smith AD and Hannon GJ: Diverse endonucleolytic cleavage sites
in the mammalian transcriptome depend upon microRNAs, Drosha, and
additional nucleases. Mol Cell. 38:781–788. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Inamura K: Lung Cancer: Understanding Its
Molecular Pathology and the 2015. WHO Classification Front Oncol.
7:1932017. View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
30
|
Chung WJ, Agius P, Westholm JO, Chen M,
Okamura K, Robine N, Leslie CS and Lai EC: Computational and
experimental identification of mirtrons in. Drosophila melanogaster
and Caenorhabditis elegans Genome Res. 21:286–300. 2011.
|
|
31
|
Lee I, Ajay SS, Yook JI, Kim HS, Hong SH,
Kim NH, Dhanasekaran SM, Chinnaiyan AM and Athey BD: New class of
microRNA targets containing simultaneous 5′-UTR and 3′-UTR
interaction sites. Genome Res. 19:1175–1183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian MicroRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Saito M, Shiraishi K, Matsumoto K,
Schetter A, Ogata-Kawata H, Tsuchiya N, Kunitoh H, Nokihara H,
Watanabe S, Tsuta K, et al: A three-microRNA signature predicts
responses to platinum-based doublet chemotherapy in patients with
lung adenocarcinoma. Clin Cancer Res. 20:4784–4793. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Li H, Hou S, Hu B, Liu J and Wang
J: The noncoding RNA expression profile and the effect of lncRNA
AK126698 on cisplatin resistance in non-small-cell lung cancer
cell. PLoS One. 8:e653092013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tabano S, Colapietro P, Cetin I, Grati FR,
Zanutto S, Mandò C, Antonazzo P, Pileri P, Rossella F, Larizza L,
et al: Epigenetic modulation of the IGF2/H19 imprinted domain in
human embryonic and extra-embryonic compartments and its possible
role in fetal growth restriction. Epigenetics. 5:313–324. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Byun HM, Wong HL, Birnstein EA, Wolff EM,
Liang G and Yang AS: Examination of IGF2 and H19 loss of imprinting
in bladder cancer. Cancer Res. 67:10753–10758. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui J, Mo J, Luo M, Yu Q, Zhou S, Li T,
Zhang Y and Luo W: c-Myc-activated long non-coding RNA H19
downregulates miR-107 and promotes cell cycle progression of
non-small cell lung cancer. Int J Clin Exp Pathol. 8:12400–12409.
2015.
|
|
40
|
Wang Q, Cheng N, Li X, Pan H, Li C, Ren S,
Su C, Cai W, Zhao C, Zhang L, et al: Correlation of long non-coding
RNA H19 expression with cisplatin-resistance and clinical outcome
in lung adenocarcinoma. Oncotarget. 8:2558–2567. 2017.
|
|
41
|
Hu Q, Wang YB, Zeng P, Yan GQ, Xin L and
Hu XY: Expression of long non-coding RNA (lncRNA) H19 in
immunodeficient mice induced with human colon cancer cells. Eur Rev
Med Pharmacol Sci. 20:4880–4884. 2016.PubMed/NCBI
|
|
42
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y,
Jia WH and Zhuang SM: Effects of microRNA-29 on apoptosis,
tumorigenicity, and prognosis of hepatocellular carcinoma.
Hepatology. 51:836–845. 2010.
|
|
44
|
Yan B, Guo Q, Nan XX, Wang Z, Yin Z, Yi L,
Wei YB, Gao YL, Zhou KQ and Yang JR: Micro-ribonucleic acid 29b
inhibits cell proliferation and invasion and enhances cell
apoptosis and chemotherapy effects of cisplatin via targeting of
DNMT3b and AKT3 in prostate cancer. Onco Targets Ther. 8:557–565.
2015.PubMed/NCBI
|
|
45
|
Wang H, Guan X, Tu Y, Zheng S, Long J, Li
S, Qi C, Xie X, Zhang H, Zhang Y, et al: MicroRNA-29b attenuates
non-small cell lung cancer metastasis by targeting matrix
metalloproteinase 2 and PTEN. J Exp Clin Cancer Res. 34:592015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li
W, Bi Z, Li L, Jiang Y, Luo Y, et al: Linc00511 acts as a competing
endogenous RNA to regulate VEGFA expression through sponging
hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med.
22:655–667. 2018. View Article : Google Scholar
|
|
47
|
Ko HJ and Kim YJ: Signal transducer and
activator of transcription proteins: Regulators of myeloid-derived
suppressor cell-mediated immunosuppression in cancer. Arch Pharm
Res. 39:1597–1608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Calò V, Migliavacca M, Bazan V, Macaluso
M, Buscemi M, Gebbia N and Russo A: STAT proteins: From normal
control of cellular events to tumorigenesis. J Cell Physiol.
197:157–168. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Siveen KS, Sikka S, Surana R, Dai X, Zhang
J, Kumar AP, Tan BK, Sethi G and Bishayee A: Targeting the STAT3
signaling pathway in cancer: Role of synthetic and natural
inhibitors. Biochim Biophys Acta. 1845:136–154. 2014.PubMed/NCBI
|
|
51
|
Zhang P, Sun Y and Ma L: ZEB1: At the
crossroads of epithelial- mesenchymal transition, metastasis and
therapy resistance. Cell Cycle. 14:481–487. 2015. View Article : Google Scholar :
|























