Introduction
Renal cell carcinoma possesses the highest mortality
rate of the genitourinary cancers, and is a heterogeneous disease
that shows molecular and genetic heterogeneity and complexity
(1). Clear cell renal cell carcinoma
is the most common histological subtype of renal cell carcinoma,
accounting for ~80% of cases of renal tumors (2). Early diagnosis and medical intervention
are vital in decreasing mortality rates and promoting quality of
life. In addition, novel molecular markers for kidney cancer are
urgently required, as markers may aid the evaluation of individual
risks, the prediction of patient outcomes, prognoses and
therapeutic effects of treatment, and the promotion of personalized
treatment.
Polo-like kinase 1 (PLK1) is one of three isoforms
of PLK, the others being PLK2 and PLK3, and has been found to be
overexpressed in several human cancers (3). The serine/threonine kinase PLK1 governs
several mitotic stages, including entry into mitosis, centrosome
maturation, bipolar spindle assembly, activation of the
anaphase-promoting complex/cyclosome by phosphorylation of early
mitotic inhibitor 1, chromosome segregation and mitotic exit
(4). The overexpression of PLK1 is
associated with a poor prognosis in a variety of cancers (5–9). Beyond
the regulation of mitosis, PLK1 possesses cancer cell-specific
functions in G1/S transition and DNA replication (10,11).
Previous studies have indicated that PLK1 overexpression may not
simply be a consequence of enhanced proliferation, and that PLK1 is
likely to actively contribute to early carcinogenesis (12,13).
The mammalian transcription factor forkhead box
protein M1 (FOXM1) is important in regulating mitotic entry and the
subsequent execution of the mitotic program by controlling the
expression of a cluster of G2/M target genes (14). Previous studies have indicated that,
during the embryonic period, FOXM1 is overexpressed in all tissues
(15); however, in adulthood, FOXM1
expression only occurs in the tissues with active proliferation and
metabolism (16). In various types of
tumor cells, FOXM1 is highly expressed in the nucleus and
cytoplasm, and causes the dysfunction of regulation processes
(17–21). In particular, FOXM1 controls mitotic
entry by the periodic upregulation of a group of genes that are
maximally expressed as cells progress through the late G2 and into
the M phase (14). Fu et al
indicated that PLK1 regulates FOXM1 transcriptional activity by
direct phosphorylation, and therefore, controls the execution of
the transcriptional program required for mitotic progression
(22). The study by Fu et al
established a novel association between PLK1 and the key mitotic
transcription factor FOXM1, and provided evidence as to how PLK1
globally regulates cell division (22). However, the precise mechanism by which
PLK1-dependent phosphorylation enhances FOXM1 transcriptional
activity remains to be determined.
The present study investigated the expression of
PLK1 and FOXM1 in the clear renal cell carcinoma 769-P and ACHN
cell lines in order to examine the coordinate association between
the two proteins. The knockdown of FOXM1 or PLK1 in renal cell
cancer cell lines caused cell cycle progression to be blocked. In
addition, the involvement of FOXM1 in PLK1-regulated cell cycle
progression was also indicated.
Materials and methods
Cell lines and reagents
The renal cell cancer 769-P and ACHN cell lines were
purchased from the Chinese Academy of Sciences Cell Bank (Shanghai,
China). All cell lines were carefully maintained in a humidified
culture incubator at 37°C in 5% CO2. The cell lines were
grown in RPMI 1640 (Invitrogen; Thermo Fisher Scientific, Waltham,
MA, USA) supplemented with 10% fetal bovine serum (Invitrogen;
Thermo Fisher Scientific).
Antibodies against PLK1 (mouse anti-human monoclonal
antibody; dilution, 1:2,000; catalog no., ab17056) and FOXM1
(rabbit anti-human polyclonal antibody; dilution, 1:2,000; catalog
no., ab180710) were purchased from Abcam (Cambridge, MA, USA).
Antibodies specific for cyclin B1 (rabbit anti-human monoclonal
antibody; dilution, 1:2,000; catalog no., #12231), aurora B (rabbit
anti-human monoclonal antibody; dilution, 1:2,000; catalog no.,
#3094) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; rabbit
anti-human monoclonal antibody; dilution, 1:2,000; catalog no.,
#5174) were obtained from Cell Signaling Technology, Inc. (Danvers,
MA, USA). V-akt murine thymoma viral oncogene homolog 1 (AKT1;
mouse anti-human monoclonal antibody; dilution, 1:2,000; catalog
no., SC-55523) and ribosomal protein S6 kinase, 70 kDa, polypeptide
1 (P70S6K1; rabbit anti-human polyclonal antibody; dilution,
1:2,000; catalog no. SC-230) antibodies were obtained from Santa
Cruz Biotechnology, Inc. (Dallas, TX, USA).
Total RNA extraction, cDNA synthesis
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was prepared by using Invitrogen TRIzol
(Thermo Fisher Scientific), and 500 ng of total RNA was reverse
transcribed into cDNA using the PrimeScript™ RT Master Mix (Perfect
Real Time; Takara Biotechnology Co. Ltd., Dalian, China). RT-qPCR
was performed under the following conditions: 95°C for 15 sec and
60°C for 1 min, for 35 cycles, with an initial denaturation at 95°C
for 10 min. The expression of the housekeeping gene β-actin was
examined as a control. The primer sequences are shown in Table I. The relative expression value of the
target gene and β-actin was calculated. The experiments were
conducted in triplicate.
 | Table I.The primer sequences of polo-like
kinase 1, forkhead box protein M1 and β-actin. |
Table I.
The primer sequences of polo-like
kinase 1, forkhead box protein M1 and β-actin.
| Gene name | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| Polo-like kinase
1 |
CAAGAAGAATGAATACAGTA |
GGATATAGCCAGAAGTAA |
| Forkhead box protein
M1 |
AGCAGTCTCTTACCTTCC |
CTGGCAGTCTCTGGATAA |
| β-actin |
TAATCTTCGCCTTAATACTT |
AGCCTTCATACATCTCAA |
Western blot analysis
Frozen cells were washed twice with ice-cold
phosphate-buffered saline (PBS) and homogenized on ice in 10 vol
(wt/vol) of lysis buffer containing 20 mM Tris-HCl, 1 mM
ethylenediaminetetraacetic acid, 50 mM NaCl, 50 mM NaF, 1 mM
Na3VO4, 1% Triton-X100, 1 mM
phenylmethanesulfonyl fluoride and phosphatase inhibitor, using a
Diax 900 homogenizer (Heidolph, Schwabach, Germany). The homogenate
was centrifuged at 10,000 × g for 30 min at 4°C. The supernatant
was collected and stored at −80°C. The protein content was
determined using a bicinchoninic acid protein assay kit (Pierce
Biotechnology, Inc., Rockford, IL, USA). From each sample
preparation, 70 µg of total protein was separated using 8% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and
the bands were then transferred to polyvinylidene fluoride blot
membranes (EMD Millipore, Billerica, MA, USA). The total protein
extracts were analyzed by immunoblotting with indicated antibodies
following the SDS-PAGE analysis. Immunoblotting was performed using
mouse monoclonal primary antibodies specific for PLK1 and GAPDH and
rabbit primary antibodies specific for cyclin B1, aurora B and
FOXM1 (Abcam, Hong Kong, China). Subsequent to blocking the
non-specific binding sites with 5% bovine serum albumin (Amresco,
Solon, OH, USA) in Tris-buffered saline (pH 7.5; Amresco)
containing 0.05% Tween-20 (Amresco) (TBST), the PLK1, FOXM1, cyclin
B1, AKT1, P70S6K1, GAPDH and aurora B1 primary antibodies were
incubated on the membranes for overnight at 4°C in TBST. Following
3 washes in TBST, the membranes were incubated for 2 h at 37°C with
goat polyclonal secondary antibodies against mouse or rabbit IgG
(dilution, 1:5,000; Abcam) labeled with horseradish peroxidase. The
proteins were detected using an enhanced chemiluminescence
detection system (Pierce ECL Plus Western Blotting Substrate;
Pierce Biotechnology, Inc.), according to the manufacturer's
protocol. Specific bands for the target protein were identified
using a MBI Fermentas prestained protein molecular weight marker
(Thermo Fisher Scientific). The EC3 Imaging System (Ultra-Violet
Products, Ltd., Cambridge, UK) was used to capture images of the
specific bands, and the optical density of each band was measured
using Image J software. The ratio between the optical densities of
target proteins of the same sample was calculated as relative
content and expressed graphically.
Knockdown of PLK1 and FOXM1
Double-stranded small interfering (si)RNA oligomers
were transfected into 769-P and ACHN cells using Lipofectamine®
2000 Transfection Reagent (Thermo Fisher Scientific), according to
the manufacturer's protocol. Briefly, cells were seeded into 6-well
plates at a density of 100,000 cells per well and grown for 12 h
prior to transfection with human PLK1 siRNA,
5′-CCCUCACAGUCCUCAAUAATT-3′, or human FOXM1 siRNA,
5′-GGAAATGCTTGTGATTCAACA-3′ (Thermo Fisher Scientific), for 24 or
48 h. The Invitrogen negative siRNA control was purchased from
Thermo Fisher Scientific.
Plasmid construction and
transfection
ACHN cells were transfected with the PLK1 pEX-2-PLK1
expression plasmid (Shanghai Genepharma Co., Ltd, Shanghai, China)
using Invitrogen Lipofectamine® 2000 (Thermo Fisher Scientific).
The expression of transfected genes was confirmed using western
blot analysis.
Analysis of the cell cycle
distribution
The 769-P and ACHN cells, with or without
transfection, were plated in 25 cm2 flasks and incubated
overnight. Briefly, the attached cells were trypsinized and
collected by centrifugation (1,000 × g), washed in PBS and fixed in
cold 70% ethanol for 1.5 h at 4°C. Subsequent to fixation, the
cells were washed in PBS again and centrifuged for 5 min at 800 ×
g. The PBS was discarded and propidium iodide (BD Biosciences,
Franklin Lakes, NJ, USA) was added to a final concentration of 50
µg/ml, which was stored in the dark at 4°C for 30 min. Flow
cytometric analysis was performed on the FACSCalibur flow cytometer
(BD Biosciences, Oxford, UK). Finally, the cell cycle was analyzed
using Cell Quest software (BD Biosciences).
Statistical analysis
SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA) was
used to perform data analysis. Student's t-test was used to
analyze the data from the RT-qPCR and western blot analysis of the
cells. Groups with or without treatment were compared using one-way
analysis of variance. The Pearson product-moment correlation
coefficient was used to determine the correlation between mRNA and
protein expression. P<0.05 was considered to indicate a
statistically significant difference.
Results
Knockdown of PLK1 may induce cell
cycle arrest in human renal cell cancer cell lines
To identify the role of PLK1 in human renal cell
cancer cells, the expression of PLK1 was examined by RT-qPCR and
western blot analysis in 2 human renal cell cancer cell lines,
769-P and ACHN. The mRNA and protein expression of PLK1 was high
and stable in the 2 cell lines (Fig.
1A–C). Then, a specific effective siRNA for PLK1 was
transfected to downregulate the expression of PLK1 (Fig. 1D and E). The effect of PLK1 knockdown
on the cell cycle was then examined. The flow cytometer was used to
analyze the cell cycle. The results revealed that the 769-P and
ACHN cells transfected with PLK1 siRNA demonstrated increased
percentages of cells (61.39 and 67.17%, respectively) in the G2/M
phase. Additionally, in the mock (without any additions) and
control siRNA groups, the numbers of cells in the G2/M phase were
significantly decreased compared with the PLK1 siRNA group
(Fig. 1F).
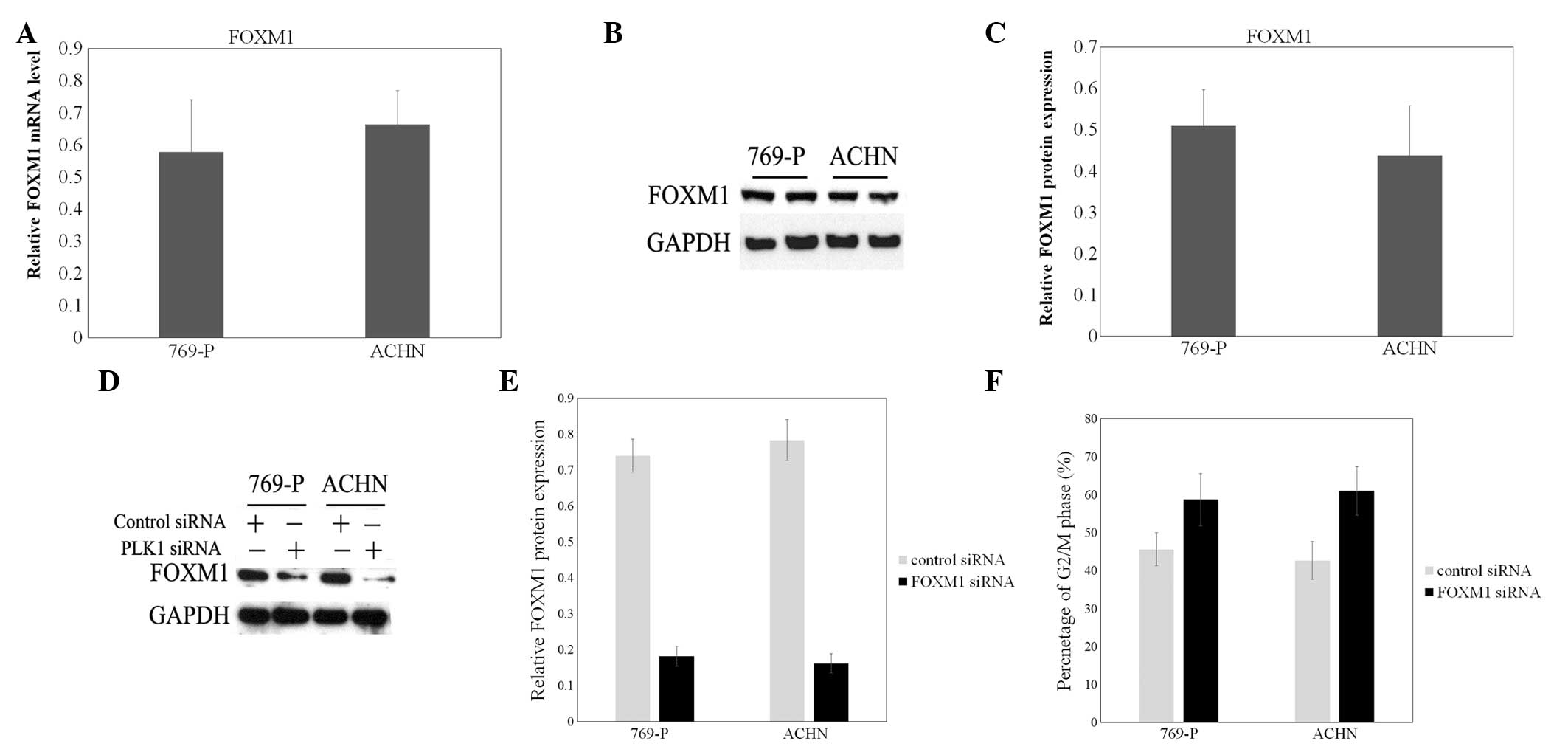
Downregulation of FOXM1 may block cell
cycle progression in human renal cell cancer cell lines
FOXM1 was previously demonstrated to be upregulated
in bladder cancer tissues and cells, compared with normal bladder
tissues and cells (23). In the
present study, FOXM1 was also highly expressed in 769-P and ACHN
cells (Fig. 2A–C). Then, a specific
effective siRNA for FOXM1 was transfected to decrease the
expression of FOXM1 (Fig. 2D and E).
To confirm the role of FOXM1 in cell division, the cell cycle
distribution of FOXM1 siRNA-treated 769-P and ACHN cells was
examined. As shown in Fig. 2F,
elevated cell numbers in the G2/M phase of the cell cycle were
indicated following FOXM1 siRNA transfection, compared with the
mock group and control siRNA group.
Expression of PLK1 and FOXM1 is
correlated in human renal cell cancer cell lines and the
suppression of PLK1 may decrease the expression of FOXM1
The aforementioned findings suggested that PLK1 and
FOXM1 were involved in cell cycle regulation in renal cell cancer
cells. Therefore, an association may exist between PLK1 and FOXM1
in cell cycle progression. To test this hypothesis, Pearson
product-moment correlation coefficient was used to determine the
correlation between the mRNA and protein expression. The results
indicated a significant positive correlation between PLK1 and FOXM1
mRNA and protein levels in 769-P and ACHN cells (Fig. 3A–D).
For additional investigation of the association
between PLK1 and FOXM1, FOXM1 expression in PLK1 siRNA-treated
769-P and ACHN cells was detected. The data demonstrated that
downregulating PLK1 may decrease the expression of FOXM1 (Fig. 3E–G). However, the knockdown of FOXM1
expression by specific siRNA did not evidently affect the
expression of PLK1 (Fig. 3H–J).
Therefore, PLK1 may regulate FOXM1, and the knockdown of PLK1 may
suppress the expression of FOXM1.
Involvement of FOXM1 in PLK1-regulated
cell cycle progression
To confirm the participation of FOXM1 in
PLK1-regulated cell cycle progression in renal cell cancer cells,
the expression of FOXM1 and associated target genes was examined
during PLK1 depletion or overexpression (Fig. 4). In PLK1-depleted ACHN cells, the
downregulation of FOXM1 and associated target genes, including
cyclin B1 and aurora B, was indicated (Fig. 4A and C). Cyclin B1 and aurora B are
required for mitotic progression. In addition, the overexpression
of wild-type PLK1 in PLK1-depleted ACHN cells was examined. The
results demonstrated that FOXM1, cyclin B1 and aurora B expression
levels in PLK1-depleted ACHN cells with overexpression of wild-type
PLK1 were restored to the level of expression in the cells without
the treatment of PLK1 siRNA (Fig. 4A and
C). Next, wild-type PLK1 was transfected into 769-P cells, and
the expression of FOXM1, cyclin B1 and aurora B was found to be
increased (Fig. 4B and D). In
addition, FOXM1 was knocked down by target siRNA in wild-type PLK1
769-P cells. The downregulation of FOXM1 induced the decreased
expression of FOXM1, cyclin B1 and aurora B (Fig. 4B and D). Overall, the data indicated
that PLK1 regulated FOXM1 and target cycle-associated genes to
control mitotic progression.
Discussion
Previously, major insights into the molecular
mechanisms of renal tumorigenesis have emerged from genetic and
molecular analyses of families with hereditary benign and malignant
kidney tumors. However, the possible heterogeneity of kidney tumors
may make the identification of specific markers more challenging. A
previous study revealed that PLK1 is overexpressed in renal cancer
and participates in the proliferation and invasion of renal cancer
cells (8). Although the evidence has
elucidated the importance of PLK1 as a tumor promoter in renal cell
carcinoma, the precise molecular mechanisms remain largely unknown.
In order to better understand the molecular mechanism of
PLK1-regulated cell proliferation, gene silencing technology was
used in the present study to knock down the expression of PLK1. The
data showed that the 769-P and ACHN cells transfected with PLK1
siRNA demonstrated an increased percentage of cells in the G2/M
phase, which supports the findings of previous studies (8,9). A
previous study reported that PLK1 was important for the
proliferation of bladder cancer cells by regulating the cancer cell
cycle between the G1/S and G2/M phases (9). Ando et al proposed that the
PLK1-mediated phosphorylation of mediator of DNA damage checkpoint
1 is involved in the regulation of G2/M transition (24). Pezuk et al indicated that PLK1
inhibition causes decreased proliferation through cell-cycle
arrest, leading to cell death in glioblastoma (25).
Previous studies have revealed that that FOXM1, a
substrate of PLK1, controls a transcriptional program that mediates
PLK1-dependent regulation of cell-cycle progression (22). Formation of the PLK1-FOXM1 complex
allows for the direct phosphorylation of FOXM1 by PLK1 at the G2/M
phase and the subsequent activation of FOXM1 activity, which is
necessary for the expression of key mitotic regulators, including
PLK1 (22). A previous study
conducted by Zhang et al revealed a working mechanism by
which PLK1 positively regulates the activity and expression levels
of FOXM1, which may greatly facilitate therapeutic interventions
that focus on targeting the PLK1- or FOXM1-mediated signaling
network (26). A study by Wang et
al indicated that FOXM1 and PLK1 were significantly associated
with certain clinicopathological indices in gallbladder cancer (GC)
(27). The evaluation of FOXM1 and
PLK1 expression may be an important factor for identifying a poor
prognosis of GC (27). FOXM1 was
previously demonstrated to be upregulated in bladder cancer tissues
and cells, compared with normal bladder tissues and cells (22). The present study demonstrated
increased cell numbers in the G2/M phase of the cell cycle
following FOXM1 siRNA transfection compared with the mock and
control siRNA groups.
To evaluate the association between PLK1 and FOXM1,
the Pearson product-moment correlation coefficient was used to
determine a significant positive correlation between PLK1 and FOXM1
in 769-P and ACHN cells. In addition, the downregulation of PLK1
was indicated to decrease the expression of FOXM1. However, the
knockdown of the FOXM1 protein by specific siRNA did not cause an
evident change in PLK1 expression levels. By contrast, a previous
study by Sharrocks et al identified that FOXM1 and the
associated PLK1 target gene are commonly overexpressed in
esophageal adenocarcinomas, and that this association may be
extended to other FOXM1 target genes, providing potentially
important biomarkers for predicting post-operative disease survival
(28). This finding may be due to the
tissue-specific presence of FOXM1.
To confirm the participation of FOXM1 in
PLK1-regulated cell cycle progression in renal cell cancer cells,
the expression of FOXM1 and its associated target genes were
assessed during PLK1 depletion or overexpression. The results
indicated that PLK1 regulated FOXM1 and the target cycle-associated
genes cyclin B1 and aurora B in order to control mitotic
progression. This finding provides an updated perspective of the
regulation of cell cycle progression by PLK1 for future
studies.
The present study identifies FOXM1 as a novel
prognostic biomarker for clear cell renal cell carcinoma, and
highlights the involvement of FOXM1 in PLK1-regulated cell cycle
progression. The results suggest that the knockdown of FOXM1 or
PLK1 in renal cell cancer cell lines caused cell cycle progression
to be blocked. Therefore, the present study indicated the
involvement of FOXM1 in PLK1-regulated cell cycle progression. This
hypothesis requires additional studies that analyze the molecular
association between the involvement of PLK1 and FOXM1 in the
development of clear renal cell carcinoma.
Acknowledgements
The present study was supported by the National
Natural Science Foundation (grant no. 81202000), Liaoning
Provincial Natural Science Foundation (grant no. 2013021066), and
Shenyang City Project of Key Laboratory (grant no.
F13-293-1-00).
References
|
1
|
Tomaszewski JJ, Uzzo RG and Smaldone MC:
Heterogeneity and renal mass biopsy: A review of its role and
reliability. Cancer Biol Med. 11:162–172. 2014.PubMed/NCBI
|
|
2
|
Farhadi A, Behzad-Behbahani A, Geramizadeh
B, Sekawi Z, Rahsaz M and Sharifzadeh S: High-risk human
papillomavirus infection in different histological subtypes of
renal cell carcinoma. J Med Virol. 86:1134–1144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van de Weerdt BC and Medema RH: Polo-like
kinases: A team in control of the division. Cell Cycle. 5:853–864.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang G, Chen Q, Zhang X, Zhang B, Zhuo X,
Liu J, Jiang Q and Zhang C: PCM1 recruits PLK1 to the
pericentriolar matrix to promote primary cilia disassembly before
mitotic entry. J Cell Sci. 126:1355–1365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stone A, Cowley MJ, Valdes-Mora F, McCloy
RA, Sergio CM, Gallego-Ortega D, Caldon CE, Ormandy CJ, Biankin AV,
Gee JM, et al: BCL-2 hypermethylation is a potential biomarker of
sensitivity to antimitotic chemotherapy in endocrine-resistant
breast cancer. Mol Cancer Ther. 12:1874–1885. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu XS, Song B, Elzey BD, Ratliff TL,
Konieczny SF, Cheng L, Ahmad N and Liu X: Polo-like kinase 1
facilitates loss of Pten tumor suppressor-induced prostate cancer
formation. J Biol Chem. 286:35795–35800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song B, Liu XS, Rice SJ, Kuang S, Elzey
BD, Konieczny SF, Ratliff TL, Hazbun T, Chiorean EG and Liu X: PLK1
phosphorylation of orc2 and hbo1 contributes to gemcitabine
resistance in pancreatic cancer. Mol Cancer Ther. 12:58–68. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang G, Zhang Z and Liu Z: Polo-like
kinase 1 is overexpressed in renal cancer and participates in the
proliferation and invasion of renal cancer cells. Tumour Biol.
34:1887–1894. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Z, Zhang G and Kong C: High
expression of polo-like kinase 1 is associated with the metastasis
and recurrence in urothelial carcinoma of bladder. Urol Oncol.
31:1222–1230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Factor VM, Seo D, Ishikawa T, Kaposi-Novak
P, Marquardt JU, Andersen JB, Conner EA and Thorgeirsson SS: Loss
of c-Met disrupts gene expression program required for G2/M
progression during liver regeneration in mice. PLoS One.
5:e127392010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu ZQ and Liu X: Role for PLK1
phosphorylation of Hbo1 in regulation of replication licensing.
Proc Natl Acad Sci USA. 105:1919–1924. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tategu M, Nakagawa H, Sasaki K, Yamauchi
R, Sekimachi S, Suita Y, Watanabe N and Yoshid K: Transcriptional
regulation of human polo-like kinases and early mitotic inhibitor.
J Genet Genomics. 35:215–224. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ito Y, Miyoshi E, Sasaki N, Kakudo K,
Yoshida H, Tomoda C, Uruno T, Takamura Y, Miya A, Kobayashi K, et
al: Polo-like kinase 1 overexpression is an early event in the
progression of papillary carcinoma. Br J Cancer. 90:414–418. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Myatt SS, Kongsema M, Man CW, Kelly DJ,
Gomes AR, Khongkow P, Karunarathna U, Zona S, Langer JK, Dunsby CW,
et al: SUMOylation inhibits FOXM1 activity and delays mitotic
transition. Oncogene. 33:4316–4329. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ustiyan V, Wang IC, Ren X, Zhang Y, Snyder
J, Xu Y, Wert SE, Lessard JL, Kalin TV and Kalinichenko VV:
Forkhead box M1 transcriptional factor is required for smooth
muscle cells during embryonic development of blood vessels and
esophagus. Dev Biol. 336:266–279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kalinichenko VV, Lim L, Shin B and Costa
RH: Differential expression of forkhead box transcription factors
following butylated hydroxytoluene lung injury. Am J Physiol Lung
Cell Mol Physiol. 280:L695–L704. 2001.PubMed/NCBI
|
|
17
|
Zhao F, Siu MK, Jiang L, Tam KF, Ngan HY,
Le XF, Wong OG, Wong ES, Gomes AR, Bella L, et al: Overexpression
of forkhead box protein M1 (FOXM1) in ovarian cancer correlates
with poor patient survival and contributes to paclitaxel
resistance. PLoS One. 9:e1134782014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gormally MV, Dexheimer TS, Marsico G,
Sanders DA, Lowe C, Matak-Vinković D, Michael S, Jadhav A, Rai G,
Maloney DJ, et al: Suppression of the FOXM1 transcriptional
programme via novel small molecule inhibition. Nat Commun.
5:51652014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang L, Wang P and Chen H: Overexpression
of FOXM1 is associated with metastases of nasopharyngeal carcinoma.
Ups J Med Sci. 119:324–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li XR, Chu HJ, Lv T, Wang L, Kong SF and
Dai SZ: miR-342-3p suppresses proliferation, migration and invasion
by targeting FOXM1 in human cervical cancer. FEBS Lett.
588:3298–3307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ning Z, Wang A, Liang J, Xie Y, Liu J,
Feng L, Yan Q and Wang Z: USP22 promotes the G1/S phase transition
by upregulating FOXM1 expression via β-catenin nuclear localization
and is associated with poor prognosis in stage II pancreatic ductal
adenocarcinoma. Int J Oncol. 45:1594–1608. 2014.PubMed/NCBI
|
|
22
|
Fu Z, Malureanu L, Huang J, Wang W, Li H,
van Deursen JM, Tindall DJ and Chen J: Plk1-dependent
phosphorylation of FOXM1 regulates a transcriptional programme
required for mitotic progression. Nat Cell Biol. 10:1076–1082.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu D, Zhang Z and Kong CZ: High FOXM1
expression was associated with bladder carcinogenesis. Tumour Biol.
34:1131–1138. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ando K, Ozaki T, Hirota T and Nakagawara
A: NFBD1/MDC1 is phosphorylated by PLK1 and controls G2/M
transition through the regulation of a TOPOIIα-mediated
decatenation checkpoint. PLoS One. 8:e827442013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pezuk JA, Brassesco MS, Morales AG, de
Oliveira JC, de Paula Queiroz RG, Machado HR, Carlotti CG Jr, Neder
L, Scrideli CA and Tone LG: Polo-like kinase 1 inhibition causes
decreased proliferation by cell cycle arrest, leading to cell death
in glioblastoma. Cancer Gene Ther. 20:499–506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang J, Yuan C, Wu J, Elsayed Z and Fu Z:
Polo-like kinase 1-mediated phosphorylation of Forkhead box protein
M1b antagonizes its SUMOylation and facilitates its mitotic
function. J Biol Chem. 290:3708–3719. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang R, Song Y, Xu X, Wu Q and Liu C: The
expression of Nek7, FoxM1, and PLK1 in gallbladder cancer and their
relationships to clinicopathologic features and survival. Clin
Transl Oncol. 15:626–632. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dibb M, Han N, Choudhury J, Hayes S,
Valentine H, West C, Ang YS and Sharrocks AD: The FOXM1-PLK1 axis
is commonly upregulated in oesophageal adenocarcinoma. Br J Cancer.
107:1766–1775. 2012. View Article : Google Scholar : PubMed/NCBI
|