Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most common type of cancer and affects an estimated 600,000
individuals worldwide annually (1). Due to its anatomical location, HNSCC
may cause problems postoperatively, such as difficulty in speaking,
swallowing and eating. Patients with advanced HNSCC require
aggressive treatment, as HNSCCs are often associated with extensive
local invasion and frequent regional lymph node metastasis
(2). Therefore, early detection,
in-depth understanding of the characteristics of cancer cells and
accurate diagnosis are crucial for successful treatment. In
addition, comprehensive analysis of HNSCC may enable the
development of novel diagnostic aids, such as molecular biomarkers,
and provide novel therapeutic targets.
The human OLFM4 gene was first cloned from
human myeloid progenitor cells (3), and it is highly expressed in
intestinal stem cells. The expression of OLFM4 is
upregulated in gastric (4,5) and pancreatic (6) cancer, whereas it is downregulated in
prostate cancer (7), colon cancer
and leukemia (8). The OLFM4
gene encodes the secreted protein OLFM4, which is normally
expressed in the prostate gland, bone marrow, small intestine,
colon and pancreas (3,9). OLFM4 expression is associated
with the differentiation and progression of gastric cancer
(10,11) and colon adenocarcinoma (12). A previous study reported that the
induction of OLFM4 expression in cancer cells exerts an
anti-apoptotic effect and promotes proliferation of cancer cells
(13). In addition, OLFM4
promotes S phase transition and proliferation of cancer cells and
regulates tumor cell adhesion and migration (6). Moreover, OLFM4 has been considered as
a novel biomarker for gastrointestinal cancers (4,11,14).
Gene expression is regulated by genetic and
epigenetic mechanisms. The effects of epigenetic modifications on
aging, growth and development, and certain diseases, have already
been reported. Epigenetic change is a type of gene abnormality that
does not involve a change in gene sequence, but rather histone
modification and DNA methylation.
Previous studies have demonstrated that DNA
hypomethylation of specific genes is associated with several types
of cancer, including HNSCC (15-18).
In addition, promoter hypermethylation of tumor suppressor genes,
such as p16, has been observed during the early stages of
HNSCC carcinogenesis (19-22). However, only few studies to date
have investigated the interconnection between epigenetics and
HNSCC.
The present study focused on gene expression
regulation via aberrant DNA methylation in HNSCC, aiming to
elucidate the effect of DNA methylation on HNSCC carcinogenesis by
comprehensive analysis of gene expression and DNA methylation using
microarray and methylation microarray. In particular, OLFM4,
a known stemness gene, was selected to elucidate the effect of DNA
methylation on stemness gene expression in cancer stem cells (CSCs)
during carcinogenesis. The correlation between OLFM4
expression and the clinical characteristics of HNSCC is discussed
below.
Materials and methods
Tissue samples
Formalin-fixed paraffin-embedded (FFPE) specimens
were collected from 59 cases of tongue SCC (40 well-differentiated,
7 moderately differentiated and 12 poorly differentiated SCCs). All
cases were treated by resection, without chemotherapy or
radiotherapy. These 59 cases, in which the tumor size was <4 cm
and there was no metastasis (T1, and T2, N0, M0), were used for
overall survival evaluation. The tumor tissues were surgically
resected between April 1998 and March 2006 at the National Cancer
Center Hospital (Tokyo, Japan). Informed consent was obtained from
the 59 adult patients (36 male and 23 female patients, mean age
60.1 years, range 28-84 years) and the Ethics Committee of the
National Cancer Center Hospital approved the study protocol
(approval no. 2010-077). The present study is retrospective and
included the use of previously stored tissue samples.
Tissue microarray (TMA) for
immunohistochemistry (IHC) analysis
Formalin-fixed paraffin-embedded specimens from the
59 cases were collected and analyzed.
The TMA blocks were cut into 4-µm sections.
The deparaffinized sections were subjected to hematoxylin and eosin
(HE) and IHC staining. IHC was performed with anti-OLFM4 primary
antibody (1:1,000; LS-B2055; Life Span Bio Sciences, Inc.). Each
section was exposed to 0.3% hydrogen peroxide for 15 min to block
endogenous peroxidase activity. An automated stainer (Dako) was
used for staining according to the manufacturer's protocol. The
ChemMate EnVision method (Dako) was used for detection. Appropriate
positive and negative controls were used. The slides were observed
under a microscope (BX53, Olympus Corporation; magnification, x200
and x400) and evaluated with the modified OLFM4 staining score. The
percentage of immunopositive cells was divided into three scores as
follows: <30% (score 0, negative); 30-69% (score 1, positive);
and >70% (score 2, diffusely positive) (23).
RNA in situ hybridization (ISH)
ISH for OLFM4 was also performed using
RNAscope FFPE assay kit (Advanced Cell Diagnostics, Inc.) as
described previously (24). In
brief, 4-µm FFPE tissue sections were pretreated with heat
and subjected to protease digestion followed by hybridization with
OLFM4 probe (Hs-OLFM4, 311041). Subsequently, an
HRP-based signal amplification system was hybrid-ized to the bound
OLFM4 probe and color was developed with
3,3′-diaminobenzidine tetrahydrochloride (DAB). The housekeeping
gene ubiquitin C (UBC) and the bacterial gene DapB served as
positive and negative controls, respectively. Samples with UBC
signals discernible with a x10 objective lens were considered to be
adequate. The present study analyzed FFPE specimens from the 59
cases collected as described above. The methylation status in
clinical samples used for ISH and IHC was not analyzed.
Tissue samples for oligonucleotide
microarray and methylation microarray
HNSCCs and corresponding non-cancerous squamous
epithelium samples were obtained from 12 consecutive patients who
underwent surgical resection at Tokai University Hospital between
April 2006 and March 2008. The patients' age at onset ranged
between 34 and 91 years (mean, 68.5; 6 male and 6 female patients).
Tissue samples sized ~5×5 mm were collected from the tumor and
non-cancerous part of the surgical specimen prior to formalin
fixation. The samples were immediately stored in RNAlater (Thermo
Fisher Scientific, Inc.) and stored at -20° C until processed.
Histological diagnosis was made according to the WHO criteria
(25). Only primary cases were
included, whereas patients who had received radiation or
chemotherapy for HNSCC were excluded. Informed consent was obtained
from all adult patients. The present study included retrospective
use of previously stored tissue samples, and the Ethics Committee
of Tokai University School of Medicine approved all the procedures
(approval no. 16 R-183).
Oligonucleotide microarray analysis for
clinical samples
Total RNA was extracted from the 12 clinical sample
pairs using RNeasy Mini Kit (Qiagen). All labeled samples were
hybridized to the Agilent 60-mer oligo microarray with an 8×15,000
probe format (the probe was designed by the Agilent Technologies
eArray website: http://earray.chem.agilent.com/eArray/; design ID:
021445). A Gene Expression Hybridization Kit and Gene Expression
Wash Buffer Kit solutions (both from Agilent Technologies, Inc.)
were used for the hybridization and washing steps, respectively.
The housekeeping genes GAPDH, β-actin and ISGF-3 (STAT1) of 100
probe sets were used as a normalization control set. Fluorescence
intensity was calculated using Feature Extraction software, version
9.5 (Agilent Technologies, Inc.) and the data were analyzed with
GeneSpring GX software, version 11.0 (Agilent Technologies, Inc.).
mRNA expression ratios between tumor and normal tissue were
calculated as tumor/normal (T/N) mRNA expression ratio in each
gene.
Methylation microarray for clinical
samples
The changes in DNA methylation in 12 clinical sample
pairs were assessed using the Illumina Infinium assay with the
HumanMethylation450K DNA Analysis BeadChip (Illumina, Inc.). DNA
methylation levels at individual 27578CpG sites represented on the
Illumina BeadChip were determined by measuring the fraction of
methylated signal over the total signal (unmethylated + methylated
fractions) in each genomic DNA sample. The OLFM4 probe is
set at a CpG region located near the ATG start site.
To compare the DNA methylation levels of CpG sites
between tumors and controls, CpG sites with a mean methylation
difference (Δβ) of >10% were considered as differentially
methylated. DNA was extracted and purified from OCT-embedded tissue
using the QIAamp DNA Mini Kit (Qiagen), including on-column RNase
digestion (Qiagen), as per the manufacturer's protocol. Bisulfite
conversion of tissue genomic DNA (500 ng) was performed using the
EZ DNA Gold methylation kit (Zymo Research Inc. A). Normalized
M-values were generated using the R package HumMeth27KQCReport
function, including the X chromosome data and using an average
probe P-value of 0.03 as the cutoff for sample inclusion.
Individual BeadChip controls (DNA sample-dependent and
sample-independent) confirmed efficient bisulfite conversion of
DNA, hybridization specificity, base extension and target removal
for all genomic DNA samples. A complete description of these
controls is available from the manufacturer. Chromosome locations,
RefSeq and GenBank accession numbers were retrieved from the
National Center for Biotechnology Information build 36 (http://www.ncbi.nlm.nih.gov/mapview/stats/BuildStats.cgi?taxid=9606&build=36&ver=1).
Integration and validation of
oligonucleotide and methylation microarray data
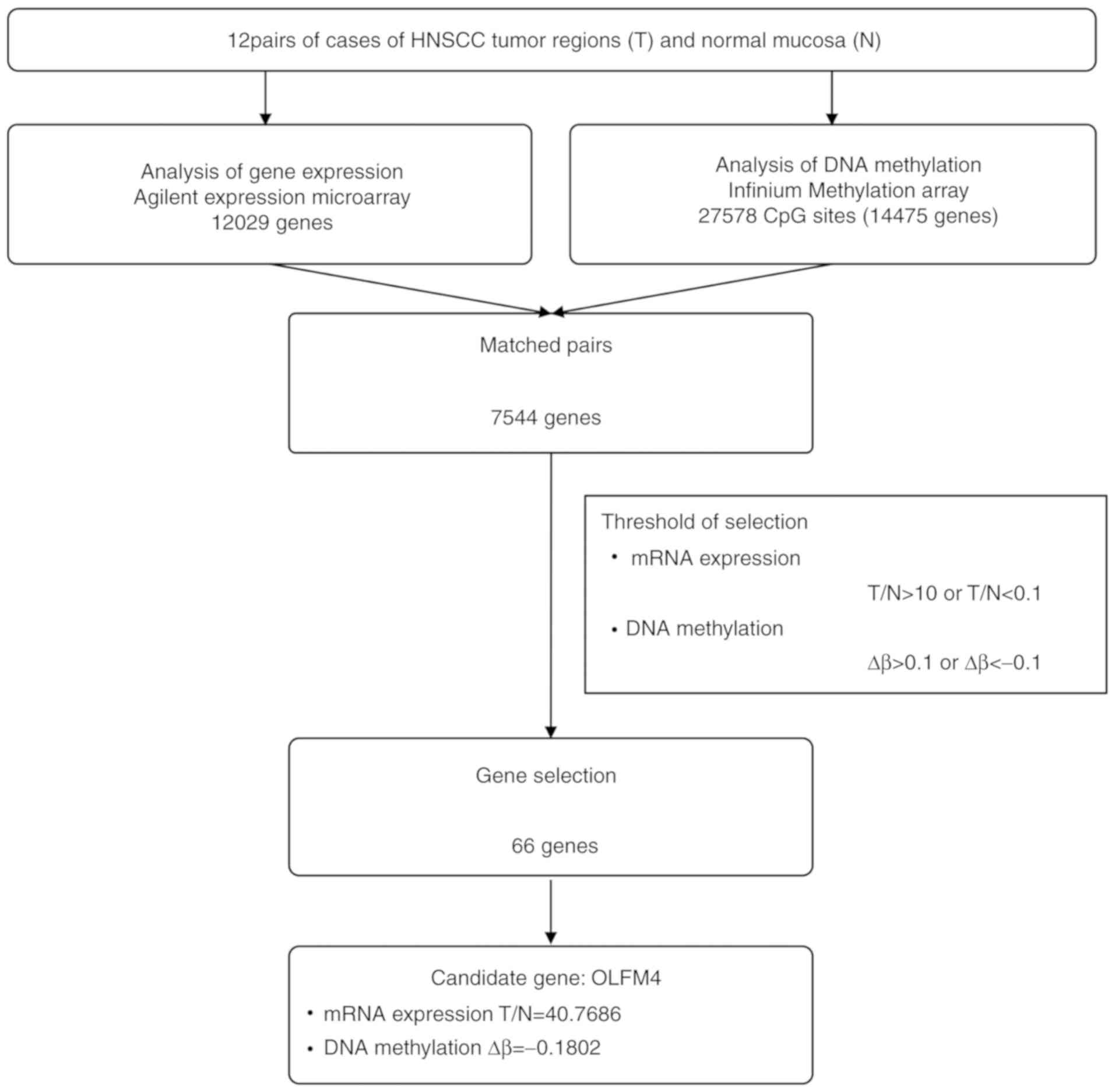
Two microarray data sets were integrated based on
each gene symbol; a total of 7,544 matched gene pairs were
identified using Microsoft Excel for Mac. For all gene pairs, T/N
and Δβ were calculated and selected according to the thresholds,
and classified into four groups as follows: Group A [T/N<0.1 and
Δβ>0.1], group B [T/N>10 and Δβ>0.1], group C [T/N>10
and Δβ<-0.1], and group D [T/N<0.1 and Δβ<-0.1]. The
UniProt protein database was used as reference for the function of
encoded proteins. The threshold for selecting candidate genes
suggested to be strongly regulated via altered DNA methylation was
set as T/N>20 and Δβ>0.15 or <-0.15.
Cell culture with 5-azacytidine
(5-aza)
An inhibitor of DNA methyltransferases, 5-aza, was
used for DNA demeth-ylation in HNSCC cell lines. FaDu/HTB-43
(26) and Detroit 562/CCL-138
(27) were obtained from the
American Type Culture Collection and cultured with 5-aza. Prior to
this analysis, 11 cell lines, including CCL-138 and HTB-43, were
cultured with 5-aza, and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis was performed to
evaluate the OLFM4 expression (data not shown). Only CCL-138
and HTB-43 cells exhibited increased expression of OLFM4
following demethylation; therefore, these two cell lines were
selected as they were considered suitable to substantiate the
correlation between DNA methylation and OLFM4
expression.
OLFM4 expression was also analyzed in a
normal fibroblast strain with/without demethylation treatment in
RT-qPCR; however, the expression of OLFM4 in fibroblasts was
extremely low and was not upregulated after demethylation
treatment. Similarly, ISH was also performed in the fibroblast
strain, and mRNA expression was very low, similar to the level in
normal tissue adjacent to the tumor. Therefore, comparison with
normal cells was not available (data not shown). Non-cancerous
cells were not considered suitable as the study control.
HTB-43 and CCL-138 cells were cultured in DMEM
(Nacalai Tesque, Inc.) with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), penicillin (100 U/ml) and streptomycin
(100 µg/ml) at 37°C in a 5% CO2 humidified
atmosphere. The cell density was adjusted to 1×106
cells/100-mm dish 24 h prior to treatment. Stock solutions of 5-aza
(Sigma-Aldrich; Merck KGaA) were dissolved in DMEM to
concentrations of 0 (negative control), 0.2 and 2 µM. Cells
were treated with 5-aza for 5 days, as previously reported
(28).
RT-qPCR analysis
Total RNA was extracted from cultured cell lines
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
and the RNeasy mini kit (Qiagen), according to the manufacturer's
instructions. Total RNA was reverse-transcribed into cDNA using the
SuperScript IV VILO Master Mix kit (Invitrogen; Thermo Fisher
Scientific, Inc.), as recommended by the manufacturer. The levels
of mRNA expression for the OLFM4 gene were analyzed using
custom TaqMan Expression Assays on the 7500 Fast Real-Time PCR
System (Thermo Fisher Scientific, Inc.) employing the relative
standard curve method. The probes and PCR primer sets employed were
TaqMan Fast Advansed Master Mix and TaqMan gene expression assays
(OLFM4 Hs00197437_m1; Applied Biosystems; Thermo Fisher
Scientific, Inc.). GAPDH served as the endogenous control.
Experiments were performed in triplicate, and the mean value for
the three experiments was used as the quantification cycle (Cq)
value. All Cq values were normalized to that of GAPDH
(GAPDH Hs0275899_g1; Applied Biosystems; Thermo Fisher
Scientific, Inc.) in the same sample. The amplification program was
according to the manufacturer's recommendations (95°C for 30 sec,
followed by 40 cycles at 95°C for 3 sec and at 60°C for 30 sec).
The data were analyzed with the 7500 system SDS (version 1.4)
software (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Oligonucleotide microarray analysis for
cell lines
Total RNA was extracted as described above.
Oligonucleotide microarray analysis was conducted using Sure Print
G3 Human Gene Expression 8×60K v3 Microarray (Agilent Technologies,
Inc.) according to the manufacturer's protocol. The slides were
scanned on the Agilent Sure Scan Microarray Scanner (G2600D;
Agilent Technologies, Inc.) using one color scan setting for 8×60k
array slides. The scanned images were analyzed with Feature
Extraction Software 11.5.1.1 (Agilent Technologies, Inc.) using
default parameters to obtain background subtracted and spatially
detrended Processed Signal intensities.
Statistical analysis
Statistical analysis was performed using SPSS
software, version 23 (IBM Corp.). The correlations between OLFM4
positivity and clinicopathological parameters were assessed using
the Chi-squared test. Between-group comparisons of the qPCR data
were performed using Kruskal-Wallis test and Bonferroni correction
as the post hoc test. Survival curves were constructed using the
Kaplan-Meier method, and the log-rank test was used to compare
groups. Survival time was calculated from the date of diagnosis to
the date of the last follow-up visit or to the date of death.
Statistical significance was set at P<0.05.
Results
Fluctuation of mRNA expression and DNA
methylation between HNSCC and normal mucosa
The mRNA expression ratio (T/N) and DNA methylation
(Δβ) of 12 clinical sample pairs were analyzed using expression
microarray and methylation microarray. The mean of the T/N and Δβ
data from the 12 samples was calculated for each gene. There were
7,544 matched gene pairs, and thresholds for the two categories
were set up as follows: T/N>10 or <0.1 and Δβ >0.1 or
<-0.1. Based on these threshold values, 66 genes were extracted
(Fig. 1) and assigned into four
groups based on the levels of T/N and Δβ as follows: Group A
[T/N<0.1 and Δβ>0.1, five genes], group B [T/N>10 and
Δβ>0.1, 22 genes], group C [T/N>10 and Δβ<-0.1, 39 genes],
and group D [T/N<0.1 and Δβ<-0.1, no genes]. Each group
included genes suggested to be regulated by methylation.
Incidentally, a small number of cancer prognostic genes were
included in these groups (representative genes are shown in
Table I; all data are included in
Table SI).
 | Table IGenes regulated by altered DNA
methylation in tumor tissue (T) compared with normal adjacent
normal mucosa (N). |
Table I
Genes regulated by altered DNA
methylation in tumor tissue (T) compared with normal adjacent
normal mucosa (N).
| Gene symbol | mRNA expression
ratio (T/N) | DNA methylation
(Δβ) | Description |
|---|
| IGLL1 | 136.4124102143 | −0.171246308 | Immunoglobulin
lambda-like polypeptide 1, transcript variant 1 |
| ACTA1 | 76.7808690487 | 0.241393352 | Actin, alpha 1,
skeletal muscle |
|
TNFRSF17 | 71.2474598502 | 0.183141233 | Tumor necrosis
factor receptor superfamily, member 17 |
| OLFM4 | 40.7686369333 | −0.180180495 | Olfactomedin 4 |
| MMP11 | 32.4300605647 | 0.159569173 | Matrix
metallopeptidase 11 (stromelysin 3) |
| ACTN2 | 30.1225359676 | 0.269785628 | Actinin, alpha
2 |
| MYH7 | 24.8015332005 | −0.186027005 | Myosin, heavy chain
7, cardiac muscle, beta |
| SOX11 | 22.5603549704 | 0.251592997 | SRY (sex
determining region Y)-box 11 |
| BST2 | 20.3898517378 | −0.348478699 | Bone marrow stromal
cell antigen 2 |
| MMP13 | 250.9824673732 | −0.111671958 | Matrix
metallopeptidase 13 (collagenase 3) |
| CXCL13 | 135.1941882092 | 0.131270067 | Chemokine (C-X-C
motif) ligand 13 (B-cell chemoattractant) |
| LHX2 | 22.183761883 | 0.110752037 | LIM homeobox 2 |
| MAGEA1 | 18.4626044 | −0.121956831 | Melanoma antigen
family A, 1 (directs expression of antigen MZ2-E) |
| DKK1 | 16.92536015 | −0.126459913 | Dickkopf homolog 1
(Xenopus laevis) |
| PIK3R5 | 14.495631 | -0.10109523 |
Phosphoinositide-3-kinase, regulatory
subunit 5 |
| CCL7 | 13.79784112 | 0.117473304 | Chemokine (C-C
motif) ligand 7 |
Next, we validated individual genes in each group.
Based on the function of the encoded protein, clustering was
performed and indicated the functional tendency in each group. In
group C, exhibiting low DNA methylation (Δβ) and high mRNA
expression (T/N), 'immune/inflammation' genes were included most
frequently (14/39 genes, respectively; 35.9% of group C).
Interestingly, this tendency is different in each group; group A
contained the most 'growth factor' members (2/5 genes, 40%), and
group B included the most 'signal' members (8/22 genes, 36.4%)
(Fig. 2, lower panel). The
methylation status in clinical samples used for ISH and IHC was not
analyzed.
Upregulation of gene expression in HNSCC
and selection of candidate genes
From these four gene groups, OLFM4 was
selected as a candidate gene regulated by DNA methylation.
OLFM4 was highly expressed, with a T/N ratio of 40.7686 and
low methylation (-18.02%), suggesting that OLFM4 was
overexpressed by promoter hypomethylation in HNSCC.
OLFM4 is a stem cell-related gene in
intestinal crypt cells (29), and
was included in group C. OLFM4 encodes a secreted protein
that is implicated in cell-cell adhesion. We hypothesized that
OLFM4 was correlated with carcinogenesis via its stemness
and function; therefore, it was selected as a candidate gene.
Demethylation treatment recovered OLFM4
expression in HNSCC cell lines
The CCL-138 and HTB-43 cell lines were cultured with
5-aza. As the clinical samples used in the microarray could not
provide the required amount for experiments, qPCR was not
performed. Therefore, experiments were conducted using cell lines
as a substitute.
RT-qPCR was performed to verify the regulation of
OLFM4 gene expression by DNA methylation. In CCL-138,
OLFM4 expression was increased by 5-aza in a
concentration-dependent manner. At 0.2 and 2 µM 5-aza, the
expression of OLFM4 was 3.49 and 9.82 times higher,
respectively, compared with the control. In HTB-43, the recovery of
OLFM4 expression was dependent on 5-aza concentration
(Fig. 3A). However, statistical
significance was only observed for CCL-138 cells (Kruskal-Wallis
test, P=0.044). In Bonferroni correction, a statistically
significant difference was only observed between 0 (negative) and 2
µM in CCL-138 cells.
Similarly, in the oligonucleotide microarray, the
two cell lines exhibited a concentration-dependent upregulation of
OLFM4. However, in this experiment, sufficient amount of
mRNA was not collected, and statistical analysis was not performed
due to the insufficient sample size (n=1, data not shown).
OLFM4 mRNA and protein expression in
tumor lesions
RNA ISH assay revealed that OLFM4 mRNA was
diffusely expressed in HNSCC cells; however, the basal layer cells
were not positive (Fig. 3B-a, HE
staining and B-b, ISH). To confirm the overexpression of the OLFM4
protein, IHC staining was performed using TMA. The OLFM4 protein
was found to be diffusely expressed in the tumor (Fig. 3B-c). The results of IHC staining
demonstrated that the OLFM4 protein was expressed in 47.5% (28/59)
of the samples. The OLFM4 protein was expressed in the cytoplasm of
tumor cells (Fig. 3B-c). The
staining scores were classified as 2+/1+/0 (negative); 2+ was
observed in only one case. The results of IHC staining and ISH
positivity and negativity were similar. However, in ISH, the result
was evaluated as + or -, as the signal tended to be weak. Scoring
by expression intensity was only performed in IHC. In this
analysis, ISH was used to confirm the expression of OLFM4,
not only at the protein but also at the mRNA level. In normal
mucosa and intratumor fibroblasts, excluding muscle fibers, both
IHC and ISH were negative (Fig. 3B-d
and B-e).
No statistically significant association between
OLFM4 protein expression and sex or clinical stage was observed. A
trend was observed between positive OLFM4 protein expression and
poor tumor differentiation; however, it was not statistically
significant (P=0.06) (Table
II).
 | Table IICharacteristics of 59 squamous cell
carcinomas based on immunostaining. |
Table II
Characteristics of 59 squamous cell
carcinomas based on immunostaining.
|
Characteristics | OLFM4 expression
| P-value |
|---|
| 2+/1+ | − |
|---|
| Number of
tumors | 28 | 31 | NA |
| Mean age (years) +
standard deviationa | 58.3+13.16 | 63.0+26.33 | NA |
| Sex | | | 0.12 |
| Female | 8 | 15 | |
| Male | 20 | 16 | |
|
Differentiation | | | 0.06 |
|
Well-differentiated (n=40) | 15 | 25 | |
| Moderately
differentiated (n=7) | 4 | 3 | |
| Poorly
differentiated (n=12) | 9 | 3 | |
| Stage | | | 0.62 |
| I | 27 | 29 | |
| II | 1 | 2 | |
The median age of the 59 patients (36 men and 23
women) who had tongue SCC <4 cm in diameter (classified as T2,
T1, and Tis, N0, M0) in the surgery alone group was 60 years
(range, 28-84 years). During a median follow-up of 2,047 days
(range, 219-3,956 days), the overall 5-year survival rate was
64.2%, with a median survival of 2,047 days [95% confidence
interval (CI): 1,720-2,372 days]. Of the 59 cases of tongue SCC, 28
(47.5%) were positive and 31 (52.5%) were negative for OLFM4
expression. χ2 and log-rank analyses revealed no
significant difference between OLFM4-positive (median survival:
1,111 days; 95% CI: 1,802-3,038 days) and -negative tumors (median
survival: 2,109 days; 95% CI: 2249-3038 days; P=0.34; Fig. 3C).
Discussion
The present study focused on the regulatory system
of gene expression via aberrant DNA methylation in HNSCC. From
integrated screening, OLFM4, which was upregulated by
promoter hypomethylation, was selected. We previously reported on
the regulatory system of stemness molecules, Bmi-1 and HMGA1, in
early-stage HNSCC (30). CSCs,
which are undifferentiated, pluripotent cells with self-renewal
ability, give rise to other malignant daughter cells and are
considered to be correlated with tumor metastasis and drug
resistance. In addition, epigenetics is deeply involved in the drug
resistance exhibited by CSCs. CSCs are resistant to conventional
chemotherapy, and can re-establish the tumor at locoregional or
distant sites, make treatment difficult. Therefore, a better
understanding of the regulatory system of CSCs may lead to the
development of new therapeutic strategies (31). The present study focused on the
correlation between DNA methylation and OLFM4, which is
known as a robust marker for human intestinal stem cells (29). OLFM4 has been widely used as
a stem cell marker for murine small intestine (32). ISH staining (29,33)
and IHC (34,35) studies revealed that
OLFM4-positive cells located at the crypt base co-expressed
Lgr5 in the small intestine. Therefore, OLFM4 was selected
as a potential marker for stemness expressed in HNSCC.
A previous study reported OLFM4 expression in
oral SCC, and IHC staining revealed overexpression of OLFM4 in 75%
of 76 HNSCC cases tested (36).
However, the regulatory mechanism of OLFM4 in HNSCC has not
been investigated. OLFM4 was confirmed to be hypomethyl-ated
and upregulated in oral SCC when compared with the surrounding
normal tissue. RT-qPCR demonstrated that demethylation treatment
induced overexpression of OLFM4. The experiment was
performed in triplicate, and statistical significance was observed
in CCL-138 cells. Although not statistically verified, the
microarray results for demethylated cell lines also demonstrated
that hypomethylation induced OLFM4 upregulation, supporting
the result of the RT-qPCR analysis. To the best of the authors'
knowledge, this is the first study to report that promoter
methylation regulates OLFM4 expression in HNSCC.
OLFM4 has been reported to be involved in
various biological processes, such as cell-to-cell interaction,
apop-tosis, migration and cell cycle regulation, in different types
of cancers (37). Aberrant
overexpression of OLFM4 has been observed in certain
cancerous lesions, particularly in cancers of the digestive tract,
including gastric (4,5), pancreatic (6) and colon cancers (38,39).
In addition, OLFM4 has been reported as a novel biomarker
for the differentiation and progression of gastrointestinal cancer
(4). Although the mechanism
regulating OLFM4 expression has only been shown in certain
types of cancer, such as gastric (40) and colorectal cancer (34), it has been revealed that the
expression of OLFM4 is associated with promoter methylation
status. However, the detailed mechanisms underlying the role of
OLFM4 in cancer remain unclear. Oue et al reported
that the expression of OLFM4 in gastric cancer tissues is observed
more frequently in stage I/II compared with stage III/IV cancers on
immunostaining. In addition, serum OLFM4 concentration in
preoperative gastric cancer patients was higher compared with that
in healthy individuals (4).
Downregulation of OLFM4 expression in advanced tumors is
associated with decreased patient survival (5). This suggests that OLFM4 not
only plays a role in the early stages of tumor initiation, but also
exerts an inhibitory effect on cancer cell invasion and metastases
in the advanced stages of tumor development (41). It has been previously reported that
the downregulation of OLFM4 expression is induced by
hypermethylation in advanced gastric cancer (41). Our results were consistent with the
findings of Guo et al (41), in that promoter methylation
regulates the expression of OLFM4. OLFM4 may exert a
cancer-promoting effect via apoptosis inhibition during the early
stages, suggesting that OLFM4 inactivation may be crucial
for tumor progression or metastasis (42). By contrast, OLFM4 was found
to be highly expressed in normal prostate tissue, moderately
expressed in benign prostatic hyperplasia tissues, and not
expressed in prostate cancer tissues, indicating that OLFM4
acted as a tumor-suppressing gene (7).
OLFM4 expression is enhanced in more highly
differentiated gastric and colon cancers, and is markedly reduced
or absent in poorly differentiated or undifferentiated cancers
(11,34). In the present study, OLFM4 tended
to be expressed more frequently in poorly differentiated tumors and
cases with poor prognosis. Although no statistical significance was
established, the cumulative overall survival rate tended to be
worse in OLFM4-positive cases. Therefore, OLFM4 cannot be
considered an independent prognostic marker for HNSCC. In addition,
no association was found between OLFM4 expression and TNM stage of
HNSCC. These results are inconsistent with those of previous
reports on other cancers. Takadate et al reported that OLFM4
expression was correlated with poor prognosis in pancreatic ductal
adenocarcinoma (PDAC) (43). In
PDAC cells, OLFM4 mRNA was highly expressed during the S
phase of the cell cycle, and the cell cycle was arrested at the S
phase by the downregulation of OLFM4 mRNA expression. This
finding demonstrated that OLFM4 promotes proliferation of
PDAC cells by favoring transition from the S to the G2/M phase.
Thus, the expression and function of OLFM4 in carcinogenesis
is organ-selective and limited by tumor size. These results
reported for PDAC are in agreement with our results, and indicate
that OLFM4 has a similar function in HNSCC.
The findings of the present study indicate that DNA
methylation reduces the expression of the stemness molecule
OLFM4, and it may affect cell heterogeneity, but does not
affect prognosis. However, DNA methylation occurs prior to
carcinogenesis in normal tissue, and OLFM4 expression would
only be involved in tumor initiation in the very early stages of
carcinogenesis, and not in progression. Thus, OLFM4
expression may not affect HNSCC prognosis. This is similar to our
previous report on Bmi1 (30),
which tends to be expressed in early and well-differentiated
cancers, and is lost in advanced/poorly differentiated HNSCCs.
Thus, it was hypothesized that Bmi1 expression arises in the
cancer-developing stage of early tumors with high plasticity. The
long half-life of the cancer 'cell of origin' allows the
accumulation of multiple mutations and epigenetic changes required
for multi-step evolution toward progression. These progressed
cancer cells exhibited decreased Bmi1 expression and gained
proliferative activity instead of loss of plasticity (30). Hence, it was concluded that Bmi1
does not affect the prognosis of HNSCC. As OLFM4 is only
expressed during the developing stage of early tumors, it may not
contribute to HNSCC prognosis. There was tendency of correlation
with poor differentiation in clinicopathological characteristics,
but was not statistically significant (P=0.06). However, we only
analyzed data from 59 cases and had no available clinical samples.
This requires further investigation by analyzing more samples.
The integrated screening analysis provided
interesting data. Some genes that are reported as a prognostic
factors in other cancers also exhibited fluctuations of T/N and Δβ
(Table I). It has been reported
that the expression of melanoma antigen encoding gene 1 (MAGEA1)
was correlated with prognosis in differentiated advanced gastric
cancer (44) and ovarian cancer
(45). Dickkopf-1 (DKK1) was also
found to be correlated with the prognosis of breast cancer
(46), laryngeal SCC (47), non-small-cell lung cancer
(48), chondrosarcoma (49),
hepatocellular carcinoma (50,51)
and cervical cancer (52).
However, the regulatory mechanism of these genes based on DNA
meth-ylation in HNSCC has not been investigated. Further studies
are required to identify their potential as novel biomarkers or
prognostic markers.
In conclusion, the aberrant stemness gene expression
caused by altered DNA methylation and its involvement in early
HNSCC characteristics was investigated, and the results revealed a
correlation with OLFM4 expression via promoter methylation
and tumor cell heterogeneity in HNSCC.
Supplementary Data
Acknowledgments
The authors would like to thank Ms. Sachiko Miura,
Ms. Toshiko Sakaguchi, Ms. Chizu Kina, Mr. Hideo Tsukamoto and Mr.
Tadayuki Sato for their skillful technical assistance.
Funding
The present study was supported in part by a
Grant-in-Aid for Scientific Research JSPS KAKENHI (grant no.
17K08710) from the Ministry of Education, Culture, Sports, Science
and Technology; partial support was received from the Research and
Study Program of Tokai University Educational System General
Research Organization.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contribution
TM conceived this project; TS and TM designed the
experiments and wrote the manuscript; TS, HY, ER KH and TM
performed the experiments and bioinformatics analysis; TS, HY, AK,
YO and TM provided clinical samples and designed the study. All
authors have read and approved the final version of this manuscript
for publication.
Ethics approval and consent to
participate
Informed consent was obtained from the patients and
the Ethics Committee of the National Cancer Center Hospital (Tokyo,
Japan) approved the procedures (approval no. 2010-077).
Patient consent for publication
Consent for publication was obtained from all
patients included in this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray and Bray F:
Cancer incidence and mortality worldwide: Sources, methods and
major patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
Patel SS, Shah KA, Shah MJ, Kothari KC and
Rawal RM: Cancer stem cells and stemness markers in oral squamous
cell carcinomas. Asian Pac J Cancer Prev. 15:8549–8556. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Liu WL, Tang DC, Chen L, Wang M,
Pack SD, Zhuang Z and Rodgers GP: Identification and
characterization of a novel member of olfactomedin-related protein
family, hGC-1, expressed during myeloid lineage development. Gene.
283:83–93. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oue N, Sentani K, Noguchi T, Ohara S,
Sakamoto N, Hayashi T, Anami K, Motoshita J, Ito M, Tanaka S, et
al: Serum olfacto-medin 4 (GW112, hGC-1) in combination with Reg IV
is a highly sensitive biomarker for gastric cancer patients. Int J
Cancer. 125:2383–2392. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo Z, Zhang Q, Zhao Z, Li B, Chen J and
Wang Y: OLFM4 is associated with lymph node metastasis and poor
prognosis in patients with gastric cancer. J Cancer Res Clin Oncol.
137:1713–1720. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kobayashi D, Koshida S, Moriai R, Tsuji N
and Watanabe N: Olfactomedin 4 promotes S-phase transition in
proliferation of pancreatic cancer cells. Cancer Sci. 98:334–340.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen L, Li H, Liu W, Zhu J, Zhao X, Wright
E, Cao L, Ding I and Rodgers GP: Olfactomedin 4 suppresses prostate
cancer cell growth and metastasis via negative interaction with
cathepsin D and SDF-1. Carcinogenesis. 32:986–994. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li H, Liu W, Chen W, Zhu J, Deng CX and
Rodgers GP: Olfactomedin 4 deficiency promotes prostate neoplastic
progression and is associated with upregulation of the
hedgehog-signaling pathway. Sci Rep. 5:169742015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulkarni NH, Karavanich CA, Atchley WR and
Anholt RR: Characterization and differential expression of a human
gene family of olfactomedin-related proteins. Genet Res. 76:41–50.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu RH, Yang MH, Xiang H, Bao LM, Yang HA,
Yue LW, Jiang X, Ang N, Wu LY and Huang Y: Depletion of OLFM4 gene
inhibits cell growth and increases sensitization to hydrogen
peroxide and tumor necrosis factor-alpha induced-apoptosis in
gastric cancer cells. J Biomed Sci. 19:382012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu W, Zhu J, Cao L and Rodgers GP:
Expression of hGC-1 is correlated with differentiation of gastric
carcinoma. Histopathology. 51:157–165. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Besson D, Pavageau AH, Valo I, Bourreau A,
Bélanger A, Eymerit-Morin C, Moulière A, Chassevent A,
Boisdron-Celle M, Morel A, et al: A quantitative proteomic approach
of the different stages of colorectal cancer establishes OLFM4 as a
new nonmet-astatic tumor marker. Mol Cell Proteomics.
10:M111.009712. 2011. View Article : Google Scholar
|
|
13
|
Zhang X, Huang Q, Yang Z, Li Y and Li CY:
GW112, a novel antiapoptotic protein that promotes tumor growth.
Cancer Res. 64:2474–2481. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mitani Y, Oue N, Matsumura S, Yoshida K,
Noguchi T, Ito M, Tanaka S, Kuniyasu H, Kamata N and Yasui W: Reg
IV is a serum biomarker for gastric cancer patients and predicts
response to 5-fluorouracil-based chemotherapy. Oncogene.
26:4383–4393. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun YW, Chen KM, Imamura Kawasawa Y,
Salzberg AC, Cooper TK, Caruso C, Aliaga C, Zhu J, Gowda K, Amin S
and El-Bayoumy K: Hypomethylated Fgf3 is a potential biomarker for
early detection of oral cancer in mice treated with the tobacco
carcinogen dibenzo[def,p]chrysene. PLoS One. 12:e01868732017.
View Article : Google Scholar
|
|
16
|
Umair A, Tarakji B, Ibrahim A, Nasser A,
Fq A and Shreen A: Quantitative study of epigenetic signature in
head and neck squamous cell carcinoma. Turk J Med Sci. 45:372–386.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Foy JP, Pickering CR, Papadimitrakopoulou
VA, Jelinek J, Lin SH, William WN Jr, Frederick MJ, Wang J, Lang W,
Feng L, et al: New DNA methylation markers and global DNA
hypomethylation are associated with oral cancer development. Cancer
Prev Res (Phila). 8:1027–1035. 2015. View Article : Google Scholar
|
|
18
|
Bakhtiar SM, Ali A and Barh D: Epigenetics
in head and neck cancer. Methods Mol Biol. 1238:751–769. 2015.
View Article : Google Scholar
|
|
19
|
Prawdzic Senkowska A, Kiczmer P,
Strzelczyk JK, Kowalski D, Krakowczyk Ł and Ostrowska Z: Impact of
HPV infection on gene expression and methylation in oral cancer
patients. J Med Microbiol. 68:440–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou C, Shen Z, Ye D, Li Q, Deng H, Liu H
and Li J: The Association and clinical significance of CDKN2A
promoter methylation in head and neck squamous cell carcinoma: A
meta-analysis. Cell Physiol Biochem. 50:868–882. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mydlarz WK, Hennessey PT, Wang H, Carvalho
AL and Califano JA: Serum biomarkers for detection of head and neck
squamous cell carcinoma. Head Neck. 38:9–14. 2016. View Article : Google Scholar
|
|
22
|
Shi H, Chen X, Lu C, Gu C, Jiang H, Meng
R, Niu X, Huang Y and Lu M: Association between P16INK4a promoter
methylation and HNSCC: A meta-analysis of 21 published studies.
PLoS One. 10:e01223022015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao J, Shu P, Duan F, Wang X, Min L, Shen
Z, Ruan Y, Qin J, Sun Y and Qin X: Loss of OLFM4 promotes tumor
migration through inducing interleukin-8 expression and predicts
lymph node metastasis in early gastric cancer. Oncogenesis.
5:e2342016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jang BG, Lee BL and Kim WH:
Olfactomedin-related proteins 4 (OLFM4) expression is involved in
early gastric carcinogenesis and of prognostic significance in
advanced gastric cancer. Virchows Arch. 467:285–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barnes L, Eveson J, Reichart P and
Sidransky D: World Health Organization classifications tumours.
Pathology and genetics of head and neck tumours Lyon: IARC;
2005
|
|
26
|
Rangan SR: A new human cell line (FaDu)
from a hypopharyngeal carcinoma. Cancer. 29:117–121. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peterson WD Jr, Stulberg CS, Swanborg NK
and Robinson AR: Glucose-6-phosphate dehydrogenase isoenzymes in
human cell cultures determined by sucrose-agar gel and cellulose
acetate zymograms. Proc Soc Exp Biol Med. 128:772–776. 1968.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Y and Hu Z: Epigenetic DNA
demethylation causes inner ear stem cell differentiation into hair
cell-like cells. Front Cell Neurosci. 10:1852016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
van der Flier LG, Haegebarth A, Stange DE,
van de Wetering M and Clevers H: OLFM4 is a robust marker for stem
cells in human intestine and marks a subset of colorectal cancer
cells. Gastroenterology. 137:15–17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamazaki H, Mori T, Yazawa M, Maeshima AM,
Matsumoto F, Yoshimoto S, Ota Y, Kaneko A, Tsuda H and Kanai Y:
Stem cell self-renewal factors Bmi1 and HMGA2 in head and neck
squamous cell carcinoma: Clues for diagnosis. Lab Invest.
93:1331–1338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Castilho RM, Squarize CH and Almeida LO:
Epigenetic modifications and Head and neck cancer: Implications for
tumor progression and resistance to therapy. Int J Mol Sci. 18:pii:
E1506. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schuijers J, van der Flier LG, van Es J
and Clevers H: Robust cre-mediated recombination in small
intestinal stem cells utilizing the olfm4 locus. Stem Cell Reports.
3:234–241. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ziskin JL, Dunlap D, Yaylaoglu M, Fodor
IK, Forrest WF, Patel R, Ge N, Hutchins GG, Pine JK, Quirke P, et
al: In situ validation of an intestinal stem cell signature in
colorectal cancer. Gut. 62:1012–1023. 2013. View Article : Google Scholar
|
|
34
|
Liu W, Liu Y, Zhu J, Wright E, Ding I and
Rodgers GP: Reduced hGC-1 protein expression is associated with
malignant progression of colon carcinoma. Clin Cancer Res.
14:1041–1049. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gersemann M, Becker S, Nuding S, Antoni L,
Ott G, Fritz P, Oue N, Yasui W, Wehkamp J and Stange EF:
Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD.
J Crohns Colitis. 6:425–434. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marimuthu A, Chavan S, Sathe G,
Sahasrabuddhe NA, Srikanth SM, Renuse S, Ahmad S, Radhakrishnan A,
Barbhuiya MA, Kumar RV, et al: Identification of head and neck
squamous cell carcinoma biomarker candidates through proteomic
analysis of cancer cell secretome. Biochim Biophys Acta.
1834:2308–2316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Grover PK, Hardingham JE and Cummins AG:
Stem cell marker olfactomedin 4: Critical appraisal of its
characteristics and role in tumorigenesis. Cancer Metastasis Rev.
29:761–775. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu W, Li H, Hong SH, Piszczek GP, Chen W
and Rodgers GP: Olfactomedin 4 deletion induces colon
adenocarcinoma in ApcMin/+ mice. Oncogene. 35:5237–5247.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sentani K, Sakamoto N, Shimamoto F, Anami
K, Oue N and Yasui W: Expression of olfactomedin 4 and claudin-18
in serrated neoplasia of the colorectum: A characteristic pattern
is associated with sessile serrated lesion. Histopathology.
62:1018–1027. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yamanoi K, Arai E, Tian Y, Takahashi Y,
Miyata S, Sasaki H, Chiwaki F, Ichikawa H, Sakamoto H, Kushima R,
et al: Epigenetic clustering of gastric carcinomas based on DNA
meth-ylation profiles at the precancerous stage: Its correlation
with tumor aggressiveness and patient outcome. Carcinogenesis.
36:509–520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo LL, He ZC, Yang CQ, Qiao PT and Yin
GL: Epigenetic silencing of olfactomedin-4 enhances gastric cancer
cell invasion via activation of focal adhesion kinase signaling.
BMB Rep. 48:630–635. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Su W, Luo L, Wu F, Lai Z, Li X, Xie Z,
Tang Z, Yang Z and Liang R: Low expression of olfactomedin 4
correlates with poor prognosis in smoking patients with non-small
cell lung cancer. Hum Pathol. 46:732–738. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Takadate T, Onogawa T, Fukuda T, Motoi F,
Suzuki T, Fujii K, Kihara M, Mikami S, Bando Y, Maeda S, et al:
Novel prognostic protein markers of resectable pancreatic cancer
identified by coupled shotgun and targeted proteomics using
formalin-fixed paraffin-embedded tissues. Int J Cancer.
132:1368–1382. 2013. View Article : Google Scholar
|
|
44
|
Ogata K, Aihara R, Mochiki E, Ogawa A,
Yanai M, Toyomasu Y, Ando H, Ohno T, Asao T and Kuwano H: Clinical
significance of melanoma antigen-encoding gene-1 (MAGE-1)
expression and its correlation with poor prognosis in
differentiated advanced gastric cancer. Ann Surg Oncol.
18:1195–1203. 2011. View Article : Google Scholar
|
|
45
|
Zhang S, Zhou X, Yu H and Yu Y: Expression
of tumor-specific antigen MAGE, GAGE and BAGE in ovarian cancer
tissues and cell lines. BMC Cancer. 10:1632010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou SJ, Zhuo SR, Yang XQ, Qin CX and Wang
ZL: Serum Dickkopf-1 expression level positively correlates with a
poor prognosis in breast cancer. Diagn Pathol. 9:1612014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shi Y, Gong HL, Zhou L, Tian J and Wang Y:
Dickkopf-1 is a novel prognostic biomarker for laryngeal squamous
cell carcinoma. Acta Otolaryngol. 134:753–759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dong LL, Qu LY, Chu LY, Zhang XH and Liu
YH: Serum level of DKK-1 and its prognostic potential in non-small
cell lung cancer. Diagn Pathol. 9:522014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen C, Zhou H, Zhang X, Ma X, Liu Z and
Liu X: Elevated levels of Dickkopf-1 are associated with β-catenin
accumulation and poor prognosis in patients with chondrosarcoma.
PLoS One. 9:e1054142014. View Article : Google Scholar
|
|
50
|
Huang Y, Yang X, Zhao F, Shen Q, Wang Z,
Lv X, Hu B, Yu B, Fan J and Qin W: Overexpression of Dickkopf-1
predicts poor prognosis for patients with hepatocellular carcinoma
after ortho-topic liver transplantation by promoting cancer
metastasis and recurrence. Med Oncol. 31:9662014. View Article : Google Scholar
|
|
51
|
Tao YM, Liu Z and Liu HL: Dickkopf-1
(DKK1) promotes invasion and metastasis of hepatocellular
carcinoma. Dig Liver Dis. 45:251–257. 2013. View Article : Google Scholar
|
|
52
|
Jiang T, Huang L and Zhang S: DKK-1 in
serum as a clinical and prognostic factor in patients with cervical
cancer. Int J Biol Markers. 28:221–225. 2013. View Article : Google Scholar : PubMed/NCBI
|