Introduction
Hypoxia inducible factors (HIFs) belong to the
family of helix-loop-helix-PAS domain transcription factors
(1,2).
It has been demonstrated that there are ~150 HIF target genes
(3). HIFs accelerate tumor
progression and cell survival by regulating a wide variety genes
that control various metabolic processes, including anaerobic
metabolism (glucose transporter 1), angiogenesis (vascular
endothelial growth factor), regulation of cell cycle and
intracellular pH (carbonic anhydrase-9), response to DNA damage,
alteration of the extracellular matrix and cell adhesion,
migration, proliferation and apoptosis [p21, p27, matrix
metalloproteinase (MMP)-2 and 9] (4–7). The HIF
pathway in hypoxia is an important therapeutic target for reducing
the size, metastatic potential and therapeutic resistance of the
primary tumor (8).
There are three isoforms of the HIF-α subunit:
HIF-1α, HIF-2α and HIF-3α. HIF-2 is dimerized by the HIF-1β subunit
and HIF-2α subunit, and the stability and transcriptional activity
of HIF-2α is accommodated by oxygen-dependent hydroxylation. In
normoxic conditions, the α subunit is constitutively expressed but
rapidly degraded. In a low-oxygen environment, the α subunit is
stabilized and translocated to the nucleus (9,10). HIF-2α
is regulated by fewer genes compared with HIF-1α; in breast
adenocarcinoma MCF-7 cells, there are only a small group of
hypoxia-associated genes that are associated with HIF-2α, while 80%
of hypoxia-regulated genes are associated with HIF-1α, including
vascular endothelial growth factor, erythropoietin and matrix
metalloproteinases (11,12). Previous studies have confirmed that
HIF-1 is associated with tumor progression in certain carcinomas,
including breast, non-small cell lung and uterine cancer, and
patients with high levels of HIF-1 have a poor response to cancer
therapies (13–19). However, little is understood
concerning the effect of HIF-2α in solid tumors. Previous studies
have demonstrated that a cell's reaction to hypoxia is primarily
regulated by HIF-1α in all cells, including breast carcinoma cells,
but is regulated by HIF-2α in gastrointestinal epithelium, heart,
kidney, and renal carcinoma cells (20).
RNA interference (RNAi) is a powerful mechanism for
targeting post-transcriptional gene silencing, in which
double-stranded RNA is successfully introduced into mammalian
cells, which downregulates the expression of target genes or
suppresses the replication and transcription of pathogens by
degrading the homologous mRNA sequences (21). In the present study, synthesized small
interfering (si) RNAs targeting the HIF-2α gene was transfected
into breast adenocarcinoma MCF-7 cells using
Lipofectamine® 2000 to knockdown the expression of the
HIF-2α on a protein and mRNA level. Results from studies regarding
the effect and impact of HIF-2α gene silencing on the cell growth
and invasion potency may contribute to additional research on the
HIF complex and its possible therapeutic applications. The present
study investigated the correlation between HIF-2α and MMP-2
expression, and the significant role of HIF-2α in breast carcinoma
cell survival and invasion.
Materials and methods
Cell lines and hypoxia treatment
The breast adenocarcinoma MCF-7 cell line was
purchased from the Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China). The cells were cultured in RPMI-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal calf serum (Invitrogen; Thermo
Fisher Scientific, Inc.), streptomycin (100 U/ml; Thermo Fisher
Scientific, Inc.) and penicillin (100 U/ml; Thermo Fisher
Scientific, Inc.) at 37°C in 5% CO2. When the cells had
reached 80% confluence, they were cultured with hypoxia-mimetic
agent, cobalt chloride (CoCl2; Sigma-Aldrich, St. Louis,
MO, USA) at various concentrations (50, 100 and 200 µmol/l) for at
37°C in 5% CO2 for 24 h. A maximum HIF-2α accumulation
was reached at 100 µmol/l CoCl2; therefore, 100 µmol/l
CoCl2 was selected for use as the standard concentration
for subsequent experiments (Fig.
1).
siRNAs and transfection
The HIF-2α siRNAs were designed according to the
study by Meade et al (22) and
were synthesized by Guangzhou Ruibo Biological Technology Co., Ltd.
(Guangzhou, China) as follows: siRNA-1, sense
5′-GCAAAUGUACCCAAUGAUADTDT-3′ and antisense
5′-UAUCAUUGGGUACAUUUGCDTDT-3′; siRNA-2, sense
5′-CAGCAUCUUUGAUAGCAGUDTDT-3′ and antisense
5′-ACUGCUAUCAAAGAUGCUGDTDT-3′; negative control siRNA, sense
5′-CAGCAGGGUUGAUAGCAUGDTDT-3′ and antisense
5′-ACUGCCCCCAAAGAUGCUGDTDT-3æ. The MCF-7 cells were transfected
with the siRNAs. Briefly, the cells were seeded into 24-well plates
and cultured in RPMI-1640 medium for 24 h (at 37°C in 5%
CO2) to reach 50–70% confluence. HIF-2α siRNA or the
negative control siRNA were transfected into the cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The cells were placed in a normoxic or hypoxic
atmosphere for 24 h following transfection. The experimental groups
were as follows: Parental cell group, MCF-7 cells without
CoCl2 treatment; control group, MCF-7 cells treated with
100 µmol/l CoCl2; hypoxia + RNAi 1 group, HIF-2α
siRNA1-transfected MCF-7 cells treated with 100 µmol/l
CoCl2; hypoxia + RNAi 2 group, HIF-2α siRNA2-transfected
MCF-7 cells treated with 100 µmol/l CoCl2.
Reverse transcription polymerase chain
reaction (PCR)
Total RNA was extracted from the cells using TRIzol
reagent (Gibco®; Thermo Fisher Scientific, Inc.). The
cDNA was synthesized from the RNA using the Reverse Transcription
System (Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. The expressions of HIF-2α and MMP-2 were
detected using the following primers: HIF-2α, forward
5′-TGAAAACAGAGTCCGAAGCC-3′ and reverse 5′-GTGGCTGACTTGAGGTTGA-3′;
MMP-2, forward 5′-TTCAAGGACCGGTTCATTTGGCGGACTGTG-3′ and reverse
5′-TTCCAAACTTCACGCTCTTCAGACTTTGGTT-3′. The following
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were also
used: forward, 5′-ATTCATCTCTCCTCTCCCA-3′ and reverse,
5′-GTTGGTGGTTGGTACTGT-3′. Primers were designed using the Primer
Premier Software version 5 (Premier Biosoft International, Palo
Alto, CA, USA) and synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.). PCR was performed using the SYBR Premix Ex Taq
II kit (Takara Biotechnology Co., Ltd., Dalian, China) and the ABI
9700 PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.), and under the following conditions: 94°C for 45 sec and 55°C
for 1 min for 35 cycles, with an initial denaturation step at 72°C
for 10 min. The PCR products were subjected to 1.5% agarose gel
electrophoresis, with GAPDH (580 bp) as an internal control. PCR
products were quantified with a TotalLab Quant Phoretix 1D Pro
software software (TotalLab Ltd., Newcastle Upon Tyne, UK).
Western blotting analysis
Protein was extracted from the cells using NP-40
Lysis Buffer (Beyotime Institute of Biotechnology, Haimen, China).
Protein concentration was determined using a bicinchoninic acid
assay (Beyotime Institute of Biotechnology), according to the
manufacturer's protocol. Total protein of breast carcinoma cells
was separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis. Proteins were displaced to polyvinylidene
difluoride (PVDF) membranes by Pharmacia Phast gel electrophoresis
system (Roche Diagnostics, Indianapolis, IN, USA). The PVDF
membrane was blocked with 5% skim milk blocking buffer (Beyotime
Institute of Biotechnology) for 1 h. The immunoblots were incubated
with the following primary antibodies: Rabbit polyclonal
anti-HIF-2α (dilution, 1:150; catalog no., ab73895; Abcam,
Cambridge, UK); mouse monoclonal anti-MMP-2 (dilution, 1:500;
catalog no., ab3158; Abcam); and mouse monoclonal anti-β-actin
(dilution, 1:500; catalog no., ab6276; Abcam). Subsequently, the
membranes were incubated at 4°C for 24 h with gentle agitation.
After washing twice with phosphate-buffered saline, the membranes
were incubated with horseradish peroxidase-conjugated goat
anti-rabbit (dilution, 1:1,000; catalog no., BA1054-1; BosterBio,
Wuhan, China) and goat anti-mouse (dilution, 1:1,000; catalog no.,
BA1051; BosterBio) IgG secondary antibodies for 1 h at room
temperature. After washing with PBS, the immuno-blotting signal was
detected using a chemiluminescence system (ECL; Thermo Fisher
Scientific, Inc.). The results were quantified using TotalLab
version 2.0 software (Total Lab, Newcastle upon Tyne, UK). β-actin
protein was used as an internal control.
Invasion assays
The invasion ability of the cells was detected using
a Boyden chamber (BD Biosciences, Franklin Lakes, NJ, USA. For
invasion assays, polycarbonate filters with 8-µm pore size (EMD
Millipore, Billerica, MA, USA) were covered with Matrigel
(Sigma-Aldrich), which was diluted to 1:20 with serum-free
RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.). The
polycarbonate filters were dried at room temperature for 24 h. In
total, ~5×106 cells in 500 µl RPMI-1640 media were
placed in the upper chamber and 400 µl RPMI-1640 medium was placed
in the lower chamber. The cells were incubated at 37°C in 5%
CO2 for 24 h. The cells on the bottom surface of the
polycarbonate filter were removed and fixed in 4% glutaraldehyde
solution and stained with hematoxylin and eosin (Enzyme-linked
Biotechnology Co., Ltd., Shanghai, China). Invasive cells were
counted using a microscope (magnification, ×400; BX51; Olympus
Corp., Tokyo, Japan). The invasion index and the cell invasion
inhibition rate were calculated as previously described (23). Briefly, the invasion index was defined
as the number of cells that migrated through the 8-µm pores of the
filter in the experimental group divided by the number of cells
that migrated through the filter in the control group × 100. The
cell invasion inhibition rate was equal to the number of cells that
migrated through the filter in the control group minus the number
of cells that migrated through the filter in the experimental group
divided by the number of cells that migrated through the filter in
the control group × 100.
Statistical analysis
The data are presented as the mean ± standard
deviation. Experiments were performed in triplicate. All
statistical analyses were performed using SPSS version 13.0
software (SPSS, Inc., Chicago, IL, USA). Independent sample t-test
was used to determine the statistical significance between the
means. P<0.05 was considered to indicate a statistically
significant difference.
Results
Hypoxia mimetic CoCl2
specifically elevated HIF-2α mRNA levels
To investigate whether the expression of HIF-2α is
regulated by a hypoxic environment, the mRNA levels of HIF-2α were
compared in cells that were cultured under normoxic (20%
O2) and hypoxia-mimetic (CoCl2) conditions.
After a 24 h incubation, HIF-2α mRNA levels clearly increased in
the cells that were treated with CoCl2 (100 µmol/l). It
has been established that CoCl2 treatment can prevent
oxygen signaling in cells, leading to chemical hypoxia. The results
revealed that the hypoxic effect increased in severity as
CoCl2 dose increased from 50 to 100 µmol/l. This
indicates that CoCl2 induces hypoxia in a dose-dependent
manner. In addition, the expression of HIF-2α also increased in a
dose-dependent manner. Notably, the toxicity of CoCl2
increased in a dose-dependent manner and thus at high doses this
led to severe hypoxia and subsequent cell death. As a result, the
expression of HIF-2α was decreased. By contrast, normoxic
conditions (20% O2) had only a slight effect on HIF-2α
transcription (Fig. 1). De
novo transcription of GAPDH was similar among treatment groups.
These results suggest that exposure of CoCl2 (100
µmol/l) promoted HIF-2α transcription, evidenced by the increase in
HIF-2α mRNA levels. A CoCl2 concentration of 100 µmol/l
was used in all subsequent experiments.
Effects of HIF-2α knockdown on the
expression of MMP-2 in MCF-7 cells
The association between HIF-2α expression and its
target gene MMP-2 was analyzed in MCF-7 cells using siRNAs against
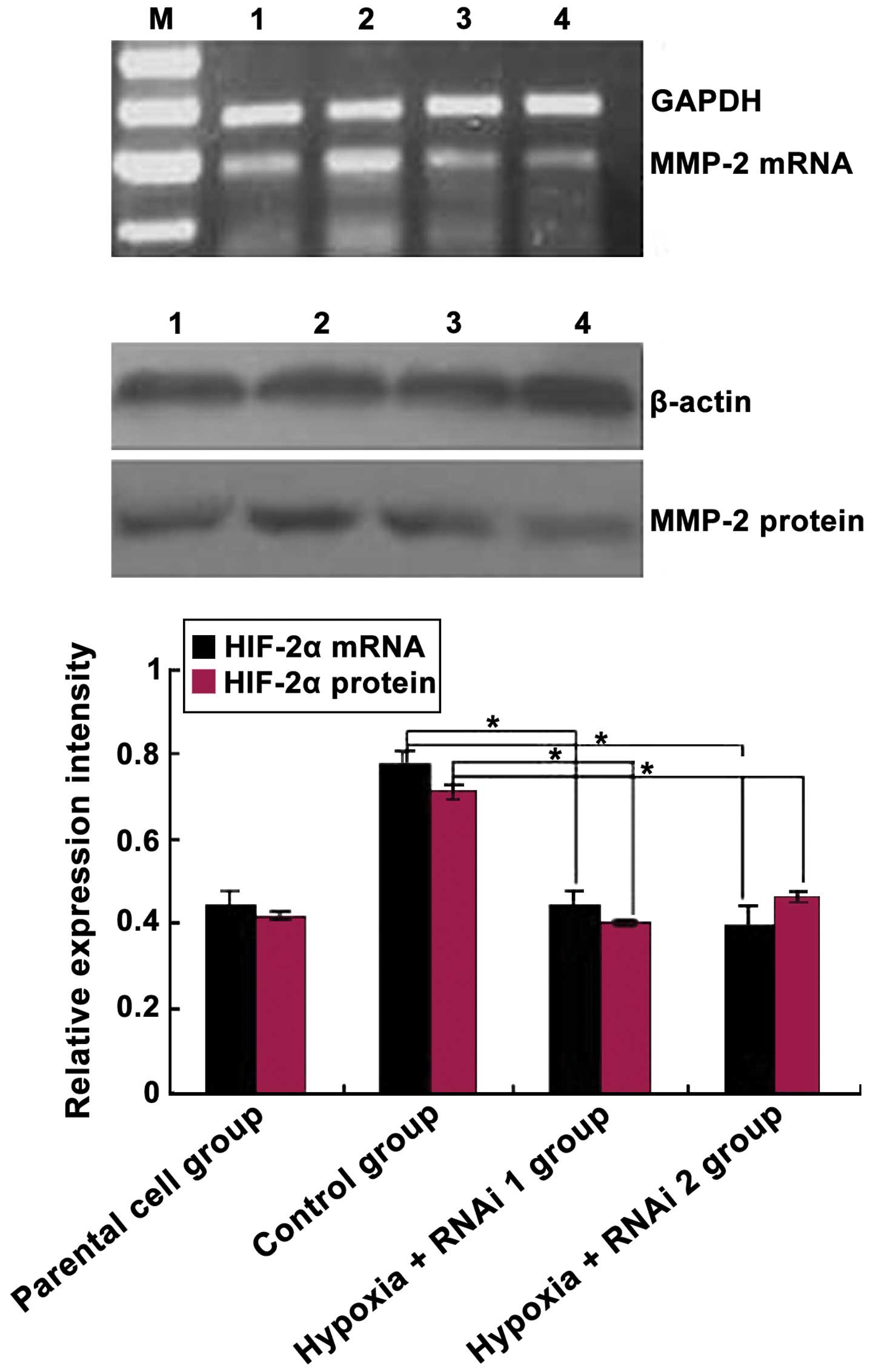
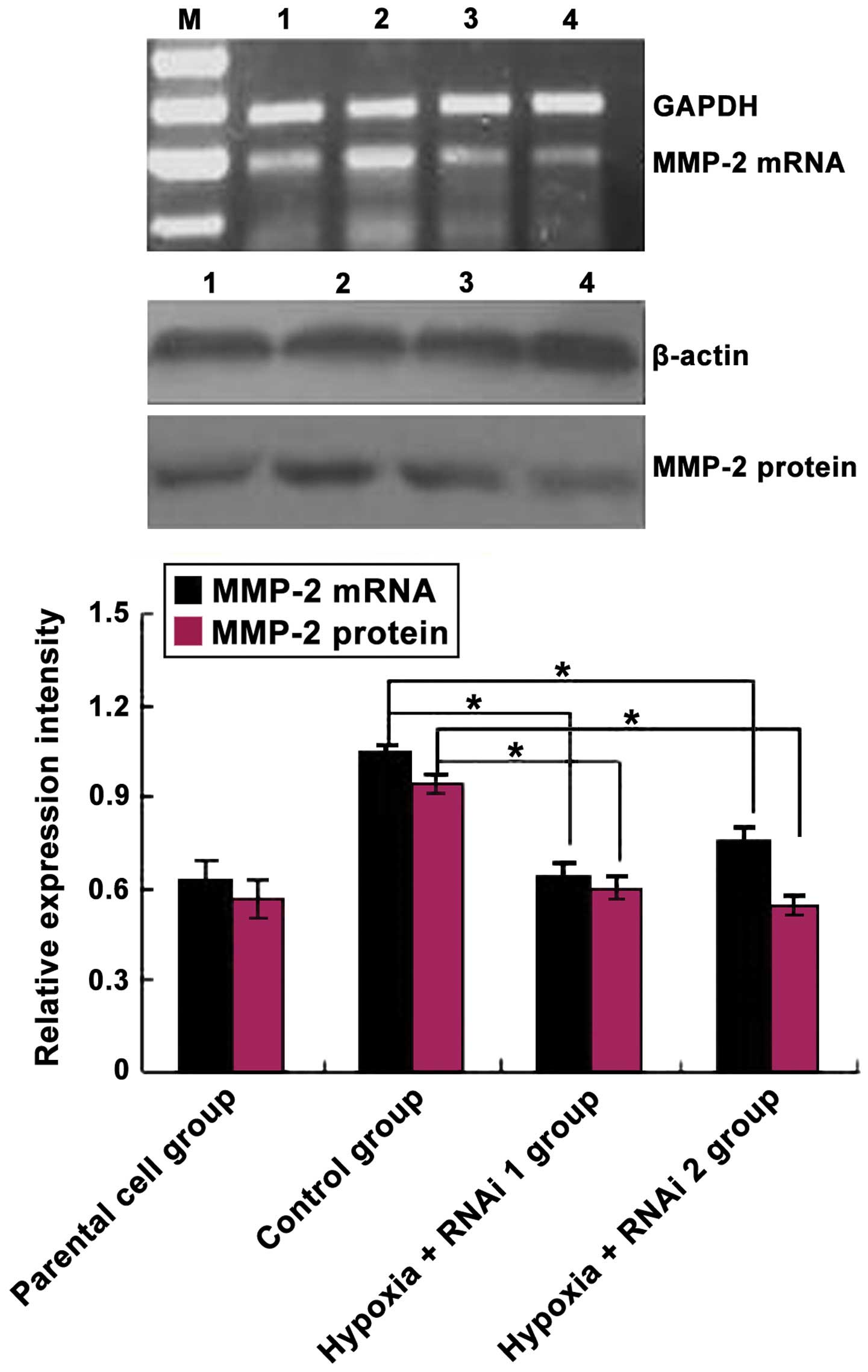
HIF-2α. As expected, HIF-2α siRNA markedly reduced the mRNA and
protein levels of the targeted HIF-2α subunit (Fig. 2). Subsequently, the effects of HIF-2α
knockdown on CoCl2-mediated induction of MMP-2
expression was analyzed. As shown in Fig.
3, MMP-2 protein and mRNA levels were downregulated
significantly in MCF-7 cells transfected with HIF-2α siRNA compared
with the control group (P=0.033). Overall, these data demonstrated
that hypoxia induces HIF-2α expression at mRNA and protein levels
in MCF-7 cells, and HIF-2α siRNA inhibited MMP-2 expression in
MCF-7 cells.
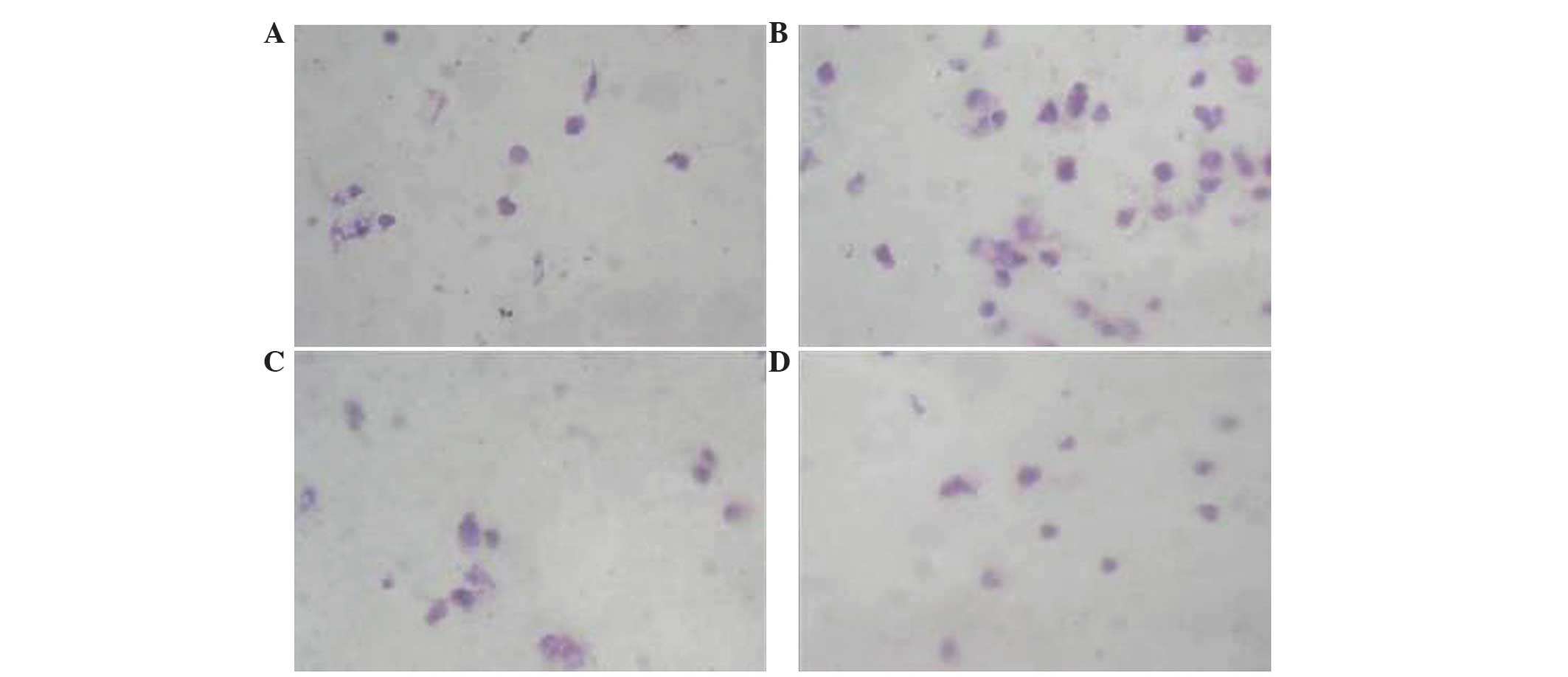
Effects of HIF-2α siRNAs on cancer
cell invasion
The effects of HIF-2α siRNAs on cell migration were
assessed using a Boyden chamber assay, which has been modified. As
shown in Fig. 4, HIF-2α siRNA
significantly inhibited cell migration compared with the control
group (hypoxia + RNAi 1, P<0.006; hypoxia + RNAi 2, P=0.004). By
contrast, the migratory ability of the control group cells were
significantly increased compared with the hypoxia + RNAi groups
(hypoxia + RNAi 1, P<0.006; hypoxia + RNAi 2, P=0.004) (Fig. 4). There was no difference on the
migratory capabilities of the hypoxia + RNAi groups compared with
the parental cell group (P=0.176). These data suggest that HIF-2α
was a positive regulator for cell invasion and HIF-2α siRNA may
inhibit the invasive capacity of MCF-7 cells (Table I).
 | Table I.Effects of HIF-2α small interfering
RNA on the invasion potency of human breast adenocarcinoma MCF-7
cells. |
Table I.
Effects of HIF-2α small interfering
RNA on the invasion potency of human breast adenocarcinoma MCF-7
cells.
| Group | No. of cells | Invasion index,
% | Cell invasion
inhibition rate, % |
|---|
| Parental cell |
16.63±11.84a |
59.06a | 40.94a |
| Control | 28.16±10.16 | 100.00 | − |
| Hypoxia + RNAi
1 |
14.44±11.12a |
51.28a | 48.72a |
| Hypoxia + RNAi
2 |
13.86±15.12a |
49.22a | 50.78a |
Discussion
Hypoxia is common in numerous types of solid tumors,
where tumor cells proliferate rapidly and form large solid tumor
masses, leading to obstruction and compression of the blood vessels
surrounding these masses. These abnormal blood vessels often do not
function properly, resulting in a poor O2 supply to the
center tumor regions. Tumor cells in this hypoxic region begin to
adapt these low oxygen tension conditions by activating several
survival pathways, including the HIF pathway (22,24,25). The
responses of cells to hypoxia are mediated primarily through HIFs,
which are critical mediators in the cellular and systemic hypoxia
response to low oxygen levels via the accommodation of numerous
genes that are induced by hypoxia (14,26–28). The
activation of the HIF transcription factor is the most commonly
identified pathway activated by hypoxic cells in harsh
microenvironments. Activated HIFs exhibit a crucial role in
adaptive responses of tumor cells to changes in O2
levels via transcriptional activation of >100 downstream genes
which regulate vital biological processes required for tumor
survival and progression. Examples include genes involved in
glucose metabolism, cell proliferation, migration and angiogenesis
(29). Currently, the role of HIF-2α
in certain solid tumors is unclear.
The present study investigated the association and
significance between the expression of HIF-2α and clinical features
of breast carcinoma tissue. The author's previous histological
studies have demonstrated that a higher expression of the HIF-2α
protein is associated with breast cancer invasion and metastasis
(30). The aim of the present study
was to reveal whether HIF-2α has effects on the invasion potency of
human breast carcinoma MCF-7 cells, and to reveal a possible
mechanism for this function under hypoxia by using the RNAi
method.
CoCl2 prevents HIF-1α from binding to
prolyl hydroxylases, so that they are not hydroxylated and are
subsequently degraded by proteasomes (31). CoCl2 has been used in in
vivo and in vitro studies to generate hypoxia by
inhibiting HIF-1 specific prolyl-hydroxylase and occupying its
iron-binding site, leading to impaired binding of the von
Hippel-Lindau protein with HIF-1α, subsequently preventing HIF-1α
proteasomal degradation (32). Since
CoCl2 stabilizes the α subunits of HIFs, the
transcription of HIF-targeted genes may be induced by HIFs in spite
of the presence of oxygen (4,33). CoCl2 has been successfully
used to mimic hypoxia in in vivo and in vitro
experimental studies (34). In the
present study, a hypoxia model was successfully established through
CoCl2 treatment of MCF-7 cells. The present results
demonstrated that CoCl2 treatment induces HIF-2α
transcription and translation and increases the mRNA and protein
expressions of HIF-2α. Additionally, transfection with HIF-2α siRNA
inhibited the expression of HIF-2α mRNA and protein, which
indicated that the silencing effect of siRNA on HIF-2α was
successful. Furthermore, the present experiments revealed that
HIF-2α siRNA inhibited breast cancer cell invasion abilities;
basement membrane assays reveled that there were a decreased number
of invading HIF-2α siRNA transfected MCF-7 cells under hypoxic
conditions in vitro compared with control cells. This
finding suggests that HIF-2α may be important in MCF-7 cell
survival and invasion. These results are similar to a previous
study that revealed that HIF-2α promotes tumor progression in
Von-Hippel-Lindau-defective renal carcinoma cells (12). By contrast in other tumor types,
HIF-2α has an important role as a tumor suppressor; embryonic stem
cells are deficient in HIF-2α and display enhanced growth in
ovarian tumors (35). In addition, a
high-expression of HIF-2α may suppress tumor growth in brain glioma
cells (36).
In the present study, in order to verify whether
HIF-2α affects MMP-2 expression, siRNA against HIF-2α was used to
downregulate the expression of HIF-2α in MCF-7 cells. The results
indicated that the expression of MMP-2 mRNA and protein decreased
once MCF-7 cells were transfected with HIF-2α siRNA under hypoxic
conditions. Collectively, the present findings demonstrate that the
regulation of MMP-2 by hypoxia leads to increased invasion of
breast cancer cells, which facilitates metastasis.
In conclusion, the present study has demonstrated
for the first time, to the best of our knowledge, that HIF-2α is
endogenously expressed in eukaryotic cells. In a hypoxia-tolerant
tumor cell line (MCF-7) HIF-2α is involved in tumor cell invasion
in vitro, which confirms the present author's hypothesis:
HIF-2α facilitates cancer cell metastasis in hypoxic environments.
Collectively, the present results provide a novel mechanism that
HIF-2α signaling is important for cancer development and cancer
cell survival by altering the expression of the downstream targets,
including MMP-2. Notably, the present study revealed that siRNA
targeting of HIF-2α may be a viable approach in the treatment of
malignant diseases.
Acknowledgements
The present study was supported by the University
Key Teacher Grant from the Ministry of Education of Henan
(Zhengzhou, China; grant no. 2012GGJS-136) and Scientific Research
Fund of Xinxiang Medical University (Xinxiang, China; grant no.
2014QN113).
References
|
1
|
Bohensky J, Terkhorn SP, Freeman TA, Adams
CS, Garcia JA, Shapiro IM and Srinivas V: Regulation of autophagy
in human and murine cartilage: Hypoxia-inducible factor 2
suppresses chondrocyte autophagy. Arthritis Rheum. 60:1406–1415.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mole DR, Blancher C, Copley RR, Pollard
PJ, Gleadle JM, Ragoussis J and Ratcliffe PJ: Genome-wide
association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA
binding with expression profiling of hypoxia-inducible transcripts.
J Biol Chem. 284:16767–16775. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kütscher C, Lampert FM, Kunze M,
Markfeld-Erol F, Stark GB and Finkenzeller G: Overexpression of
hypoxia-inducible factor-1 alpha improves vasculogenesis-related
functions of endothelial progenitor cells. Microvasc Res.
105:85–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rankin EB, Rha J, Unger TL, Wu CH, Shutt
HP, Johnson RS, Simon MC, Keith B and Haase VH: Hypoxia-inducible
factor-2 regulates vascular tumorigenesis in mice. Oncogene.
27:5354–5358. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Copple BL, Bai S and Moon JO:
Hypoxia-inducible factor-dependent production of profibrotic
mediators by hypoxic Kupffer cells. Hepatol Res. 40:530–539. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wykoff CC, Sotiriou C, Cockman ME,
Ratcliffe PJ, Maxwell P, Liu E and Harris AL: Gene array of VHL
mutation and hypoxia shows novel hypoxia-induced genes and that
cyclin D1 is a VHL target gene. Br J Cancer. 90:1235–1243. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu W, Xin H, Eckert DT, Brown JA and
Gnarra JR: Hypoxia and cell cycle regulation of the von
Hippel-Lindau tumor suppressor. Oncogene. 30:21–31. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao S, Jin C, Zhao X, Jin B, Hui L, Zhou
W, Niu G and Tao S: Expression and clinical significance of ING4
and HIF-1 alpha in brain astrocytoma. Zhonghua Yi Xue Za Zhi.
95:3533–3536. 2015.(In Chinese). PubMed/NCBI
|
|
9
|
Burrows N, Babur M, Resch J, Williams KJ
and Brabant G: Hypoxia-inducible factor in thyroid carcinoma. J
Thyroid Res. 2011:7629052011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Trisciuoglio D, Gabellini C, Desideri M,
Ziparo E, Zupi G and Del Bufalo D: Bcl-2 regulates HIF-1α protein
stabilization in hypoxic melanoma cells via the molecular chaperone
HSP90. PLoS One. 5:e117722010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mardilovich K and Shaw LM: Hypoxia
regulates insulin receptor substrate-2 expression to promote breast
carcinoma cell survival and invasion. Cancer Res. 69:8894–8901.
2009.10.1158/0008-5472.CAN-09-1152. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Raval RR, Lau KW, Tran MGB, Sowter HM,
Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL and Ratcliffe
PJ: Contrasting properties of hypoxia-inducible factor 1 (HIF-1)
and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol
Cell Biol. 25:5675–5686. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mazumdar J, Hickey MM, Pant DK, Durham AC,
Sweet-Cordero A, Vachani A, Jacks T, Chodosh LA, Kissil JL, Simon
MC and Keith B: HIF-2α deletion promotes Kras-driven lung tumor
development. Proc Natl Acad Sci USA. 107:14182–14187. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Florczyk U, Czauderna S, Stachurska A,
Tertil M, Nowak W, Kozakowska M, Poellinger L, Jozkowicz A, Loboda
A and Dulak J: Opposite effects of HIF-1α and HIF-2α on the
regulation of IL-8 expression in endothelial cells. Free Radic Biol
Med. 1:1882–1892. 2011. View Article : Google Scholar
|
|
15
|
Rankin EB, Rha J, Selak MA, Unger TL,
Keith B, Liu Q and Haase VH: Hypoxia-inducible factor 2 regulates
hepatic lipid metabolism. Mol Cell Biol. 29:4527–4538. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi YK, Kim CK, Lee H, Jeoung D, Ha KS,
Kwon YG, Kim KW and Kim YM: Carbon monoxide promotes VEGF
expression by increasing HIF-1α protein level via two distinct
mechanisms, translational activation and stabilization of HIF-1α
protein. J Biol Chem. 285:32116–32125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kapitsinou PP, Liu Q, Unger TL, Rha J,
Davidoff O, Keith B, Epstein JA, Moores SL, Erickson-Miller CL and
Haase VH: Hepatic HIF-2 regulates erythropoietic responses to
hypoxia in renal anemia. Blood. 116:3039–3048. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rankin EB, Biju MP, Liu Q, Unger TL, Rha
J, Johnson RS, Simon MC, Keith B and Haase VH: Hypoxia-inducible
factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin
Invest. 117:1068–1077. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei K, Piecewicz SM, McGinnis LM,
Taniguchi CM, Wiegand SJ, Anderson K, Chan CW, Mulligan KX, Kuo D,
Yuan J, et al: A liver Hif-2α-Irs2 pathway sensitizes hepatic
insulin signaling and is modulated by Vegf inhibition. Nat Med.
19:1331–1337. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qiu Y, Zheng H, Sun LH, Peng K, Xiao WD
and Yang H: Hypoxia-inducible factor-1 modulates upregulation of
mutT homolog-1 in colorectal cancer. World J Gastroenterol.
21:13447–13456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fukasawa T, Enomoto A and Miyagawa K:
Serine-Threonine Kinase 38 regulates CDC25A stability and the DNA
damage-induced G2/M checkpoint. Cell Signal. 27:1569–1575. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meade ES, Ma YY and Guller S: Role of
hypoxia-inducible transcription factors 1α and 2α in the regulation
of plasminogen activator inhibitor-1 expression in a human
trophoblast cell line. Placenta. 28:1012–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li N, Wang HX, Zhang A, Ye YP and He GY:
KISS-1 inhibits the proliferation and invasion of gastric carcinoma
cells. World J Gastroenterol. 18:1827–1833. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Semenza GL: Defining the role of
hypoxia-inducible factor 1 in cancer biology and therapeutics.
Oncogene. 29:625–634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bensellam M, Duvillié B, Rybachuk G,
Laybutt DR, Magnan C, Guiot Y, Pouysségur J and Jonas JC:
Glucose-induced O2 consumption activates hypoxia
inducible factors 1 and 2 in rat insulin-secreting pancreatic
beta-cells. PLoS One. 7:e298072012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jing SW, Wang YD, Kuroda M, Su JW, Sun GG,
Liu Q, Cheng YJ and Yang CR: HIF-1α contributes to hypoxia-induced
invasion and metastasis of esophageal carcinoma via inhibiting
E-cadherin and promoting MMP-2 expression. Acta Med Okayama.
66:399–407. 2012.PubMed/NCBI
|
|
27
|
Leonard MO, Howell K, Madden SF, Costello
CM, Higgins DG, Taylor CT and McLoughlin P: Hypoxia selectively
activates the CREB family of transcription factors in the in vivo
lung. Am J Respir Crit Care Med. 178:977–983. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gordan JD, Bertout JA, Hu CJ, Diehl JA and
Simon MC: HIF-2α promotes hypoxic cell proliferation by enhancing
c-myc transcriptional activity. Cancer Cell. 11:335–347. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang KT, Takano EA, Mikeska T, Byrne DJ,
Dobrovic A and Fox SB: Aberrant DNA methylation but not mutation of
CITED4 is associated with alteration of HIF-regulated genes in
breast cancer. Breast Cancer Res Treat. 130:319–329. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li N, Wang HX, Qin C, Wang XH and Han FY:
Relationship between clinicopathological features and HIF-2α in
gastric adenocarcinoma. Genet Mol Res. 14:1404–1413. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao H, Gu Z, Wang G and Zhao T: The
possible mechanisms underlying the impairment of HIF-1α pathway
signaling in hyperglycemia and the beneficial effects of certain
therapies. Int J Med Sci. 10:1412–1421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yokoe S, Nakagawa T, Kojima Y, Higuchi K
and Asahi M: Indomethacin-induced intestinal epithelial cell damage
is mediated by pVHL activation through the degradation of collagen
I and HIF-1α. Biochem Biophys Res Commun. 468:671–676. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu P, Ning Y, Yao L, Chen M and Xu C: The
proliferation, apoptosis, invasion of endothelial-like epithelial
ovarian cancer cells induced by hypoxia. J Exp Clin Cancer Res.
29:1242010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yoo HI, Moon YH and Kim MS: Effects of
CoCl2 on multi-lineage differentiation of C3H/10T1/2 mesenchymal
stem cells. Korean J Physiol Pharmacol. 20:53–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Q, Peng K and He L: Expression of the
HIF-2α in epithelial ovarian cancer and clinical significance.
Zhong Nan Da Xue Xue Bao Yi Xue Ban. 39:889–893. 2014.PubMed/NCBI
|
|
36
|
Anelli V, Gault CR, Cheng AB and Obeid LM:
Sphingosine kinase 1 is up-regulated during hypoxia in U87MG glioma
cells. Role of hypoxia-inducible factors 1 and 2. J Biol Chem.
283:3365–3375. 2008. View Article : Google Scholar : PubMed/NCBI
|