Introduction
Chronic myelogenous leukemia (CML) is a
myeloproliferative neoplasm that originates from abnormal
pluripotent bone marrow stem cells and is consistently associated
with the breakpoint cluster region/Abelson (BCR/ABL) fusion gene,
which is located in the Philadelphia (Ph) chromosome (1–4). CML is
diagnosed based on the presence of splenomegaly, increased
peripheral white blood cells and the expression of BCR-ABL.
Usually, 90–95% of CML cases in the chronic phase of disease have
the characteristic t(9;22)(q34;q11.2) reciprocal translocation that
results in the Ph chromosome (2,5). In such
cases, the BCR/ABL fusion gene is present and may be identified
using fluorescence in situ hybridization (FISH) analysis,
reverse transcription-polymerase chain reaction (RT-PCR) or
Southern blot analysis techniques (6). In Ph+ CML patients, increased
tyrosine kinase activity and the presence of the BCR/ABL chimeric
protein p210 are required for multiple pathways to confer the
leukemia phenotype (7). CML cases
with complex chromosomal aberrations that involve additional
chromosomes have been reported (8).
Imatinib, also termed Gleevec or Glivec, is a
tyrosine kinase inhibitor that inhibits the tyrosine kinase
activity of the BCR/ABL protein. Imatinib is widely used as an
initial treatment for newly-diagnosed CML patients in the chronic
phase of disease. In >95% patients, a complete hematological
response may be induced by imatinib and a complete cytogenetic
response may be induced in >75% of patients. Patients that are
treated with imatinib reported an improved quality of life
(9). However, resistance to imatinib
remains to be a challenging obstacle to achieving a better clinical
outcome. Therefore, optimized combinations of drugs are required to
be developed to improve the treatment of CML.
The present study reports the case of a CML patient
with novel complex aberrations that involved 5 chromosome
translocations, the symptoms of which were improved by treatment
with imatinib and hydroxyurea.
Case report
A 37-year-old male patient was admitted to Jining
No. 1 People's Hospital (Jining, China) on June 24, 2013, with
progressive weight loss and a cough that had lasted for two months.
The ultrasonic examination (MyLab™ClassC; Esaote China Ltd., Hong
Kong, China) revealed splenomegaly, a white blood cell (WBC) count
of 361.0×109 cells/l (normal range,
3.5–9.5×109 cells/l), 0.64% eosinophils (normal range,
0.4–8.0%) and a platelet count of 226×109 platelets/l
(normal range, 150–400×109 platelets/l). The serum
parameters of the patient were as follows: Serum lactic
dehydrogenase, 1092.0 units/l (normal range, 218.0–458.0 units/l);
γ-glutamyltranspeptadase, 61.8 units/l (normal range, 3.0–50.0
units/l); hydroxybutyrate dehydrogenase, 876.0 units/l (normal
range, 61.0–155.0 units/l); triglyceride, 2.49 mmol/l (normal
range, 0.45–1.81 mmol/l); β2-microglobulin probe 3, 82 mg/l (normal
range, 0.8–2.4 mg/l); and blood sugar, 2.23 mmol/l (normal range,
3.9–6.1 mmol/l), as measured using an chemistry analyzer (AU680,
Beckman Coulter, Inc., Brea, CA, USA).
The Giemsa (GTG)-banding technique was performed for
chromosome analysis, according to the manufacturer's protocol
(10). A total of 20 metaphases that
were obtained from the unstimulated bone marrow of the patient were
analyzed, and the karyotypes were described according to the
International System for Human Cytogenetic Nomenclature (11). Karyotyping was performed prior to the
initiation of chemotherapy treatment and the t(1;6)(p36.1;q25) and
t(9;22;11)(q34;q11.2;q11) karyotype changes were observed (Fig. 1). Amplification of the BCR-ABL gene
was performed and the results were as follows: BCR-ABL/ABL, 48.97%;
ABL gene copy, 2.63×105; and major-BCR (p210) copy,
1.29×105.
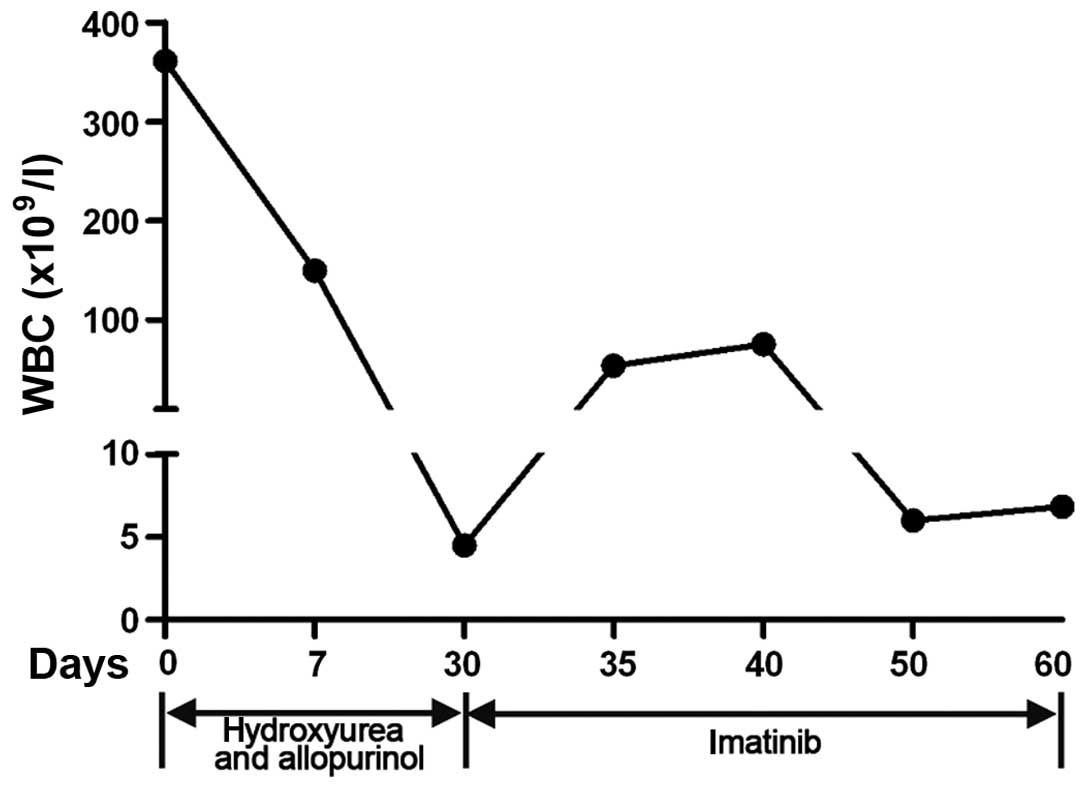
The patient was treated with hydroxyurea (3,000
mg/day) and allopurinol (3,000 mg/day). Subsequent to 1 week of
treatment, the WBC count of the patient had decreased to
149.9×109 cells/l, and the other hematological
parameters were 1.34% monocytes and 1.84% eosinophils. The
concentration of hemoglobin was 86 g/dl (normal range, 120–160
g/dl) and the platelet count was 172×109 platelets/l.
One month later, the WBC count dropped to 4.49×109
cells/l, the hemoglobin B concentration was 91 g/dl and the
platelet count was 258×109 platelets/l. In addition, the
splenomegaly became less evident. Following these improvements, 400
mg/day imatinib was administered for an additional 10 days.
Notably, the WBC count had increased to 54×109 cells/l
with 6.84% lymphocytes, 8.64% monocytes and 1.14% eosinophils 5
days subsequent to treatment with imatinib. The hemoglobin and
platelet counts were 103 g/dl and 260×109 platelets/l,
respectively. Following 10 days of treatment, the WBC count was
75.29×109 cells/l. Subsequent to the continued use of
imatinib for an additional 30 days, the WBC count and spleen
returned to normal (Fig. 2). As the
spleen had returned to normal, the patient continued treatment at
home with imitinib for 6 months. A follow-up appointment 6 months
later confirmed that the patient remained disease-free. Informed
consent was obtained from the present patient for the publication
of the present case report.
Discussion
The cytogenetic hallmark of CML, such as the Ph
chromosome and complex chromosomal rearrangements that involve
additional chromosomes, have also been described in numerous
studies (8,10–12). In
the present study, the case of a rare Ph chromosome-positive
patient with CML and a novel complex variant translocation t(1;6),
t(9;22;11) was reported. Following treatment with imatinib and
hydroxyurea, the WBC count and the condition of the spleen returned
to normal, which indicated that the combined treatment was an
effective strategy.
In total, ~5% of CML patients demonstrate the
involvement of one or more chromosomal translocations, in addition
to the Ph chromosome (5). In the
present study, the involvement of chromosomes 1, 6, 9, 11 and 22
was detected. The typical Ph chromosome demonstrating BCBCR/ABL
fusion was detected using GTG-binding and PCR. In addition,
t(1;6)(p36.1;q25) and t(9;22;11)(q34;q11.2;q11) were also detected
using GTG-binding analysis. Notably, a rare translocation of 11q11
between chromosomes 9 and 22 was indicated in the present study. To
the best of our knowledge, the translocation with t(1;6)(p36.1;q25)
and t(9;22;11)(q34;q11.2;q11) has never been described in the
literature. The formation of variant translocations that involve
various chromosomes may have prognostic importance (13).
Following the diagnosis, the present patient was
initially treated with hydroxyurea and allopurinol and then treated
with imatinib continually to maintain the improvement. The
hematological parameters of the patient were significantly improved
and the WBC count returned to normal. The signs of splenomegaly
disappeared, indicating that the combination treatment strategy was
effective.
In conclusion, the present study reported the rare
case of a Ph chromosome-positive patient with CML in the chronic
phase of disease and novel complex aberrations that involved the
t(1;6)(p36.1;q25) and t(9;22;11)(q34;q11.2;q11) translocations.
According to the outcome of treatment, hydroxyurea in combination
with imatinib is recommended for use in similar CML cases.
References
|
1
|
Melo JV and Barnes DJ: Chronic myeloid
leukaemia as a model of disease evolution in human cancer. Nat Rev
Cancer. 7:441–453. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nowell PC: The minute chromosome (Phl) in
chronic granulocytic leukemia. Blut. 8:65–66. 1962. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rowley JD: Letter: A new consistent
chromosomal abnormality in chronic myelogenous leukaemia identified
by quinacrine fluorescence and Giemsa staining. Nature.
243:290–293. 1973. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Al-Achkar W, Wafa A and Nweder MS: A
complex translocation t(5;9;22) in Philadelphia cells involving the
short arm of chromosome 5 in a case of chronic myelogenous
leukemia. J Exp Clin Cancer Res. 26:411–415. 2007.PubMed/NCBI
|
|
5
|
La Starza R, Testoni N, Lafage-Pochitaloff
M, Ruggeri D, Ottaviani E, Perla G, Martelli MF, Marynen P and
Mecucci C: Complex variant Philadelphia translocations involving
the short arm of chromosome 6 in chronic myeloid leukemia.
Haematologica. 87:143–147. 2002.PubMed/NCBI
|
|
6
|
Melo JV: The diversity of BCR-ABL fusion
proteins and their relationship to leukemia phenotype. Blood.
88:2375–2384. 1996.PubMed/NCBI
|
|
7
|
Lugo TG, Pendergast AM, Muller AJ and
Witte ON: Tyrosine kinase activity and transformation potency of
bcr-abl oncogene products. Science. 247:1079–1082. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Johansson B, Fioretos T and Mitelman F:
Cytogenetic and molecular genetic evolution of chronic myeloid
leukemia. Acta Haematol. 107:76–94. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iqbal N and Iqbal N: Imatinib: A
breakthrough of targeted therapy in cancer. Chemother Res Pract.
2014:3570272014.PubMed/NCBI
|
|
10
|
Al-Achkar W, Wafa A and Almedani S: BCR
translocation to derivative chromosome 2: A new case of chronic
myeloid leukemia with a complex variant translocation and
Philadelphia chromosome. Oncol Lett. 1:445–447. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaffer LG, McGowan-Jordan J and Schmid M:
ISCN 2013: An International System for Human Cytogenetic
Nomenclature (2013). Karger Medical and Scientific Publishers.
9–14. 2013.
|
|
12
|
Al Achkar W, Wafa A, Mkrtchyan H, Moassass
F and Liehr T: Novel complex translocation involving 5 different
chromosomes in a chronic myeloid leukemia with Philadelphia
chromosome: A case report. Mol Cytogenet. 2:212009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Al-Achkar W, Wafa A and Liehr T: A new
t(9;11;20;22)(q34;p11.2;q11.21;q11) in a Philadelphia-positive
chronic myeloid leukemia case. Oncol Lett. 5:605–608.
2013.PubMed/NCBI
|