Introduction
As the most common primary intraocular malignant
tumor in adults, uveal melanoma (UM) has an incidence of 7 cases in
every million individuals (1,2). Due to its drug resistance and the high
likelihood of metastasis to other organs, UM has a poor prognosis
(3). Despite successful treatment of
the primary tumor, nearly 40% of patients succumb to metastatic
disease (4). In the clinic, the
conventional treatments for UM are chemotherapy, radiotherapy and
surgical excision, however, the efficacy of these treatments is
limited. To overcome this problem, extensive studies have been
performed with regard to immunotherapy, which have shown that this
therapy is a significant constituent of the treatment for malignant
melanomas (5,6).
Tyrosinase (TYR) is a copper-containing enzyme and a
type I membrane protein that is involved in the generation of
melanin, the main pigment in vertebrates. TYR is encoded at the Tyr
locus (formerly the albino or C locus). The protein catalyzes the
initial conversion of tyrosine to 3,4-dihydroxyphenylalanine
(DOPA), and may also catalyze the oxidation of DOPA to DOPA quinone
and 5,6-dihydroxyindole to indole-5,6-quinone (7,8). TYR is
able to catalyze the oxylation of tyrosine to DOPA quinone directly
(9–12). The TYR-related protein 1 (TYRP1) locus
encodes a 75-kDa glycoprotein that exhibits an amino acid sequence
homology to TYR of 43% (13).
Furthermore, TYRP1 has been proposed to be a second, less efficient
tyrosine hydroxylase (14),
catalyzing the generation of DOPA from tyrosine. TYRP1 is believed
to primarily stabilize and maintain the protein levels of TYR in
humans (15,16). As a marker, TYRP1 plays a crucial role
in the immunotherapy of melanoma, and it has been widely studied
for a number of years (17–19). Nonetheless, the complete mechanisms of
TYRP1 activity have yet to be illuminated.
The present study examined the expression of TYRP1
in four human UM cell lines and one retinal pigment epithelium cell
line at the mRNA, protein and morphological levels. Differential
expression of TYRP1 by these UM cells was observed, providing novel
insights with regard to TYRP1, which may be significant in further
research and may be crucial for the development of treatments for
UM.
Materials and methods
Cell lines and culture
The human UM cell lines, SP6.5, OM431, OCM1 and
OCM290, were kindly provided by Professor John F. Marshall (Tumor
Biology Laboratory, Cancer Research UK Clinical Center, John Vane
Science Centre, London, UK) (20).
The human retinal pigment epithelium (RPE) cell line was generously
provided by the Department of Ophthalmology, Ruijin Hospital,
Shanghai Jiao Tong University School of Medicine (Shanghai, China).
The UM and RPE cells were cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
All culture media were supplemented with 10% fetal bovine serum,
and all cells were incubated at 37°C in a humidified incubator with
5% CO2. The study was approved by the Animal Care and
Use Committee at Shanghai Jiaotong University School of Medicine
(Shanghai, China).
Reverse transcription-polymerase chain
reaction (RT-PCR) analysis
Total cellular RNA was extracted using the TRIzol®
Plus RNA Purification System (Gibco; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. The RNA were
reverse-transcribed using the Prime-Script 1st Strand cDNA
Synthesis kit (Takara Bio, Inc., Otsu, Japan) in a 20 µl volume
with 1 µl reverse transcriptase (MBI Fermentas, Inc.; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The PCR
reaction system (20 µl) contained 1 µg cDNA, 1 µM of each primer
and 10 µl Ex Taq solution (SYBR Premix Ex Taq™ II kit; Takara Bio,
Inc.). RT-PCR was performed on the ABI 9700 PCR machine (Applied
Biosystems; Thermo Fisher Scientific, Inc.). RT-PCR was performed
under the following conditions: 95°C for 10 min for 1 cycle,
followed by 40 cycles of denaturation at 94°C for 30 sec, annealing
at 60°C for 30 sec and extension at 72°C for 30 sec. Premier Primer
5 software (Premier Biosoft, Palo Alto, CA, USA) was used to assess
the PCR primers. The PCR primers were as follows: TYRP1 sense,
5′-GTAACAGCACCGAGGATGG-3′ and antisense, 5′-TCCAAGCACTGAGCGACAT-3′;
and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sense,
5′-TGGGGAAGGTGAAGGTCGG-3′ and antisense, 5′-CTGGAAGATGGTGATGGG-3′.
GAPDH was used as the internal control in PCR amplification. After
staining with ethidium bromide and visualization under ultraviolet
light, the amplified products were analyzed by electrophoresis on
2% agarose gels.
Quantitative (q)PCR
qPCR was performed using the aforementioned PCR
program, with SYBR Premix Ex Taq II (Takara Bio, Inc.), on a
Rotor-Gene 3000 Real-Time Thermo Cycler (Corbett Research, New
South Wales, Australia) following the manufacturer's protocols
(Takara Bio, Inc.), and the data were standardized against the
quantification cycle of the GAPDH control. The extension steps were
manipulated as follows: 95°C for 30 sec for 1 cycle, followed by
95°C for 5 sec, 60°C for 30 sec and 72°C for 15 sec for 40
cycles.
Western blot analysis
As previously described (21), western blot analysis was performed to
examine TYRP1 protein expression in the UM and RPE cells. Firstly,
the cells were harvested, washed in cold phosphate-buffered saline
(PBS) and then lysed with lysis buffer. Using the Bicinchoninic
Acid Protein assay kit (Thermo Fisher Scientific Inc.), the protein
samples were divided equally. In total, ~30 mg of protein extracts
were generated and immunoblotting was performed according to
standard protocols. Cell lysates were electrophoresed on 10%
polyacrylamide slab gels and transferred to a nitrocellulose
membrane. The membrane was incubated with the TYRP1 (mouse
anti-human monoclonal; dilution, 1:1,000; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) antibodies. An Odyssey Infrared Imaging
System (LI-COR Biosciences, Lincoln, NE, USA) was used to visualize
the signals. Monoclonal mouse TYRP1 (rabbit polyclonal; 1:1,000;
Santa Cruz Biotechnology, Inc.) and primary monoclonal mouse
anti-β-actin (1:5,000; Sigma-Aldrich, St. Louis, MO, USA) were used
as antibodies.
Immunocytochemical analysis
The collected cells were attached to glass slides,
the smears of which were then fixed by 4% paraformaldehyde for 30
min and incubated with 0.1% Triton X-100 and 5% dimethylsulfoxide
in PBS for 30 min. Subsequent to being washed 3 times with cold
PBS, the cells were subsequently blocked with 3% bovine serum
albumin (BSA; Sigma-Aldrich) at 37°C for 30 min. The cells were
incubated with TYRP1 (rabbit anti-human polyclonal; dilution,
1:1,000; catalog no., sc-25543; Santa Cruz Biotechnology, Inc.,)
antibody overnight at 4̊C, and then subjected to the secondary
antibody (goat anti-rabbit polyclonal; dilution, 1:500; catalog
no., A0545; Sigma-Aldrich) for 30 min. Following another wash in
PBS for 15 min, the cells were stained with 3,3′-diaminobenzidine
(Dako, Carpinteria, CA, USA). Nuclear counterstaining with Harris
stain was then performed for 3 min.
Immunofluorescence staining
The cells were prepared as aforementioned, and then
incubated overnight with rabbit polyclonal TYRP1 antibody at 4̊C.
Subsequently, the cells were gently rinsed 3 times for 15 min each
time in cold PBS, followed by incubation with goat anti-rabbit
immunoglobulin G secondary antibody (1:300 dilution in PBS/5% BSA;
Invitrogen; Thermo Fisher Scientific, Inc.) and
4′,6-diamidino-2-phenylindole (1:1,000 dilution in PBS/5% BSA) for
10 min at 37°C in the dark. Finally the cell smears were placed on
coverslips and images were captured under a fluorescence microscope
at 490–520 nm.
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiments. Statistical significance was
assessed using the Student's two-tailed t-test and all statistical
analyses were performed using SPSS 19.0 software (SPSS Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of TYRP1 in UM cells at the
mRNA level
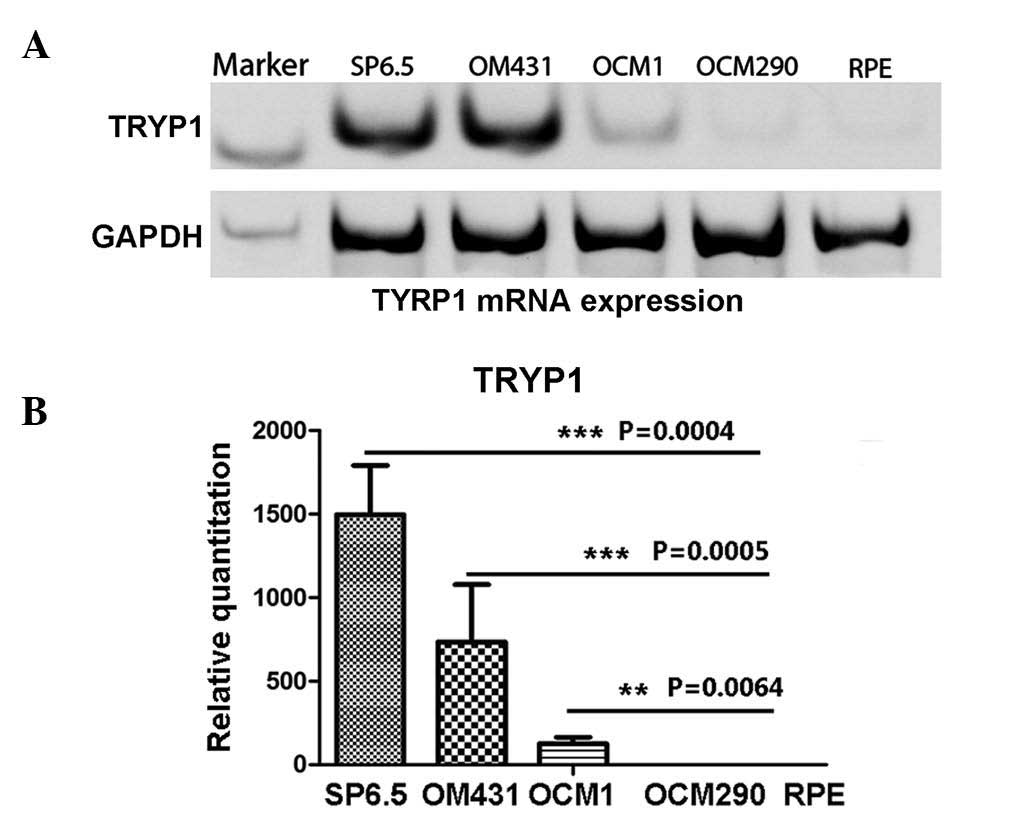
The present study first investigated the mRNA
expression of TYRP1. Experiments were performed on four human UM
cell lines (SP6.5, OM431, OCM1 and OCM290), using the human retinal
pigment epithelium cell line, RPE, as the control. The RT-PCR
results clearly showed that the control RPE cells did not express
TYRP1, while the UM cells expressed TYRP1 mRNA, with the exception
of OCM290 cells (P<0.01). SP6.5 cells expressed the highest
level of TYRP1 mRNA, while OCM1 and OM431 cells produced less in
comparison (Fig. 1A). qPCR was
performed to confirm the differences in the expression of TYRP1
mRNA in these cell lines. The results were consistent with those of
the RT-PCR, and showed that the SP6.5 cells expressed the highest
level of TYRP1 mRNA compared with the OM431 and OCM1 cells (P=0.015
and P=0.019, respectively).
Expression of TYRP1 in UM cells at the
protein level
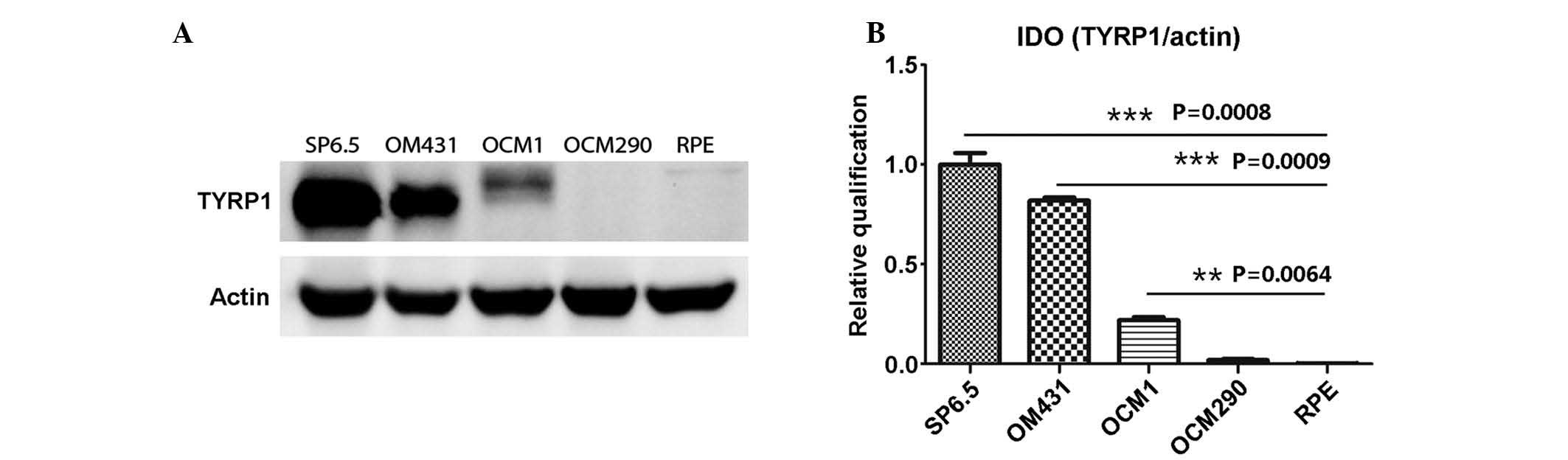
TYRP1 protein expression in the UM and RPE cells was
determined by western blot analysis. The results were consistent
with those of the PCR, and showed that the SP6.5 cells strongly
expressed TYRP1, while the OCM1 and OM431 cells expressed
relatively lower protein levels (P<0.01). In the RPE and OCM290
cells, TYRP1 protein expression was not found (Fig. 2).
Expression of TYRP1 in UM cells at the
cellular level
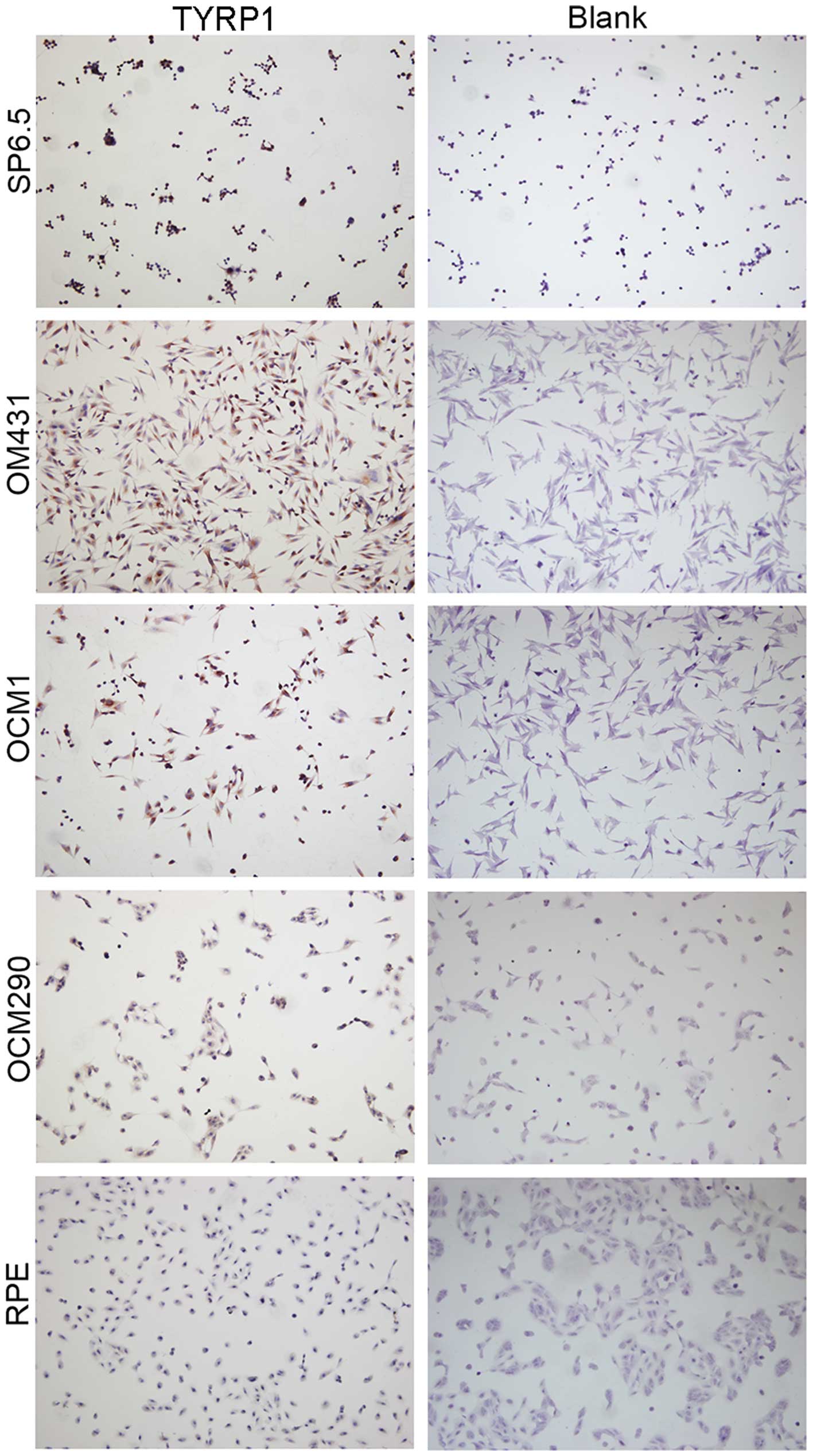
Immunocytochemistry was used to determine the
expression of TYRP1 in the UM and RPE cells. In comparison to the
blank control group, the SP6.5, OM431 and OCM1 cells were markedly
stained (P=0.005, P=0.008 and P=0.032, respectively) (Fig. 3), while no staining was observed in
the OCM290 and RPE cells. Moreover, positive staining was found in
the OCM1 and OM431 cells. The presence of TYRP1 protein expression
in these cells was confirmed by immunofluorescence staining
(Fig. 4).
Discussion
UM exhibits the highest incidence among adult
primary intraocular tumors (1,2). Treatment
strategy design for UM is associated with the two main issues of
drug resistance and a high metastatic rate. With the development of
gene research, gene therapy and immunotherapy have become more
important and their utility has also been improved. Immunotherapy
has been extensively studied, and is currently a significant
feature of the treatment regimen for malignant melanomas (5,6). Extensive
studies have been performed on UM, including studies for a series
of other proteins besides TYRP1.
In melanocytes, TYRP1 is significantly expressed and
has been identified during melanin synthesis in the melanosome.
TYRP1 is specifically expressed in melanocytes and is involved in
melanin synthesis within melanosomes, as are the other members of
the tyrosinase related protein family, which includes tyrosinase
(TYR), and dopachrome tautomerase (TYRP2). RPE cells in human
adults do not generate melanin in vitro under ordinary
culture conditions (22–26). Smith-Thomas et al reported that
TYRP1 and TYRP2 protein was not detected in cultured human RPE
cells, however, this study only used a non-quantitative
immunostaining method (23).
In the present study, it was demonstrated that the
expression of TYRP1 was markedly different in four UM cell lines.
Thus, the effects of immunotherapy mediated by the antigenicity of
TYRP1 in patients with UM can differ. These findings can clearly
lead to distinctly varied prognoses. The expression of TYRP1 in the
present study was as follows: TYRP1 mRNA expression in the OCM431
and OCM1 cells occurred at similar levels, while expression was
slightly lower in the SP6.5 cells, and almost no TYPR1 mRNA was
expressed in the OCM290 cells. The TYRP1 protein level corresponded
with the TYRP1 mRNA level. These findings represent the initial
stages of understanding TYRP1 expression in UM cells. The
functional significance and regulatory mechanisms of TYRP1 have yet
to be defined, however, findings such as aforementioned may result
in the development of immunotherapy for UM
Acknowledgements
This study was supported by the Scientific Research
Program of National Health and Family Planning Commission of China
(grant no. 201402014), The National Natural Science Foundation of
China (grant nos. 81372469 and 81372909), and The Science and
Technology Commission of Shanghai (grant nos. 13JC14006202,
12ZR1417300 and 13ZR1423600).
References
|
1
|
Stang A and Jöckel KH: Trends in the
incidence of ocular melanoma in the United States, 1974–1998.
Cancer Causes Control. 15:95–96. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singh AD, Bergman L and Seregard S: Uveal
melanoma: Epidemiologic aspects. Ophthalmol Clin North Am.
18:75–84. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Flaherty LE, Unger JM, Liu PY, Mertens WC
and Sondak VK: Metastatic melanoma from intraocular primary tumors:
The southwest oncology group experience in phase II advanced
melanoma clinical trials. Am J Clin Oncol. 21:568–572. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kujala E, Tuomaala S, Eskelin S and Kivelä
T: Mortality after uveal and conjunctival melanoma: Which tumour is
more deadly? Acta Ophthalmol. 87:149–153. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alexandrescu DT, Ichim TE, Riordan NH,
Marincola FM, Di Nardo A, Kabigting FD and Dasanu CA: Immunotherapy
for melanoma: Current status and perspectives. J Immunother.
33:570–590. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jandus C, Speiser D and Romero P: Recent
advances and hurdles in melanoma immunotherapy. Pigment Cell
Melanoma Res. 22:711–723. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tripathi RK, Hearing VJ, Urabe K, Aroca P
and Spritz RA: Mutational mapping of the catalytic activities of
human tyrosinase. J Biol Chem. 267:23707–23712. 1992.PubMed/NCBI
|
|
8
|
Korner A and Pawelek J: Mammalian
tyrosinase catalyzes three reactions in the biosynthesis of
melanin. Science. 217:1163–1165. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cooksey CJ, Garratt PJ, Land EJ, Pavel S,
Ramsden CA, Riley PA and Smit NP: Evidence of the indirect
formation of the catecholic intermediate substrate responsible for
the autoactivation kinetics of tyrosinase. J Biol Chem.
272:26226–26235. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hearing VJ and Jiménez M: Mammalian
tyrosinase-the critical regulatory control point in melanocyte
pigmentation. Int J Biochem. 19:1141–1147. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiménez M, Maloy WL and Hearing VJ:
Specific identification of an authentic clone for mammalian
tyrosinase. J Biol Chem. 264:3397–3403. 1989.PubMed/NCBI
|
|
12
|
Jimenez M, Tsukamoto K and Hearing VJ:
Tyrosinases from two different loci are expressed by normal and by
transformed melanocytes. J Biol Chem. 266:1147–1156.
1991.PubMed/NCBI
|
|
13
|
Jackson IJ: A cDNA encoding
tyrosinase-related protein maps to the brown locus in mouse. Proc
Natl Acad Sci USA. 85:4392–4396. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boissy RE, Zhao H, Oetting WS, Austin LM,
Wildenberg SC, Boissy YL, Zhao Y, Sturm RA, Hearing VJ, King RA and
Nordlund JJ: Mutation in and lack of expression of
tyrosinase-related protein-1 (TRP-1) in melanocytes from an
individual with brown oculocutaneous albinism: A new subtype of
albinism classified as 'OCA3′. Am J Hum Genet. 58:1145–1156.
1996.PubMed/NCBI
|
|
15
|
Kobayashi T, Imokawa G, Bennett DC and
Hearing VJ: Tyrosinase stabilization by TRYP1 (the brown locus
protein). J Biol Chem. 273:31801–31805. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Manga P, Sato K, Ye L, Beermann F,
Lamoreux ML and Orlow SJ: Mutational analysis of the modulation of
tyrosinase by tyrosinase-related proteins 1 and 2 in vitro. Pigment
Cell Res. 13:364–374. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Vries TJ, Trancikova D, Ruiter DJ and
van Muijen GN: High expression of immunotherapy candidate proteins
gp100, MART-1, tyrosinase and TRP-1 in uveal melanoma. Br J Cancer.
78:1156–1161. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shidham VB, Qi D, Rao RN, Acker SM, Chang
CC, Kampalath B, Dawson G, Machhi JK and Komorowski RA: Improved
immunohistochemical evaluation of micrometastases in sentinel lymph
nodes of cutaneous melanoma with ‘MCW melanoma cocktail’-a mixture
of monoclonal antibodies to MART-1, Melan-A, and tyrosinase. BMC
Cancer. 3:152003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kawakami Y, Robbins PF, Wang RF, Parkhurst
M, Kang X and Rosenberg SA: The use of melanosomal proteins in the
immunotherapy of melanoma. J Immunother. 21:237–246. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jia R, Jiao Z, Xu X, Wang J, Zhou Y, Song
X, Ge S and Fan X: Functional significance of B7-H1 expressed by
human uveal melanoma cells. Mol Med Rep. 4:163–167. 2011.PubMed/NCBI
|
|
21
|
Song X, Zhou Y, Jia R, Xu X, Wang H, Hu J,
Ge S and Fan X: Inhibition of retinoblastoma in vitro and in vivo
with conditionally replicating oncolytic adenovirus H101. Invest
Ophthalmol Vis Sci. 51:2626–2635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boulton M and Dayhaw-Barker P: The role of
the retinal pigment epithelium: Topographical variation and ageing
changes. Eye (Lond). 15:384–389. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Smith-Thomas L, Richardson P, Thody AJ,
Graham A, Palmer I, Flemming L, Parsons MA, Rennie IG and MacNeil
S: Human ocular melanocytes and retinal pigment epithelial cells
differ in their melanogenic properties in vivo and in vitro. Curr
Eye Res. 15:1079–1091. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Flood MT, Gouras P and Kjeldbye H: Growth
characteristics and ultrastructure of human retinal pigment
epithelium in vitro. Invest Ophthalmol Vis Sci. 19:1309–1320.
1980.PubMed/NCBI
|
|
25
|
Newsome DA: Retinal pigmented epithelium
culture: Current applications. Trans Ophthalmol Soc UK.
103:458–466. 1983.PubMed/NCBI
|
|
26
|
Albert DM, Ruzzo MA, McLaughlin MA,
Robinson NL, Craft JL and Epstein J: Establishment of cell lines of
uveal melanoma. Methodology and characteristics. Invest Ophthalmol
Vis Sci. 25:1284–1299. 1984.PubMed/NCBI
|